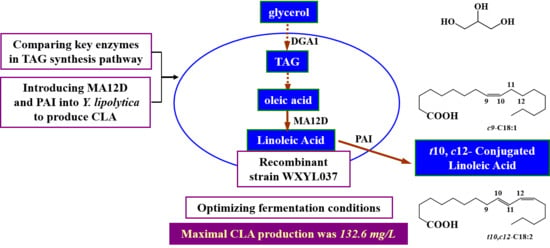
Modulating Heterologous Pathways and Optimizing Culture Conditions for Biosynthesis of trans-10, cis-12 Conjugated Linoleic Acid in Yarrowia lipolytica
Abstract
1. Introduction
2. Results and Discussion
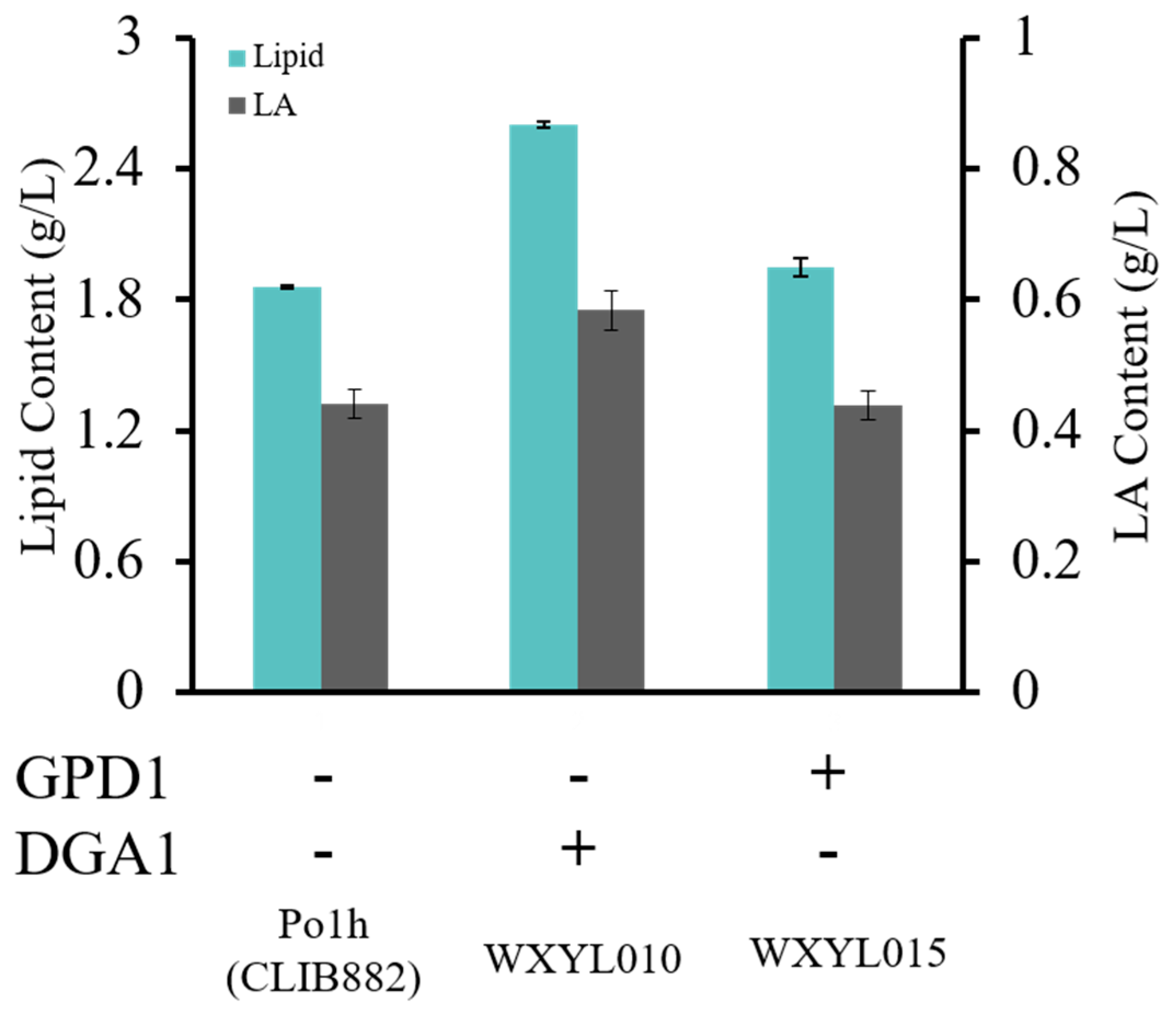
2.1. Effects of DGA1 and GPD1 on Lipid Production in Y. lipolytica
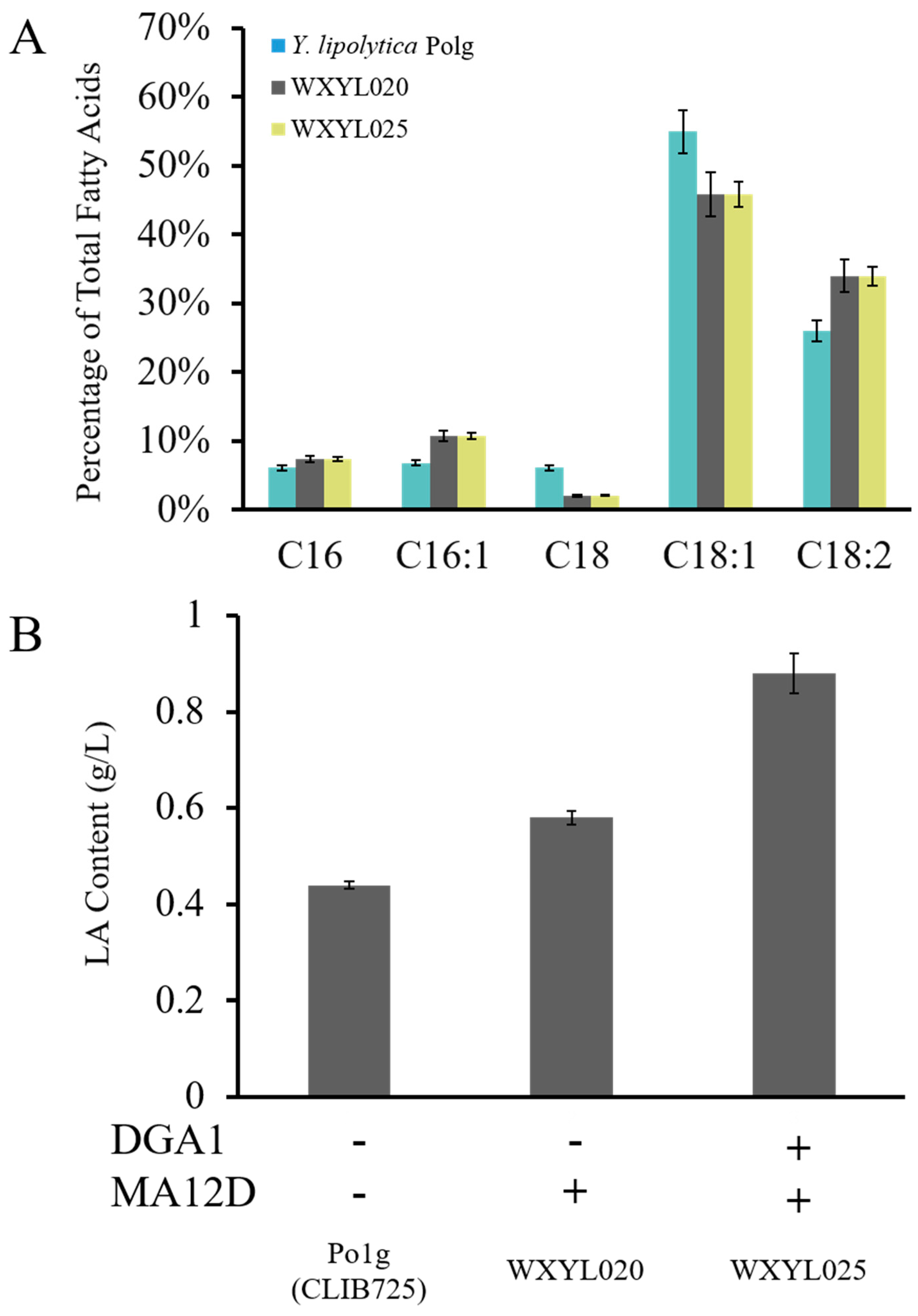
2.2. Establishment of a LA Biosynthesis Pathway in Y. lipolytica
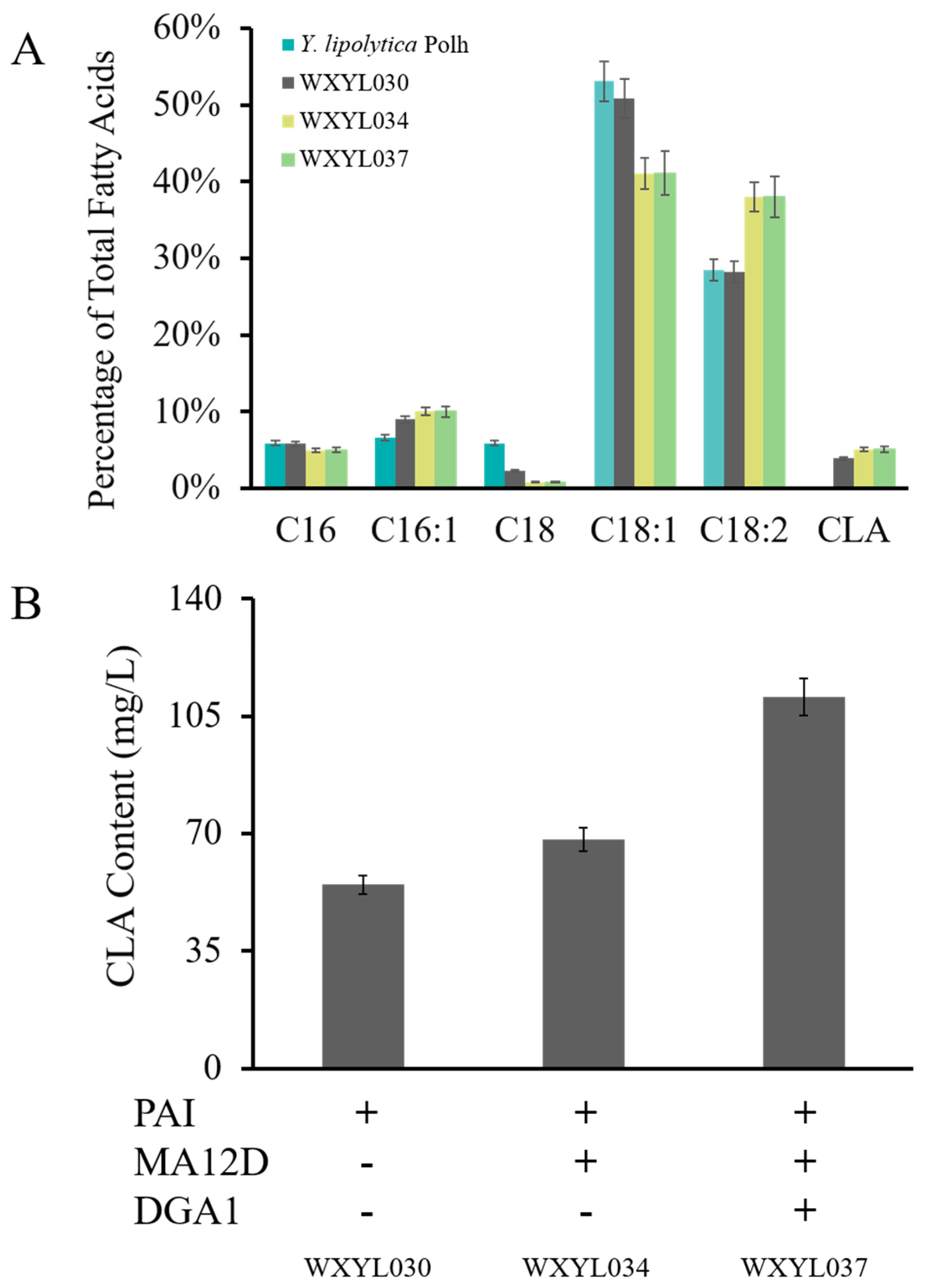
2.3. Establishment of a CLA Biosynthesis Pathway in Y. lipolytica
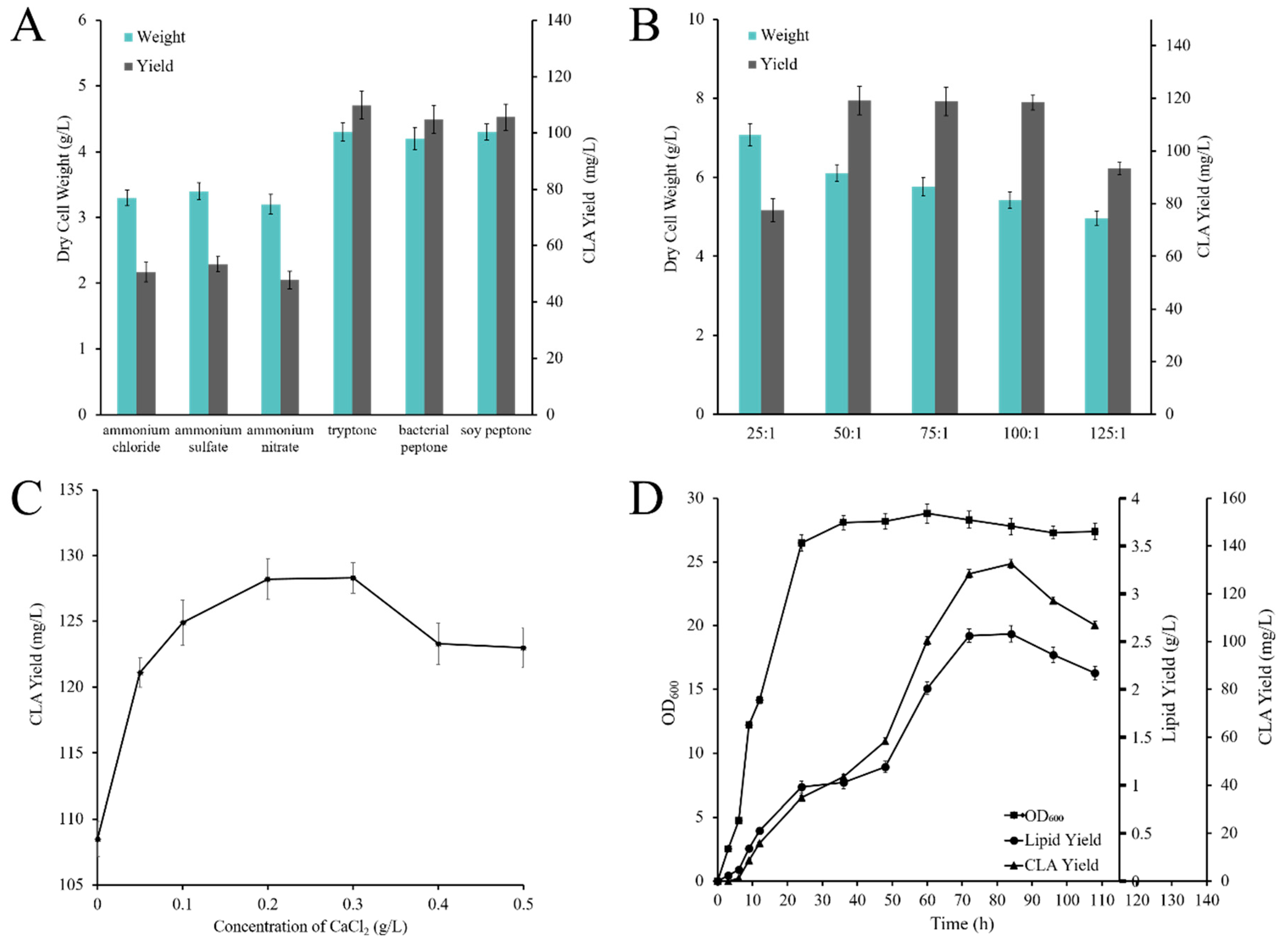
2.4. Optimization of Flask Culture Conditions
2.4.1. Effect of Carbon and Nitrogen Sources on CLA Production
2.4.2. Effect of C/N Ratio on CLA Production
2.4.3. Effect of CaCl2 Concentration on CLA Production
2.5. CLA Production by Recombinant Strain
3. Conclusions
4. Materials and Methods
4.1. Plasmids and Strains
4.2. Growth and Culture Conditions
4.3. Lipid Extraction and Quantification
4.4. Fluorescence and Electron Microscopy
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lin, T.Y.; Lin, C.-W.; Lee, C.-H. Conjugated linoleic acid concentration as affected by lactic cultures and added linoleic acid. Food Chem. 1999, 67, 1–5. [Google Scholar] [CrossRef]
- Alonso, L.; Cuesta, E.P.; Gilliland, S.E. Production of free conjugated linoleic acid by Lactobacillus acidophilus and Lactobacillus casei of human intestinal origin. J. Dairy Sci. 2003, 86, 1941–1946. [Google Scholar] [CrossRef]
