A Combined ELONA-(RT)qPCR Approach for Characterizing DNA and RNA Aptamers Selected against PCBP-2
Abstract
1. Introduction
2. Results
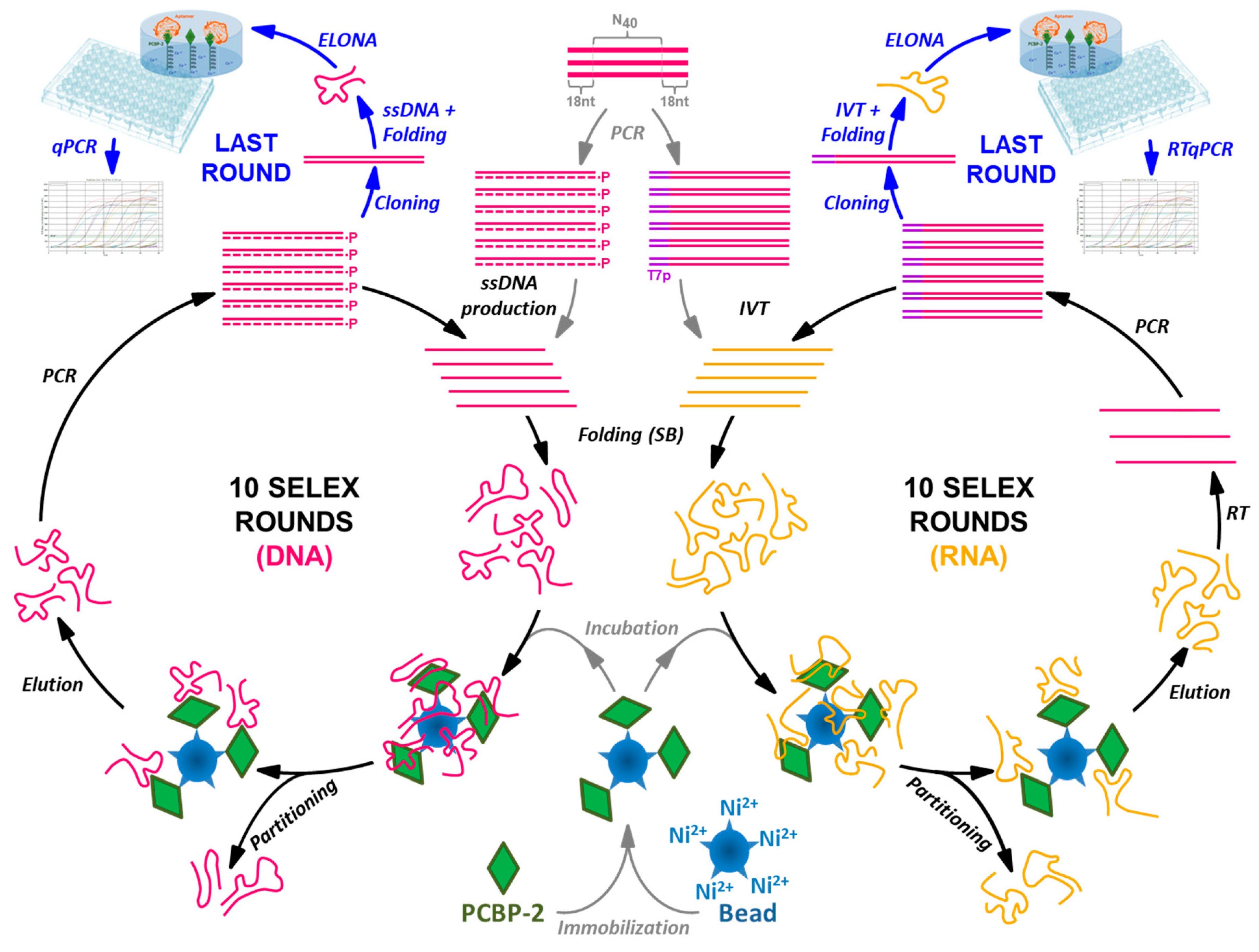
2.1. DNA and RNA Aptamer Selection
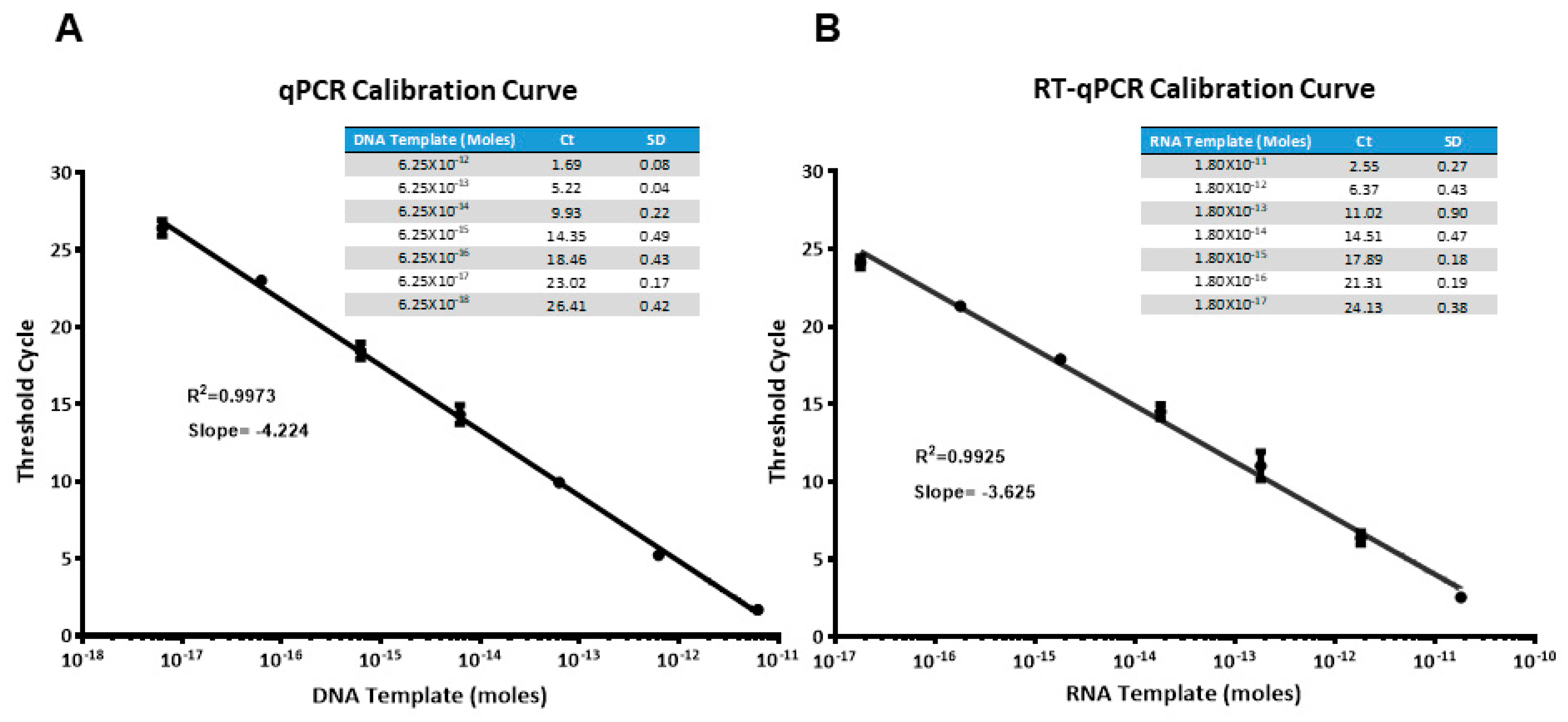
2.2. (RT)qPCR Calibration Curves
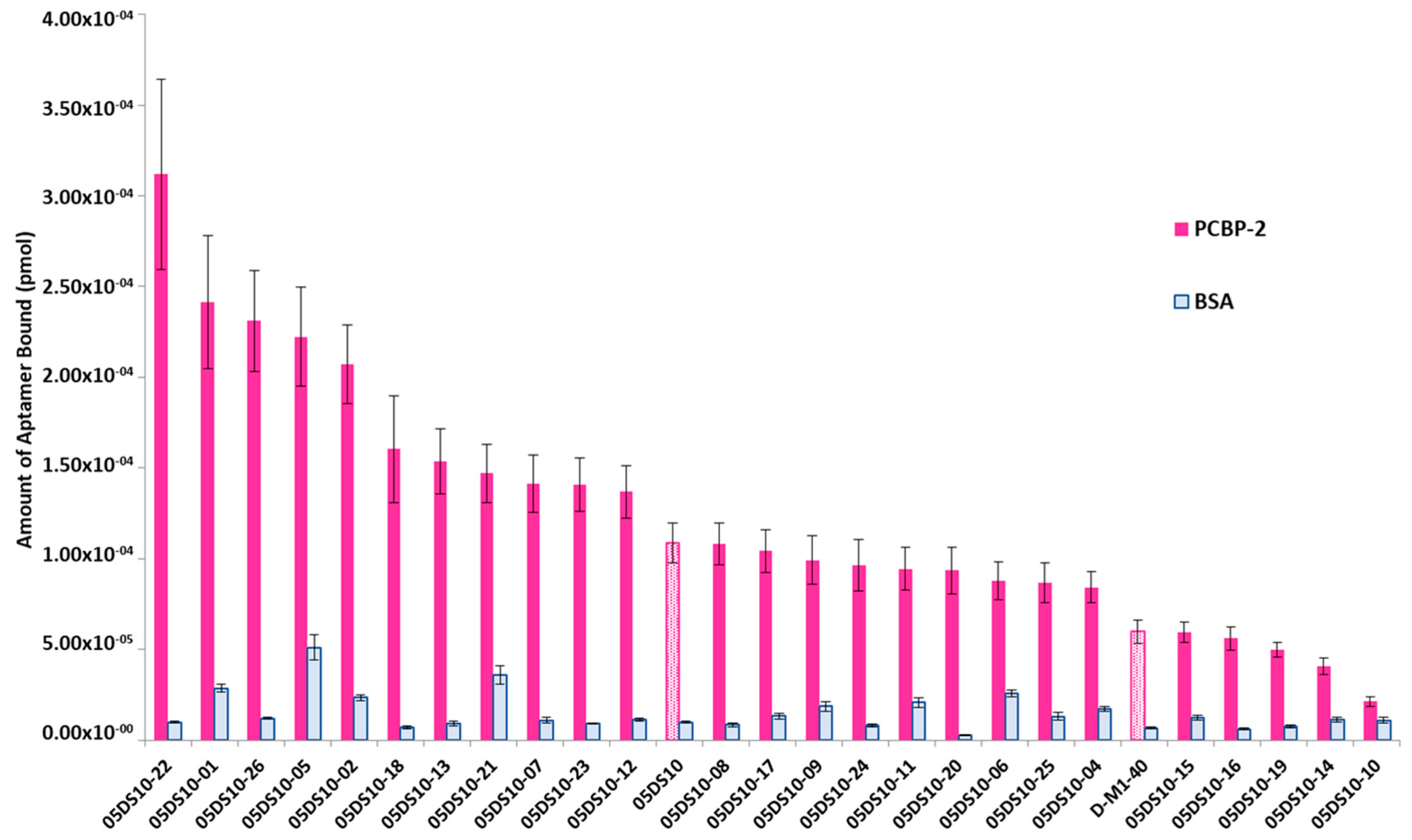
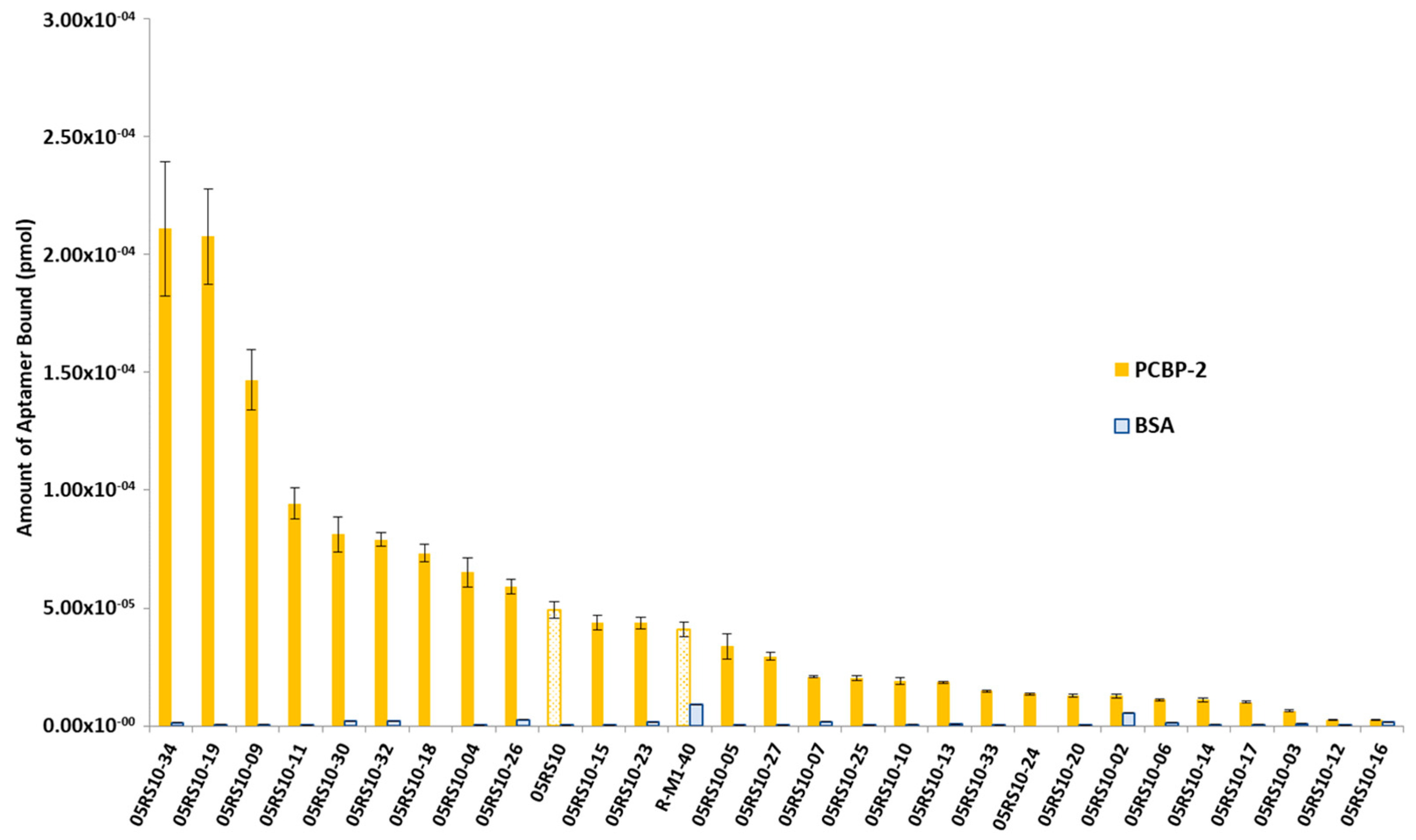
2.3. Affinity Analyses Using ELONA-(RT)qPCR
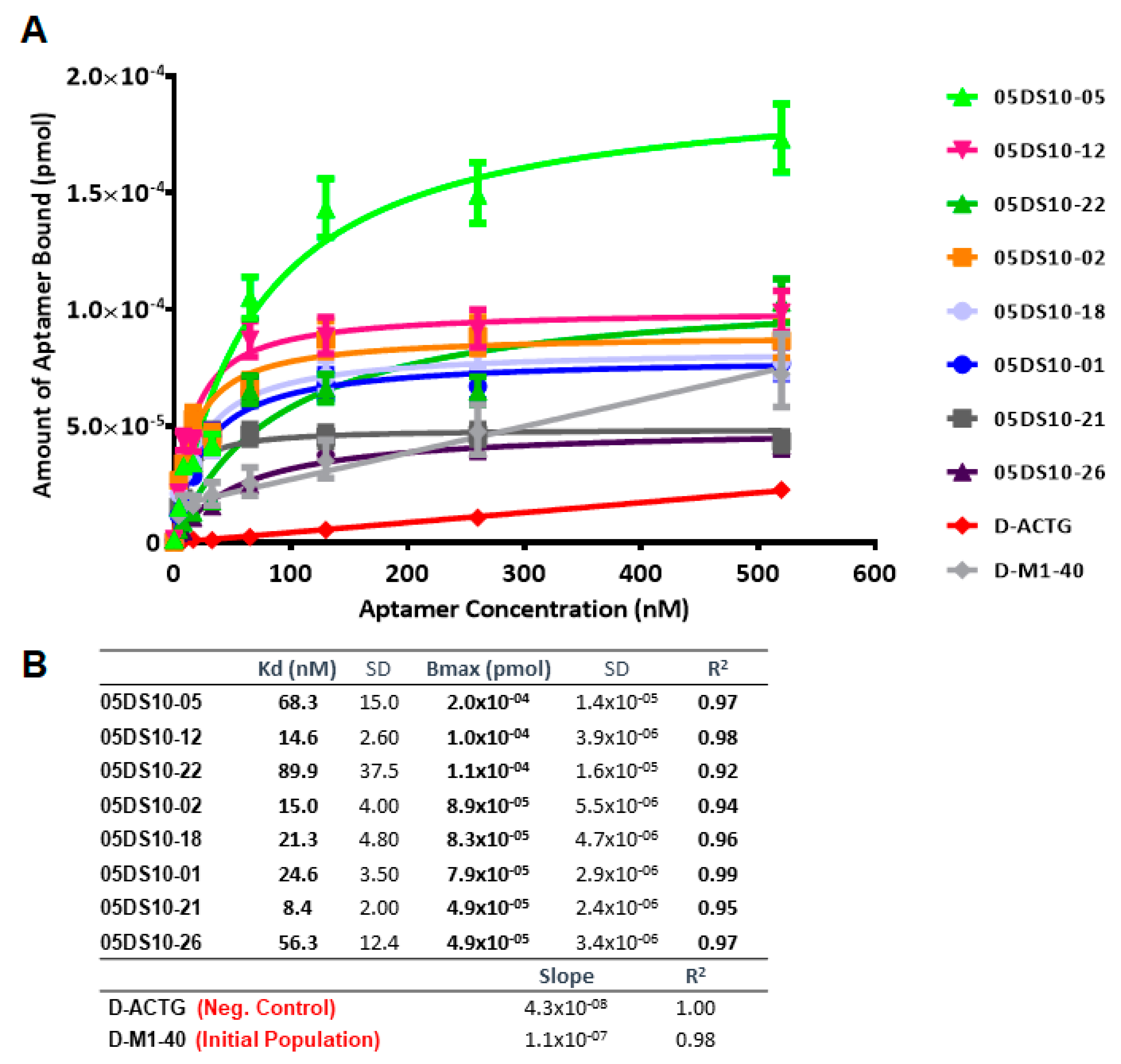
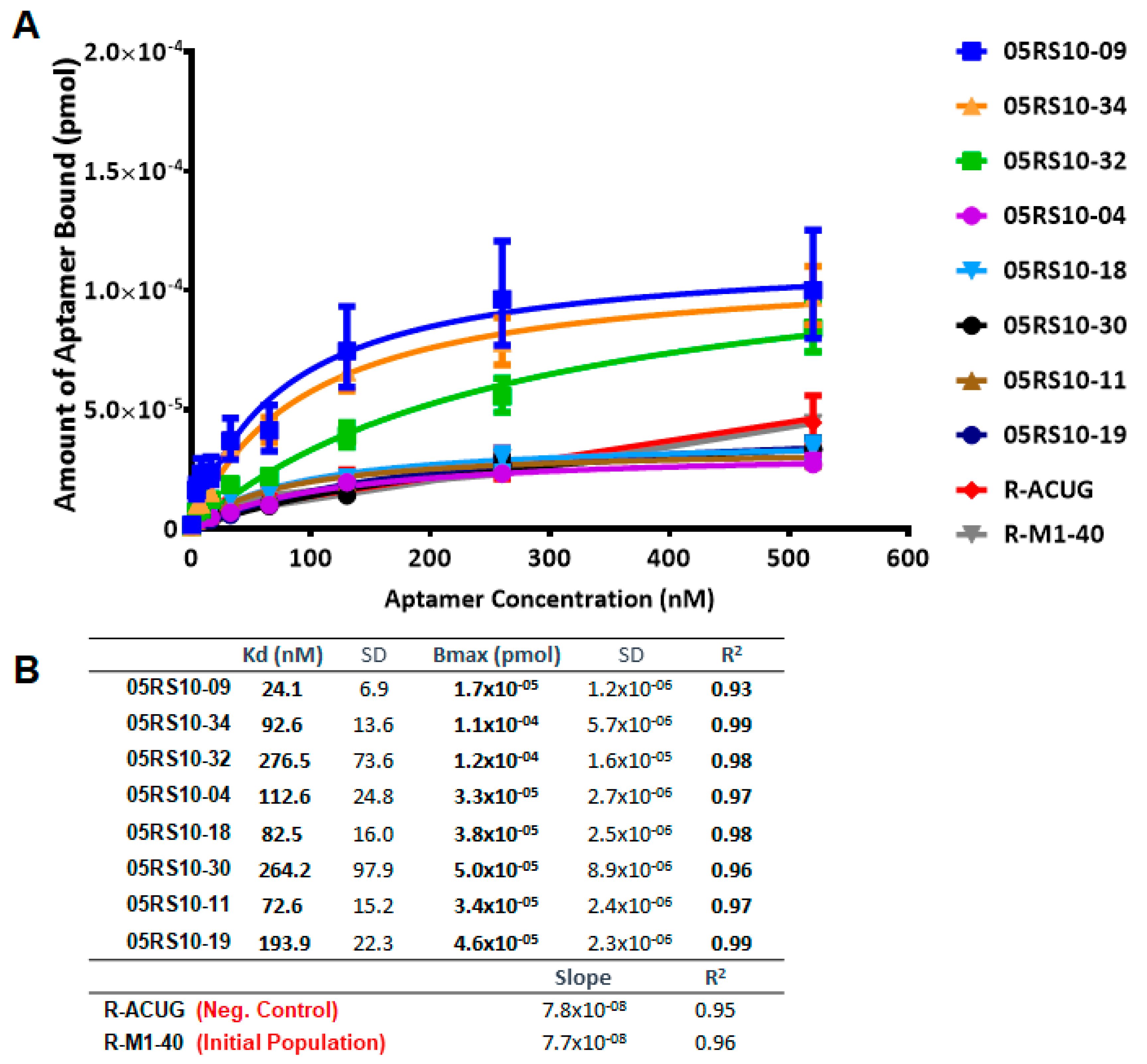
2.4. Determination of Kd and Bmax Using ELONA-(RT)qPCR
2.5. Secondary Structure Prediction of the High Affinity DNA and RNA Aptamers
2.6. Colorimetric ELONA as an Alternative Technique to Analyse Selected DNA Aptamers
