High-Sensitivity Determination of Nutrient Elements in Panax notoginseng by Laser-induced Breakdown Spectroscopy and Chemometric Methods
Abstract
:1. Introduction
2. Materials and Methods
2.1. Panax notoginseng Samples
2.2. Spectral Acquisition
2.3. Reference Method for Nutrient Elements’ Content Determination
2.4. Data Analysis
2.4.1. Data Preprocessing
2.4.2. Multivariate Analysis Methods
2.4.3. Performance Evaluation
2.4.4. Software Tools
3. Results
3.1. Nutritive Elements Content of Panax Notoginseng
3.2. LIBS Spectra Analysis
3.3. Univariate Analysis
3.4. Multivariate Analysis
3.4.1. Modeling Using Full Spectra
3.4.2. Modeling Using Selected Variables
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ng, T.B. Pharmacological activity of sanchi ginseng (Panax notoginseng). J. Pharm. Pharmacol. 2006, 58, 1007–1019. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Mu, R.; Dou, M.; Yan, D.; Liu, B.; Wei, Q.; Wan, J.; Tang, Y.; Hu, Y. Epigenetic modification in histone deacetylase deletion strain of Calcarisporium arbuscula leads to diverse diterpenoids. N.a. Pharm. Sin. B 2018, 8, 687–697. [Google Scholar] [CrossRef]
- Sun, S.; Wang, C.Z.; Tong, R.; Li, X.L.; Fishbein, A.; Wang, Q.; He, T.C.; Du, W.; Yuan, C.S. Effects of steaming the root of Panax notoginseng on chemical composition and anticancer activities. N.a. Chem. 2010, 118, 307–314. [Google Scholar] [CrossRef]
- Yang, W.; Qiao, X.; Li, K.; Fan, J.; Bo, T.; Guo, D.A.; Ye, M. Identification and differentiation of Panax ginseng, Panax quinquefolium, and Panax notoginseng by monitoring multiple diagnostic chemical markers. N.a. Pharm. Sin. B 2016, 6, 568–575. [Google Scholar] [CrossRef]
- Zhao, G.R.; Xiang, Z.J.; Ye, T.X.; Yuan, Y.J.; Guo, Z.X. Antioxidant activities of Salvia miltiorrhiza and Panax notoginseng. Food Chem. 2006, 99, 767–774. [Google Scholar] [CrossRef]
- Fan, Z.Y.; Miao, C.P.; Qiao, X.G.; Zheng, Y.K.; Chen, H.H.; Chen, Y.W.; Xu, L.H.; Zhao, L.X.; Guan, H.L. Diversity, distribution, and antagonistic activities of rhizobacteria of Panax notoginseng. J. Ginseng Res. 2016, 40, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Toh, D.F.; New, L.S.; Koh, H.L.; Chan, E.C.Y. Ultra-high performance liquid chromatography/time-of-flight mass spectrometry (UHPLC/TOFMS) for time-dependent profiling of raw and