- Baba, Y.; Kallas, Z.; Costa-Font, M.; Maria Gil, J.; Realini, C.E. Impact of hedonic evaluation on consumers’ preferences for beef attributes including its enrichment with n-3 and CLA fatty acids. Meat Sci. 2016, 111, 9–17. [Google Scholar] [CrossRef]
- Miguel Rodriguez-Alcala, L.; Fontecha, J.; de la Hoz, L.; Nunes da Silva, V.S.; Carvalho, J.E.; Bertoldo Pacheco, M.T. CLA-enriched milk powder reverses hypercholesterolemic risk factors in hamsters. Food Res. Int. 2013, 51, 244–249. [Google Scholar] [CrossRef]
- Churruca, I.; Fernandez-Quintela, A.; Portillo, M.P. Conjugated linoleic acid isomers: Differences in metabolism and biological effects. Biofactors 2009, 35, 105–111. [Google Scholar] [CrossRef]
- Griinari, J.M.; Corl, B.A.; Lacy, S.H.; Chouinard, P.Y.; Nurmela, K.V.; Bauman, D.E. Conjugated linoleic acid is synthesized endogenously in lactating dairy cows by Δ9-desaturase. J. Nutr. 2000, 130, 2285–2291. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Du, W.; Liu, D. Perspectives of microbial oils for biodiesel production. Appl. Microbiol. Biotechnol. 2008, 80, 749–756. [Google Scholar] [CrossRef] [PubMed]
- Hennessy, A.A.; Barrett, E.; Ross, R.P.; Fitzgerald, G.F.; Devery, R.; Stanton, C. The Production of Conjugated α-Linolenic, γ-Linolenic and Stearidonic Acids by Strains of Bifidobacteria and Propionibacteria. Lipids 2012, 47, 313–327. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, E.S.; Kermanshahi, R.K.; Hosseinkhani, S.; Shojaosadati, S.A.; Nazari, M. Conjugated linoleic acid production from various substrates by probiotic Lactobacillus plantarum. Ann. Microbiol. 2015, 65, 27–32. [Google Scholar] [CrossRef]