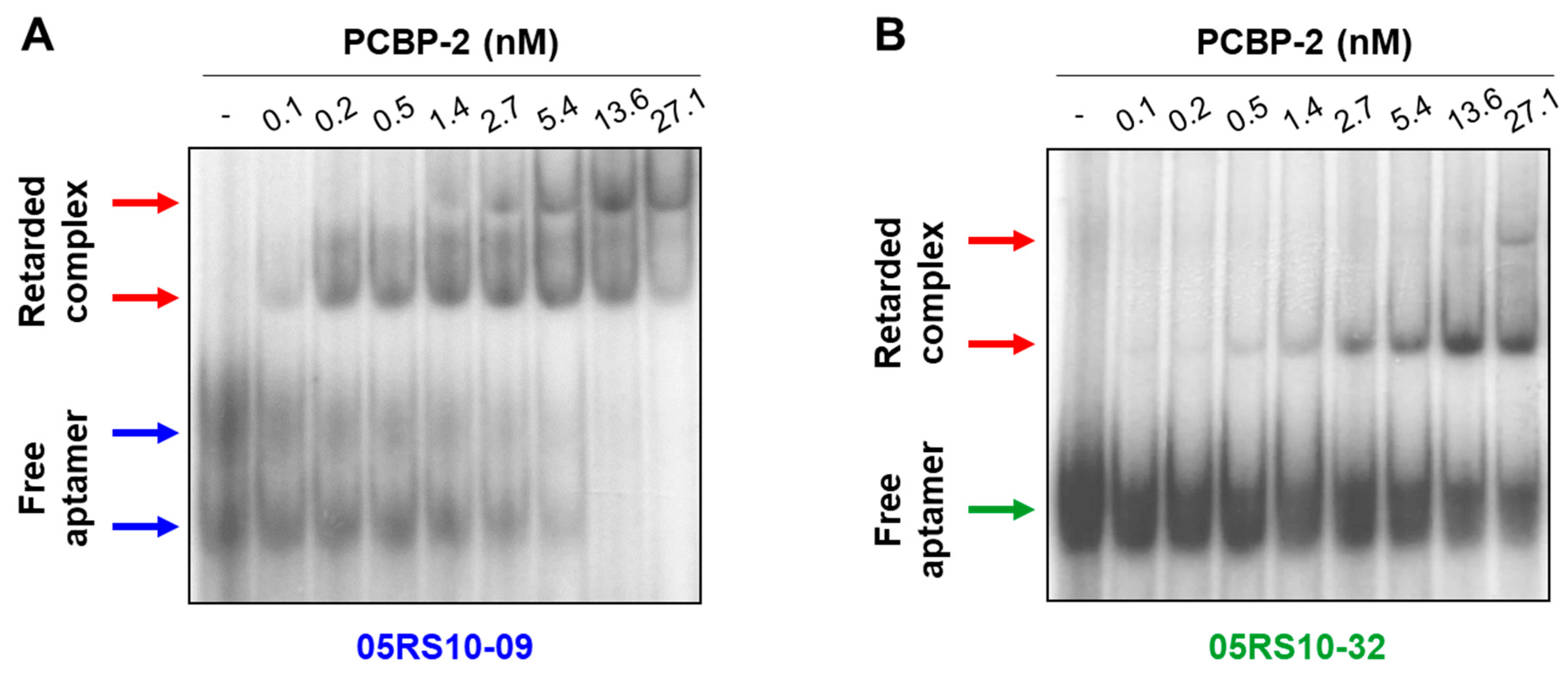
2.7. Binding of RNA Aptamers to PCBP-2 in Solution
3. Discussion
4. Materials and Methods
4.1. Expression and Purification of PCBP-2
4.2. DNA and RNA Selection Protocol
4.3. Molecular Cloning and Sequencing of Individual Aptamers
4.4. Real Time qPCR and RTqPCR
4.5. ELONA-(RT)qPCR for Affinity Analysis
4.6. ELONA-(RT)qPCR for the Functional Characterization of Aptamers
4.7. Colorimetric ELONA for the Functional Characterization of ssDNA Aptamers
4.8. Electrophoretic Mobility Shift Assays (EMSA) of Aptamer–PCBP-2 Complexes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage t4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Yuce, M.; Ullah, N.; Budak, H. Trends in aptamer selection methods and applications. Analyst 2015, 140, 5379–5399. [Google Scholar] [CrossRef] [PubMed]