steamed Panax notoginseng. J. Pharm. Biomed. Anal. 2010, 52, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Shan, S.M.; Luo, J.G.; Huang, F.; Kong, L.Y. Chemical characteristics combined with bioactivity for comprehensive evaluation of Panax ginseng C.A. Meyer in different ages and seasons based on HPLC-DAD and chemometric methods. J. Pharm. Biomed. Anal. 2014, 89, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Pytlakowska, K.; Kita, A.; Janoska, P.; Połowniak, M.; Kozik, V. Multi-element analysis of mineral and trace elements in medicinal herbs and their infusions. Food Chem. 2012, 135, 494–501. [Google Scholar] [CrossRef]
- Maiti, R.; Rodriguez, H.G.; Kumari, A.; Sarkar, N. Macro and micro-nutrient contents of 18 medicinal plants used traditionally to alleviate diabetes in nuevo leon, northeast of Mexico. Pak. J. Bot. 2016, 48, 271–276. [Google Scholar]
- Maiti, R.; Rodriguez, H.G.; Degu, H.D.; Kumari, C.A.; Sarkar, N.C.; Leon, F.D.C.F.U.A.D.N. Macro and micronutrients of 44 medicinal plant species used traditionally in nuevo leon, Mexico. Int. J. Bio-resource N.A. Manag. 2016, 7, 1054–1062. [Google Scholar] [CrossRef]
- McCall, K.A.; Huang, C.C.; Fierke, C.A. Function and mechanism of zinc metalloenzymes. J. Nutr. 2000, 130, 1437S–1446S. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.H.; Yu, X.J.; Wang, D.; Qi, X.M.; Wang, H.P.; Yang, T.T.; Xu, X.H. Alterations of trace elements (Zn, Se, Cu, Fe) and related metalloenzymes in rabbit blood after severe trauma. J. Elements Med. Boil. 2007, 21, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Karadaş, C.; Kara, D. Chemometric approach to evaluate trace metal concentrations in some spices and herbs. Food Chem. 2012, 130, 196–202. [Google Scholar] [CrossRef]
- Tokalıoğlu, Ş. Determination of trace elements in commonly consumed medicinal herbs by ICP-MS and multivariate analysis. Food Chem. 2012, 134, 2504–2508. [Google Scholar] [CrossRef]
- Mirosławski, J.; Paukszto, A. Determination of the Cadmium, Chromium, Nickel, and Lead Ions relays in selected polish medicinal plants and their infusion. Biol. Trace Elem. Res. 2018, 182, 147–151. [Google Scholar] [CrossRef]