- Hornung, E.; Krueger, C.; Pernstich, C.; Gipmans, M.; Porzel, A.; Feussner, I. Production of (10E,12Z)-conjugated linoleic acid in yeast and tobacco seeds. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2005, 1738, 105–114. [Google Scholar] [CrossRef]
- Zhang, B.; Rong, C.; Chen, H.; Song, Y.; Zhang, H.; Chen, W. De novo synthesis of trans-10, cis-12 conjugated linoleic acid in oleaginous yeast Yarrowia lipolytica. Microb. Cell Factories 2012, 11, 51. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gu, H.; Shi, H.; Wang, F.; Li, X. Green Synthesis of Conjugated Linoleic Acids from Plant Oils Using a Novel Synergistic Catalytic System. J. Agric. Food Chem. 2017, 65, 5322–5329. [Google Scholar] [CrossRef] [PubMed]
- Beopoulos, A.; Cescut, J.; Haddouche, R.; Uribelarrea, J.-L.; Molina-Jouve, C.; Nicaud, J.-M. Yarrowia lipolytica as a model for bio-oil production. Prog. Lipid Res. 2009, 48, 375–387. [Google Scholar] [CrossRef] [PubMed]
- Blazeck, J.; Hill, A.; Liu, L.; Knight, R.; Miller, J.; Pan, A.; Otoupal, P.; Alper, H.S. Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production. Nat. Commun. 2014, 5, 3131. [Google Scholar] [CrossRef]
- Dujon, B.; Sherman, D.; Fischer, G.; Durrens, P.; Casaregola, S.; Lafontaine, I.; de Montigny, J.; Marck, C.; Neuveglise, C.; Talla, E.; et al. Genome evolution in yeasts. Nature 2004, 430, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Tai, M.; Stephanopoulos, G. Engineering the push and pull of lipid biosynthesis in oleaginous yeast Yarrowia lipolytica for biofuel production. Metab. Eng. 2013, 15, 1–9. [Google Scholar] [CrossRef]