- Gold, L.; Walker, J. Directed evolution ☆. In Reference Module in Life Sciences; Elsevier: Amsterdam, The Netherlands, 2017. [Google Scholar]
- Blank, M. Next-generation analysis of deep sequencing data: Bringing light into the black box of selex experiments. In Nucleic Acid Aptamers; Mayer, G., Ed.; Springer: New York, NY, USA, 2016; Volume 1380, pp. 85–95. [Google Scholar]
- Alam, K.K.; Chang, J.L.; Burke, D.H. Fastaptamer: A bioinformatic toolkit for high-throughput sequence analysis of combinatorial selections. Mol. Ther. Nucleic Acids 2015, 4, e230. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Cowley, A.; Uludag, M.; Gur, T.; McWilliam, H.; Squizzato, S.; Park, Y.M.; Buso, N.; Lopez, R. The embl-ebi bioinformatics web and programmatic tools framework. Nucleic Acids Res. 2015, 43, W580–W584. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal w and clustal x version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Hoinka, J.; Berezhnoy, A.; Sauna, Z.E.; Gilboa, E.; Przytycka, T.M. Aptacluster—A method to cluster ht-selex aptamer pools and lessons from its application. Res. Comput. Mol. Biol. 2014, 8394, 115–128. [Google Scholar]
- Hoinka, J.; Dao, P.; Przytycka, T.M. Aptagui[mdash]a graphical user interface for the efficient analysis of ht-selex data. Mol. Ther. Nucleic Acids 2015, 4, e257. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, R.; Bernhart, S.H.; Höner zu Siederdissen, C.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. Viennarna package 2.0. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Sharma, T.K.; Bruno, J.G.; Dhiman, A. Abcs of DNA aptamer and related assay development. Biotechnol. Adv. 2017, 35, 275–301. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.Y.; Acquah, C.; Sidhu, A.; Ongkudon, C.M.; Yon, L.S.; Danquah, M.K. Selex modifications and bioanalytical techniques for aptamer-target binding characterization. Crit. Rev. Anal. Chem. 2016, 46, 521–537. [Google Scholar] [CrossRef] [PubMed]
- Ruscito, A.; DeRosa, M.C. Small-molecule binding aptamers: Selection strategies, characterization, and applications. Front. Chem. 2016, 4, 14. [Google Scholar] [CrossRef] [PubMed]
- Drolet, D.W.; Moon-McDermott, L.; Romig, T.S. An enzyme-linked oligonucleotide assay. Nat. Biotechnol. 1996, 14, 1021–1025. [Google Scholar] [CrossRef] [PubMed]
- Ljungstrom, I.; Engvall, E.; Ruitenberg, E.J. Proceedings: Elisa, enzyme linked immunosorbent assay—A new technique for sero-diagnosis of trichinosis. Parasitology 1974, 69, xxiv. [Google Scholar] [PubMed]
- Vivekananda, J.; Kiel, J.L. Anti-francisella tularensis DNA aptamers detect tularemia antigen from different subspecies by aptamer-linked immobilized sorbent assay. Lab. Investig. 2006, 86, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Jee, M.H.; Kwon, O.S.; Keum, S.J.; Jang, S.K. Infectivity