- Peng, J.; He, Y.; Ye, L.; Shen, T.; Liu, F.; Kong, W.; Liu, X.; Zhao, Y. Moisture influence reducing method for heavy metals detection in plant materials using laser-induced breakdown spectroscopy: A case study for chromium content detection in rice leaves. Anal. Chem. 2017, 89, 7593–7600. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Ye, L.; Shen, T.; Liu, F.; Song, K.; He, Y. Fast determination of copper content in tobacco (Nicotina tabacum L.) leaves using laser-induced breakdown spectroscopy with univariate and multivariate analysis. Trans. ASABE 2018, 61, 821–829. [Google Scholar] [CrossRef]
- Peng, J.Y.; Liu, F.; Shen, T.T.; Ye, L.H.; Kong, W.W.; Wang, W.; Liu, X.D.; He, Y. Comparative study of the detection of Chromium content in rice leaves by 532 nm and 1064 nm laser-induced breakdown spectroscopy. Sensors 2018, 18, 621. [Google Scholar] [CrossRef]
- Juvé, V.; Portelli, R.; Boueri, M.; Baudelet, M.; Yu, J. Space-resolved analysis of trace elements in fresh vegetables using ultraviolet nanosecond laser-induced breakdown spectroscopy. Spectrochim. N.a. N.a. B: At. Spectrosc. 2008, 63, 1047–1053. [Google Scholar] [CrossRef]
- Liu, F.; Wang, W.; Shen, T.; Peng, J.; Kong, W. Rapid identification of Kudzu powder of different origins using laser-induced breakdown spectroscopy. Sensors 2019, 19, 1453. [Google Scholar] [CrossRef] [PubMed]
- Braga, J.W.B.; Trevizan, L.C.; Nunes, L.C.; Rufini, I.A.; Santos, D.; Krug, F.J. Comparison of univariate and multivariate calibration for the determination of micronutrients in pellets of plant materials by laser-induced breakdown spectrometry. Spectrochim. Acta Part B: At. Spectrosc. 2010, 65, 66–74. [Google Scholar] [CrossRef]
- De Carvalho, G.G.A.; Moros, J.; Santos, D.; Krug, F.J.; Laserna, J.J.; Santos, J.D. Direct determination of the nutrient profile in plant materials by femtosecond laser-induced breakdown spectroscopy. Anal. Chim. Acta 2015, 876, 26–38. [Google Scholar] [CrossRef] [PubMed]
- Gilon, N.; El-Haddad, J.; Stankova, A.; Lei, W.; Ma, Q.; Motto-Ros, V.; Yu, J. A matrix effect and accuracy evaluation for the determination of elements in milk powder LIBS and laser ablation/ICP-OES spectrometry. Anal. Bioanal. Chem. 2011, 401, 2681–2689. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Liu, X.; Lv, Y.; Liu, F.; Peng, J.; Shen, T.; Zhao, Y.; Tang, Y.; Luo, S. Quantitative analysis of nutrient elements in soil using single and double-pulse laser-induced breakdown spectroscopy. Sensors 2018, 18, 1526. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, Y.; Gu, Y.H.; Guo, H.; Li, N. LIBS quantitative analysis of Cr and Ni in iron alloys with support vector machine (SVM). Spectrosc. Spectr. Anal. 2016, 36, 2244–2248. [Google Scholar]