- Fan, X.; Zhu, Q.; Hong, R.; Sharpe, P.; Xue, Z.; Yadav, N.; Tyreus, B.; Boonyaratanakornkit, B.; Dellomonaco, C.; Short, D.; et al. Metabolic engineering of Yarrowia lipolytica to produce polyunsaturated fatty acids. Yeast 2013, 30, 39. [Google Scholar]
- Juszczyk, P.; Tomaszewska, L.; Kita, A.; Rymowicz, W. Biomass production by novel strains of Yarrowia lipolytica using raw glycerol, derived from biodiesel production. Bioresour. Technol. 2013, 137, 124–131. [Google Scholar] [CrossRef]
- Easterling, E.R.; French, W.T.; Hernandez, R.; Licha, M. The effect of glycerol as a sole and secondary substrate on the growth and fatty acid composition of Rhodotorula glutinis. Bioresour. Technol. 2009, 100, 356–361. [Google Scholar] [CrossRef]
- Hajjari, M.; Tabatabaei, M.; Aghbashlo, M.; Ghanavati, H. A review on the prospects of sustainable biodiesel production: A global scenario with an emphasis on waste-oil biodiesel utilization. Renew. Sustain. Energy Rev. 2017, 72, 445–464. [Google Scholar] [CrossRef]
- Kennedy, E.P. Biosynthesis of complex lipids. Fed. Proc. 1961, 20, 934–940. [Google Scholar] [PubMed]
- Albertyn, J.; Hohmann, S.; Thevelein, J.M.; Prior, B.A. GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol. Cell. Biol. 1994, 14, 4135–4144. [Google Scholar] [CrossRef]
- Athenstaedt, K.; Daum, G. The life cycle of neutral lipids: Synthesis, storage and degradation. Cell. Mol. Life Sci. 2006, 63, 1355–1369. [Google Scholar] [CrossRef] [PubMed]
- Liavonchanka, A.; Hornung, E.; Feussner, I.; Rudolph, M.G. Structure and mechanism of the Propionibacterium acnes polyunsaturated fatty acid isomerase. Proc. Natl. Acad. Sci. USA 2006, 103, 2576–2581. [Google Scholar] [CrossRef]