- Dyar, M.D.; Carmosino, M.L.; Breves, E.A.; Ozanne, M.V.; Clegg, S.M.; Wiens, R.C. Comparison of partial least squares and lasso regression techniques as applied to laser-induced breakdown spectroscopy of geological samples. Spectrochim. Acta Part B: At. Spectrosc. 2012, 70, 51–67. [Google Scholar] [CrossRef]
- Liu, F.; Shen, T.T.; Kong, W.W.; Peng, J.Y.; Zhang, C.; Song, K.L.; Wang, W.; Zhang, C.; He, Y. Quantitative analysis of cadmium in tobacco roots using laser-induced breakdown spectroscopy with variable index and chemometrics. Front. Plant Sci. 2018, 9, 1316. [Google Scholar] [CrossRef]
- Divya, B.; Harish, S.; Ramaswamy, K.; Kishorebabu, M.; Govindaiah, R.; Rambabu, U.; Munirathnam, N.R.; Raju, N. Estimation of Cd, Pb and flame retardants in electric mosquito bat using EDXRF, ICP-OES, AAS and GC–MS. Int. J. Environ. Sci. Technol. 2017, 14, 2603–2612. [Google Scholar] [CrossRef]
- Yang, W.; Li, B.; Zhou, J.; Han, Y.; Wang, Q. Continuous-wavelet-transform-based automatic curve fitting method for laser-induced breakdown spectroscopy. Appl. Opt. 2018, 57, 7526–7532. [Google Scholar] [CrossRef]
- Tripathi, M.M.; Eseller, K.E.; Yueh, F.-Y.; Singh, J.P. Multivariate calibration of spectra obtained by laser induced breakdown spectroscopy of plutonium oxide surrogate residues. Spectrochim. Acta Part B: At. Spectrosc. 2009, 64, 1212–1218. [Google Scholar] [CrossRef]
- Liu, F.; Ye, L.; Peng, J.; Song, K.; Shen, T.; Zhang, C.; He, Y. Fast detection of copper content in rice by laser-induced breakdown spectroscopy with uni- and multivariate analysis. Sensors 2018, 18, 705. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, Q.; Liu, F.; He, Y.; Xiao, Y. Rapid and non-destructive measurement of spinach pigments content during storage using hyperspectral imaging with chemometrics. Measurement 2017, 97, 149–155. [Google Scholar] [CrossRef]
- Vapnik, V.N. The Nature of Statistical Learning Theory; Springer-Verlag: Berlin/Heidelberg, Germany, 1995. [Google Scholar]