- Zhang, B.; Chen, H.; Li, M.; Gu, Z.; Song, Y.; Ratledge, C.; Chen, Y.Q.; Zhang, H.; Chen, W. Genetic engineering of Yarrowia lipolytica for enhanced production of trans-10, cis-12 conjugated linoleic acid. Microb. Cell Factories 2013, 12, 70. [Google Scholar] [CrossRef]
- Madzak, C.; Treton, B.; Blanchin-Roland, S. Strong hybrid promoters and integrative expression/secretion vectors for quasi-constitutive expression of heterologous proteins in the yeast Yarrowia lipolytica. J. Mol. Microbiol. Biotechnol. 2000, 2, 207–216. [Google Scholar] [PubMed]
- Chuang, L.-T.; Chen, D.-C.; Nicaud, J.-M.; Madzak, C.; Chen, Y.-H.; Huang, Y.-S. Co-expression of heterologous desaturase genes in Yarrowia lipolytica. New Biotechnol. 2010, 27, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Cui, W.; Wang, Q.; Zhang, F.; Zhang, S.-C.; Chi, Z.-M.; Madzak, C. Direct conversion of inulin into single cell protein by the engineered Yarrowia lipolytica carrying inulinase gene. Process Biochem. 2011, 46, 1442–1448. [Google Scholar] [CrossRef]
- Gasmi, N.; Ayed, A.; Nicaud, J.-M.; Kallel, H. Design of an efficient medium for heterologous protein production in Yarrowia lipolytica: Case of human interferon alpha 2b. Microb. Cell Factories 2011, 10, 38. [Google Scholar] [CrossRef]
- Sun, M.-L.; Madzak, C.; Liu, H.-H.; Song, P.; Ren, L.-J.; Huang, H.; Ji, X.-J. Engineering Yarrowia lipolytica for efficient gamma-linolenic acid production. Biochem. Eng. J. 2017, 117, 172–180. [Google Scholar] [CrossRef]