- Liu, F.; He, Y. Use of visible and near infrared spectroscopy and least squares-support vector machine to determine soluble solids content and pH of cola beverage. J. Agric. Food Chem. 2007, 55, 8883–8888. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; He, M.; Shen, T.; Liu, F.; He, Y.; Liu, X.; Qiu, Z. Multi-element analysis of heavy metal content in soils using laser-induced breakdown spectroscopy: A case study in eastern China. Spectrochim. Acta Part B: At. Spectrosc. 2018, 149, 300–312. [Google Scholar] [CrossRef]
- Ytsma, C.R.; Dyar, M.D. Effects of univariate and multivariate regression on the accuracy of hydrogen quantification with laser-induced breakdown spectroscopy. Spectrochim. Acta Part B: At. Spectrosc. 2018, 139, 27–37. [Google Scholar] [CrossRef]
- Li, X.; Wang, Z.; Lui, S.L.; Fu, Y.; Li, Z.; Liu, J.; Ni, W. A partial least squares based spectrum normalization method for uncertainty reduction for laser-induced breakdown spectroscopy measurements. Spectrochim. Acta Part B: At. Spectrosc. 2013, 88, 180–185. [Google Scholar] [CrossRef]
- Moncayo, S.; Manzoor, S.; Rosales, J.; Anzano, J.; Caceres, J. Qualitative and quantitative analysis of milk for the detection of adulteration by Laser-induced breakdown spectroscopy (LIBS). Food Chem. 2017, 232, 322–328. [Google Scholar] [CrossRef]
- Bricklemyer, R.S.; Brown, D.J.; Turk, P.J.; Clegg, S.M. Improved intact soil-core carbon determination applying regression shrinkage and variable selection techniques to complete spectrum laser-induced breakdown spectroscopy (LIBS). Appl. Spectrosc. 2013, 67, 1185–1199. [Google Scholar] [CrossRef]
Sample Availability: Samples of the Panax notoginseng are not available from the authors. |
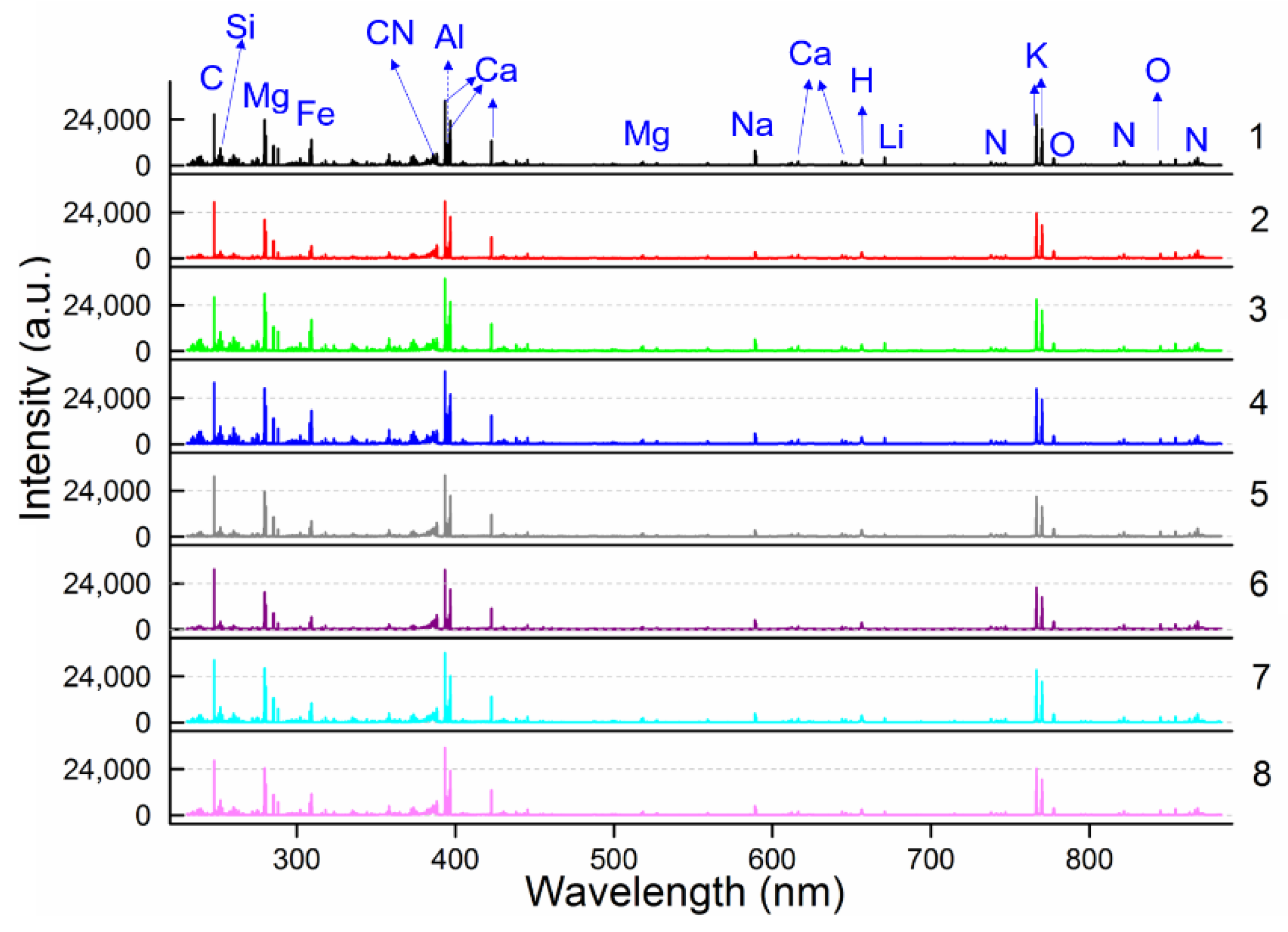
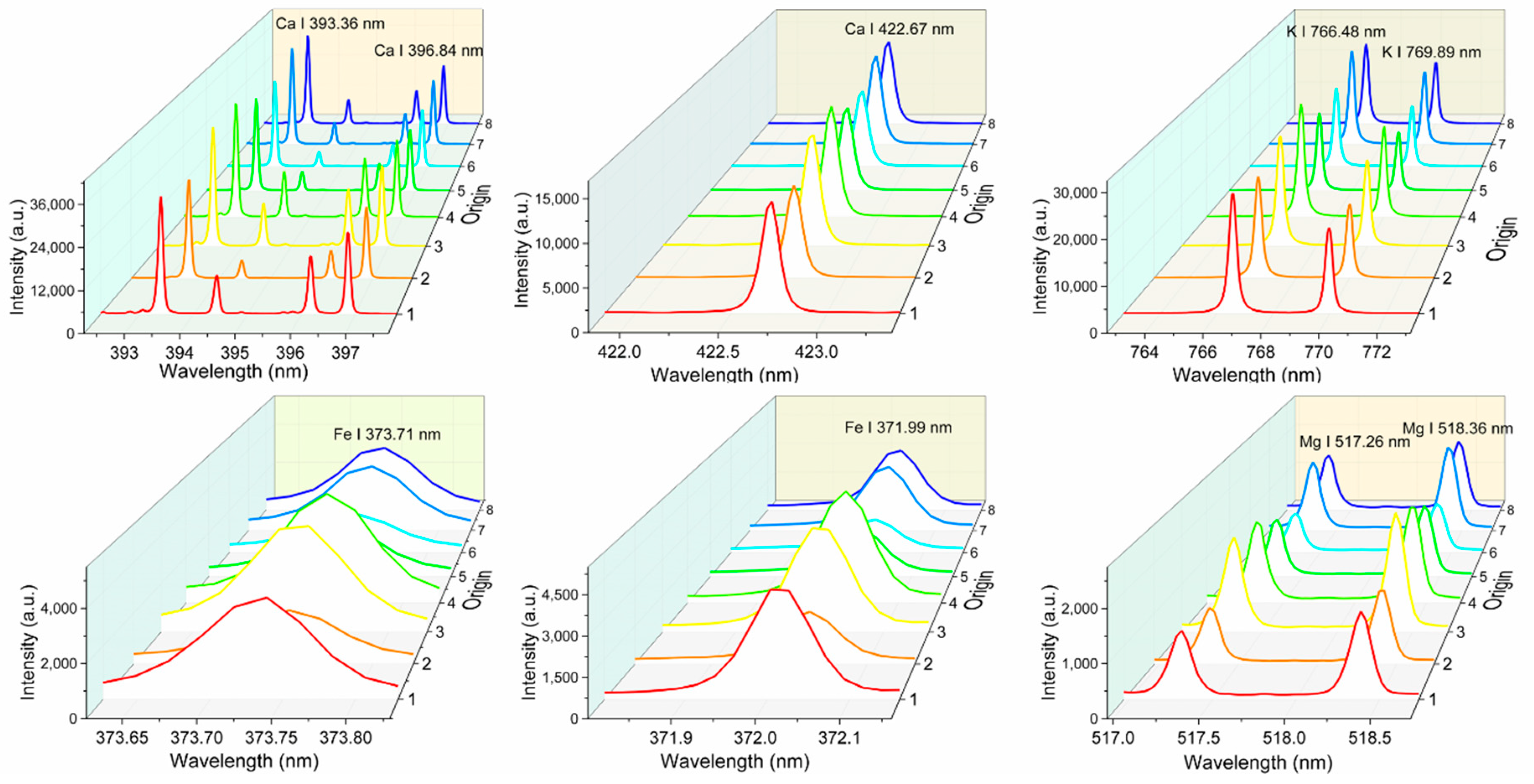

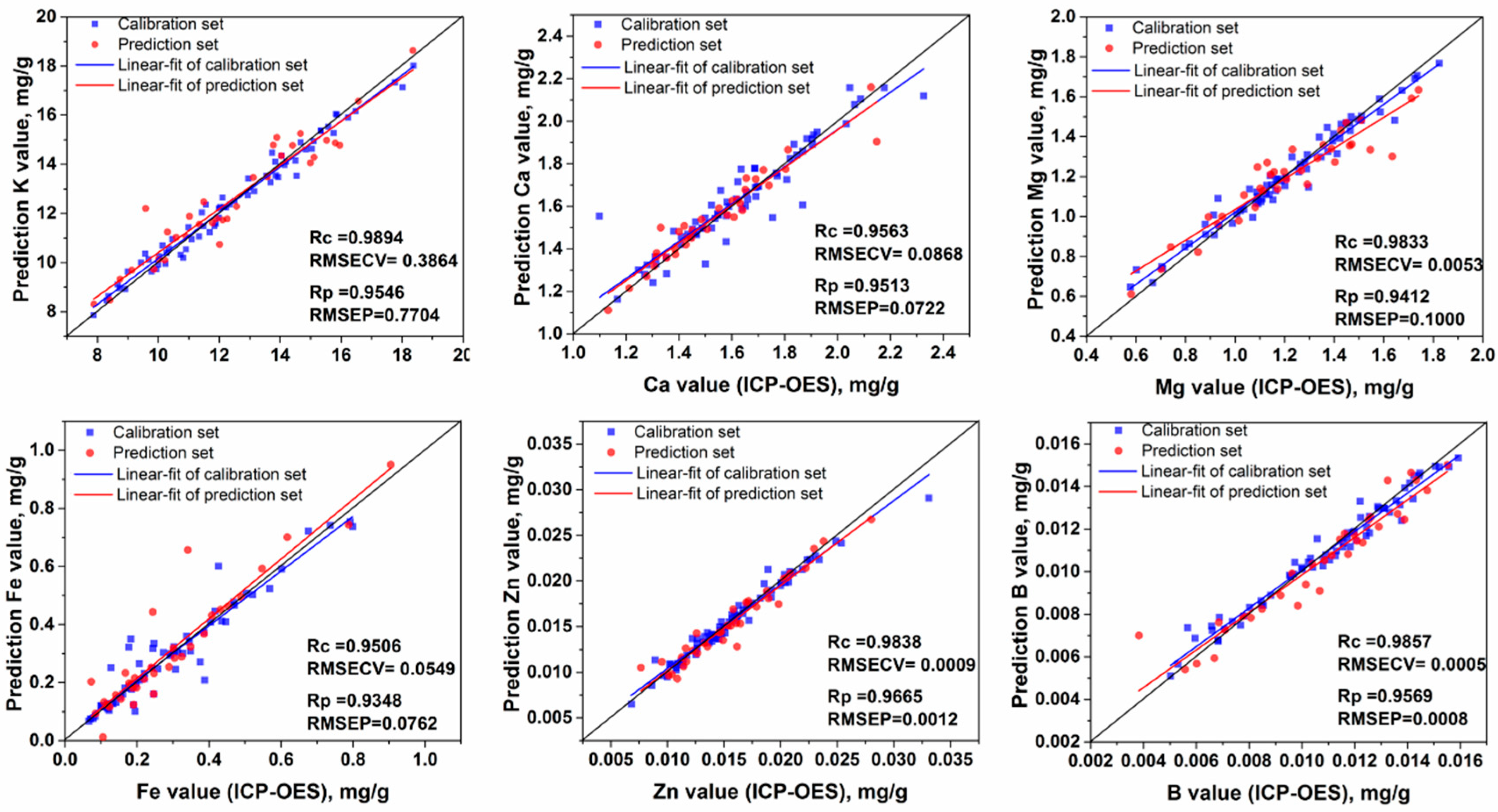
| Element | Groups a | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|---|
| Number | 13 | 13 | 13 | 13 | 13 | 13 | 13 | 13 | |
| K | Min | 11.5682 | 8.2990 | 12.1523 | 11.0964 | 7.8709 | 6.4402 | 10.2981 | 8.7456 |
| Max | 17.4899 | 15.4417 | 19.7162 | 18.3589 | 10.8974 | 13.8608 | 15.8312 | 18.0112 | |
| Mean | 14.0558 | 11.7255 | 16.0186 | 14.7329 | 9.3546 | 10.0683 | 12.8228 | 13.9661 | |
| S.D. | 1.6676 | 1.9894 | 2.5840 | 2.0317 | 0.9795 | 1.8793 | 1.4315 | 2.71071 | |
| Ca | Min | 1.4194 | 1.1814 | 1.5225 | 1.2717 | 1.2216 | 1.0807 | 1.0997 | 1.4007 |
| Max | 2.1659 | 1.8081 | 2.4316 | 2.3112 | 1.8509 | 2.3226 | 1.9342 | 2.3260 | |
| Mean | 1.7756 | 1.4200 | 1.9667 | 1.6736 | 1.4758 | 1.3450 | 1.4921 | 1.8509 | |
| S.D. | 0.2292 | 0.2062 | 0.4276 | 0.3137 | 0.1993 | 0.3683 | 0.2203 | 0.4987 | |
| Mg | Min | 0.8821 | 0.8153 | 1.0908 | 1.1779 | 1.0813 | 0.5774 | 0.9143 | 1.1373 |
| Max | 1.2918 | 1.4632 | 1.9072 | 1.6739 | 1.7435 | 1.1640 | 1.8240 | 1.8797 | |
| Mean | 1.1343 | 1.0583 | 1.5977 | 1.4081 | 1.3567 | 0.7984 | 1.2961 | 1.3825 | |
| S.D. | 0.1194 | 0.1824 | 0.2170 | 0.1559 | 0.2113 | 0.1802 | 0.2730 | 0.2240 | |
| Fe | Min | 0.0288 | 0.0711 | 0.0837 | 0.1003 | 0.0661 | 0.0781 | 0.1154 | 0.0783 |
| Max | 0.8145 | 0.3023 | 1.0317 | 0.9004 | 0.3862 | 0.5021 | 0.7329 | 0.7258 | |
| Mean | 0.2401 | 0.1598 | 0.5550 | 0.4918 | 0.1885 | 0.1903 | 0.3003 | 0.3785 | |
| S.D. | 0.0938 | 0.0668 | 0.1039 | 0.0546 | 0.0869 | 0.0779 | 0.0803 | 0.0840 | |
| Zn | Min | 0.0147 | 0.0075 | 0.0122 | 0.0116 | 0.0103 | 0.0085 | 0.0069 | 0.0121 |
| Max | 0.0351 | 0.0225 | 0.0303 | 0.0250 | 0.0159 | 0.0217 | 0.0159 | 0.0242 | |
| Mean | 0.0203 | 0.0130 | 0.0213 | 0.0192 | 0.0129 | 0.0131 | 0.0113 | 0.0183 | |
| S.D. | 0.0073 | 0.0037 | 0.0089 | 0.0044 | 0.0016 | 0.0041 | 0.0022 | 0.0041 | |
| B | Min | 0.0091 | 0.0061 | 0.0057 | 0.0035 | 0.0038 | 0.0027 | 0.0074 | 0.0047 |
| Max | 0.0154 | 0.0148 | 0.0165 | 0.0159 | 0.0134 | 0.0167 | 0.0147 | 0.0156 | |
| Mean | 0.0138 | 0.0105 | 0.0131 | 0.0130 | 0.0074 | 0.0079 | 0.0105 | 0.0132 | |
| S.D. | 0.0032 | 0.0024 | 0.0066 | 0.0057 | 0.0026 | 0.0036 | 0.0023 | 0.0038 |
| Elements | Wavelength (nm) |
|---|---|
| C I | 247.86, 296.72 |
| Si I | 250.68, 251.43, 251.61, 251.92, 252.41, 288.15 |
| Fe I | 302.06, 371.99, 385.99, 293.69, 498.24, 499.41 |
| Fe II | 253.54, 257.60, 259.37, 260.54, 263.08, |
| Mg I | 277.98, 382.94, 383.23, 383.83, 389.19, 516.73, 517.27, 518.36 |
| Mg II | 279.55, 279.80, 280.27 |
| Ca I | 299.50, 300.09, 300.69, 422.67, 428.30, 428.94, 429.90, 430.25, 430.77, 431.87, 442.54, 443.50, 458.15, 458.60, 527.03, 558.87, 559.45, 559.85, 585.75, 610.27, 612.22, 616.22, 643.91, 644.98, 646.26, 649.38, 671.77, 714.82, 854.21 |
| Ca II | 315.89, 317.93, 373.69, 393.37, 396.85, 866.21 |
| Sc II | 364.37 |