- Da Silva, G.P.; Mack, M.; Contiero, J. Glycerol: A promising and abundant carbon source for industrial microbiology. Biotechnol. Adv. 2009, 27, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Dharmadi, Y.; Murarka, A.; Gonzalez, R. Anaerobic fermentation of glycerol by Escherichia coli: A new platform for metabolic engineering. Biotechnol. Bioeng. 2006, 94, 821–829. [Google Scholar] [CrossRef]
- Ratledge, C. Regulation of lipid accumulation in oleaginous micro-organisms. Biochem. Soc. Trans. 2002, 30, 1047–1050. [Google Scholar] [CrossRef] [PubMed]
- Pourmortazavi, S.M.; Rahimi-Nasrabadi, M.; Fazli, Y.; Mohammad-Zadeh, M. Taguchi method assisted optimization of electrochemical synthesis and structural characterization of copper tungstate nanoparticles. Int. J. Refract. Met. Hard Mater. 2015, 51, 29–34. [Google Scholar] [CrossRef]
- Schwartz, C.M.; Hussain, M.S.; Blenner, M.; Wheeldon, I. Synthetic RNA Polymerase III Promoters Facilitate High-Efficiency CRISPR-Cas9-Mediated Genome Editing in Yarrowia lipolytica. ACS Synth. Biol. 2016, 5, 356–359. [Google Scholar] [CrossRef] [PubMed]
- Madzak, C.; Gaillardin, C.; Beckerich, J.M. Heterologous protein expression and secretion in the non-conventional yeast Yarrowia lipolytica: A review. J. Biotechnol. 2004, 109, 63–81. [Google Scholar] [CrossRef] [PubMed]
- Chong, L. Molecular cloning—A laboratory manual, 3rd edition. Science 2001, 292, 446. [Google Scholar] [CrossRef]
- Mark, E.; Sharon, H.; Lisa, L.; Jeff, P.; Greg, W. Process for the Biological Production of 1,3-Propanediol. U.S. Patent US09/641,652, 18 August 2000. [Google Scholar]
- Le Dall, M.T.; Nicaud, J.M.; Gaillardin, C. Multiple-copy integration in the yeast Yarrowia lipolytica. Curr. Genet. 1994, 26, 38–44. [Google Scholar] [CrossRef]