| CN | 385.01 (CN 4-4), 385.44 (CN 3-3), 386.15 (CN 2-2), 387.12 (CN 1-1), 388.32 (CN 0-0) |
| Al I | 394.40, 396.15 |
| K I | 693.87, 766.49, 769.90 |
| Sr I | 460.73 |
| Sr II | 407.77, 421.55 |
| Na I | 589.00, 589.59 |
| H | 656.28 |
| O I | 777.42, 844.67 |
| Li I | 670.79 |
| N I | 742.36, 744.23, 746.83, 818.48, 821.63, 824.23, 862.92, 868.02 |
| Emission Lines nm | Calibration Set | Prediction Set | ||
|---|---|---|---|---|
| Rc | RMSECV mg/g | Rp | RMSEP mg/g | |
| K I 766.49 | 0.8324 | 1.4002 | 0.7476 | 1.7601 |
| K I 769.90 | 0.8413 | 1.3707 | 0.7836 | 1.6103 |
| Ca II 393.37 | 0.6872 | 0.1284 | 0.6327 | 0.1282 |
| Ca II 396.85 | 0.7764 | 0.1104 | 0.7118 | 0.1160 |
| Ca I 422.67 | 0.7941 | 0.1062 | 0.7779 | 0.1074 |
| Mg I 517.27 | 0.8403 | 0.1519 | 0.7564 | 0.1927 |
| Mg I 518.36 | 0.7520 | 0.1840 | 0.7168 | 0.2025 |
| Fe I 373.71 | 0.7509 | 0.1217 | 0.8378 | 0.1027 |
| Fe I 371.99 | 0.8944 | 0.0820 | 0.8577 | 0.0973 |
| Element | Model | Parameter | Calibration Set | Prediction Set | ||
|---|---|---|---|---|---|---|
| Rc | RMSECV mg/g | Rp | RMSEP mg/g | |||
| K | PLS d | 10 a | 0.9558 | 0.8120 | 0.9505 | 0.7152 |
| LS-SVM | (992.5, 799,024.9) b | 0.9800 | 0.3120 | 0.9391 | 0.8721 | |
| Lasso | 53 c | 0.9547 | 0.7740 | 0.9496 | 0.7956 | |
| Ca | PLSd | 13 a | 0.9563 | 0.0868 | 0.9513 | 0.0722 |
| LS-SVM | (111.5, 16,929,970) b | 0.9799 | 0.0357 | 0.9135 | 0.1101 | |
| Lasso | 54 c | 0.9533 | 0.0872 | 0.9508 | 0.0798 | |
| Mg | PLS | 11 a | 0.9270 | 0.1066 | 0.9171 | 0.1182 |
| LS-SVM | (236.1, 344,300.9) b | 0.9601 | 0.0986 | 0.9011 | 0.1246 | |
| Lassod | 51 c | 0.9294 | 0.1022 | 0.9207 | 0.1110 | |
| Fe | PLS | 4 a | 0.9234 | 0.0791 | 0.9334 | 0.0906 |
| LS-SVM | (311.9, 4,680,480) b | 0.9799 | 0.0451 | 0.9284 | 0.0854 | |
| Lassod | 51 c | 0.9506 | 0.0549 | 0.9348 | 0.0762 | |
| Zn | PLSd | 4 a | 0.9503 | 0.0017 | 0.9460 | 0.0016 |
| LS-SVM | (289.3, 1,593,133.6) b | 0.9886 | 0.0009 | 0.9060 | 0.0021 | |
| Lasso | 54 c | 0.9406 | 0.0015 | 0.9228 | 0.0019 | |
| B | PLSd | 4 a | 0.9566 | 0.0008 | 0.9475 | 0.0010 |
| LS-SVM | (244.1, 48,672.5) b | 0.9866 | 0.0007 | 0.9036 | 0.0014 | |
| Lasso | 52 c | 0.9502 | 0.0009 | 0.9348 | 0.0009 | |
| Element (number) | Model | Parameter | Calibration Set | Prediction Set | ||
|---|---|---|---|---|---|---|
| Rc | RMSECV mg/g | Rp | RMSEP mg/g | |||