- Nicaud, J.M.; Madzak, C.; van den Broek, P.; Gysler, C.; Duboc, P.; Niederberger, P.; Gaillardin, C. Protein expression and secretion in the yeast Yarrowia lipolytica. FEMS Yeast Res. 2002, 2, 371–379. [Google Scholar]
- Madzak, C.; Pandalai, S.G. New tools for heterologous protein production in the yeast Yarrowia lipolytica. Recent Res. Dev. Microbiol. 2003, 7, 453–479. [Google Scholar]
- Barth, G.; Gaillardin, C. Physiology and genetics of the dimorphic fungus Yarrowia lipolytica. FEMS Microbiol. Rev. 1997, 19, 219–237. [Google Scholar] [CrossRef]
Sample Availability: No samples are available from the authors. |

| Strains (Host Strain) | Genotype or Plasmid | Source or Reference |
|---|---|---|
| E. coli | ||
| Top10 | F-mcrA Δ(mrr-hsdRMS-mcrBC) φ80 lacZ ΔM15 ΔlacX74 recA1 araΔ139 Δ(ara-leu)7697 galU galK rpsL (StrR) endA1 nupG | Invitrogen |
| Plasmid | ||
| pINA1312 | hp4d, XPR2t, ura3d4, KanR | [40] |
| pINA1267 | hp4d, XPR2 prepro, XPR2t, LEU2, AmpR | [41] |
| pWX010 | pINA1312 hp4d-DGA1 | This work |
| pWX015 | pINA1312 hp4d-GPD1 | This work |
| pWX020 | pINA1267 hp4d-MA12D | This work |
| pWX025 | pWX020 hp4d-DGA1 | This work |
| pWX030 | pINA1312 hp4d-PAI | This work |
| pWX034 | pWX030 hp4d-MA12D | This work |
| pWX037 | pWX034 hp4d-DGA1 | This work |
| Y. lipolytica | ||
| Po1g (CLIB725) | MatA, leu2-270, ura3-302::URA3, xpr2-332, axp1-2, Leu−, ΔAEP, ΔAXP, Suc+, pBR322 | [26,41] |
| Po1h (CLIB882) | MatA, ura3-302, xpr2-232, axp1-2, Ura−, ΔAEP, ΔAXP, Suc+ | [36,41] |
| WXYL010 | MatA, ura3-302, xpr2-232, axp1-2 hp4d-DGA1-URA3 | This work |
| WXYL015 | MatA, ura3-302, xpr2-232, axp1-2 hp4d-GPD1-URA3 | This work |
| WXYL020 | MatA, leu2-270, ura3-302::URA3, xpr2-332, axp1-2 hp4d-MA12D-LEU2 | This work |
| WXYL025 | MatA, leu2-270, ura3-302::URA3, xpr2-332, axp1-2 hp4d-MA12D+hp4d-DGA1-LEU2 | This work |
| WXYL030 | MatA, ura3-302, xpr2-232, axp1-2 hp4d-PAI-URA3 | This work |
| WXYL034 | MatA, ura3-302, xpr2-232, axp1-2 hp4d-PAI+hp4d-MA12D-URA3 | This work |
| WXYL037 | MatA, ura3-302, xpr2-232, axp1-2 hp4d-PAI+hp4d-MA12D+hp4d-DGA1-URA3 | This work |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Xia, Q.; Wang, F.; Zhang, Y.; Li, X. Modulating Heterologous Pathways and Optimizing Culture Conditions for Biosynthesis of trans-10, cis-12 Conjugated Linoleic Acid in Yarrowia lipolytica. Molecules 2019, 24, 1753. https://doi.org/10.3390/molecules24091753
Wang X, Xia Q, Wang F, Zhang Y, Li X. Modulating Heterologous Pathways and Optimizing Culture Conditions for Biosynthesis of trans-10, cis-12 Conjugated Linoleic Acid in Yarrowia lipolytica. Molecules. 2019; 24(9):1753. https://doi.org/10.3390/molecules24091753
Chicago/Turabian StyleWang, Xun, Qianjun Xia, Fei Wang, Yu Zhang, and Xun Li. 2019. "Modulating Heterologous Pathways and Optimizing Culture Conditions for Biosynthesis of trans-10, cis-12 Conjugated Linoleic Acid in Yarrowia lipolytica" Molecules 24, no. 9: 1753. https://doi.org/10.3390/molecules24091753
APA StyleWang, X., Xia, Q., Wang, F., Zhang, Y., & Li, X. (2019). Modulating Heterologous Pathways and Optimizing Culture Conditions for Biosynthesis of trans-10, cis-12 Conjugated Linoleic Acid in Yarrowia lipolytica. Molecules, 24(9), 1753. https://doi.org/10.3390/molecules24091753