| K (64) | PLS | 8 a | 0.9655 | 0.6852 | 0.9530 | 0.7853 |
| LS-SVM d | (192.1, 6,274.1) b | 0.9894 | 0.3864 | 0.9546 | 0.7704 | |
| Lasso | 78 c | 0.9689 | 0.6491 | 0.9482 | 0.8239 | |
| Ca (73) | PLS | 13 a | 0.9420 | 0.0638 | 0.9047 | 0.0757 |
| LS-SVM d | (691.7, 19,536.2) b | 0.9890 | 0.0299 | 0.9176 | 0.0712 | |
| Lasso | 66 c | 0.9416 | 0.0639 | 0.9012 | 0.0776 | |
| Mg (61) | PLS | 7 a | 0.9405 | 0.0957 | 0.9365 | 0.0979 |
| LS-SVM d | (146.2, 3,195.7) b | 0.9833 | 0.0053 | 0.9412 | 0.1000 | |
| Lasso | 60 c | 0.9236 | 0.1080 | 0.9291 | 0.1034 | |
| Fe (66) | PLS d | 6 a | 0.9299 | 0.0684 | 0.9169 | 0.0724 |
| LS-SVM | (2585.9, 20,694.3) b | 0.9999 | 0.0002 | 0.9159 | 0.0891 | |
| Lasso | 100 c | 0.9070 | 0.0784 | 0.9034 | 0.0801 | |
| Zn (73) | PLS | 6 a | 0.9158 | 0.0018 | 0.9613 | 0.0012 |
| LS-SVM d | (81.3, 3,389.1) b | 0.9838 | 0.0009 | 0.9665 | 0.0012 | |
| Lasso | 100 c | 0.9561 | 0.0013 | 0.9100 | 0.0019 | |
| B (62) | PLS | 6 a | 0.9579 | 0.0008 | 0.9432 | 0.0009 |
| LS-SVM d | (569.8, 16,582.3) b | 0.9857 | 0.0005 | 0.9569 | 0.0008 | |
| Lasso | 70 c | 0.9515 | 0.0009 | 0.9195 | 0.0011 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, T.; Li, W.; Zhang, X.; Kong, W.; Liu, F.; Wang, W.; Peng, J. High-Sensitivity Determination of Nutrient Elements in Panax notoginseng by Laser-induced Breakdown Spectroscopy and Chemometric Methods. Molecules 2019, 24, 1525. https://doi.org/10.3390/molecules24081525
Shen T, Li W, Zhang X, Kong W, Liu F, Wang W, Peng J. High-Sensitivity Determination of Nutrient Elements in Panax notoginseng by Laser-induced Breakdown Spectroscopy and Chemometric Methods. Molecules. 2019; 24(8):1525. https://doi.org/10.3390/molecules24081525
Chicago/Turabian StyleShen, Tingting, Weijiao Li, Xi Zhang, Wenwen Kong, Fei Liu, Wei Wang, and Jiyu Peng. 2019. "High-Sensitivity Determination of Nutrient Elements in Panax notoginseng by Laser-induced Breakdown Spectroscopy and Chemometric Methods" Molecules 24, no. 8: 1525. https://doi.org/10.3390/molecules24081525
APA StyleShen, T., Li, W., Zhang, X., Kong, W., Liu, F., Wang, W., & Peng, J. (2019). High-Sensitivity Determination of Nutrient Elements in Panax notoginseng by Laser-induced Breakdown Spectroscopy and Chemometric Methods. Molecules, 24(8), 1525. https://doi.org/10.3390/molecules24081525