of hepatitis c virus correlates with the amount of envelope protein e2: Development of a new aptamer-based assay system suitable for measuring the infectious titer of hcv. Virology 2013, 439, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Farrell, R.E. Chapter 12—Nucleic acid probe technology. In RNA Methodologies, 4th ed.; Farrell, R.E., Ed.; Academic Press: San Diego, CA, USA, 2010; pp. 261–282. [Google Scholar]
- Pauff, S.; Withers, J.M.; McKean, I.J.W.; Mackay, S.P.; Burley, G.A. Synthetic biological approaches for RNA labelling and imaging: Design principles and future opportunities. Curr. Opin. Biotechnol. 2017, 48, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Sive, H.L.; Grainger, R.M.; Harland, R.M. Synthesis and purification of digoxigenin-labeled RNA probes for in situ hybridization. CSH Protoc. 2007, 2007, pdb.prot4778. [Google Scholar] [CrossRef]
- Guerra-Perez, N.; Ramos, E.; Garcia-Hernandez, M.; Pinto, C.; Soto, M.; Martin, M.E.; Gonzalez, V.M. Molecular and functional characterization of ssdna aptamers that specifically bind leishmania infantum pabp. PLoS ONE 2015, 10, e0140048. [Google Scholar] [CrossRef]
- Thisted, T.; Lyakhov, D.L.; Liebhaber, S.A. Optimized RNA targets of two closely related triple kh domain proteins, heterogeneous nuclear ribonucleoprotein k and ±cp-2kl, suggest distinct modes of RNA recognition. J. Biol. Chem. 2001, 276, 17484–17496. [Google Scholar] [CrossRef] [PubMed]
- Flynn, R.A.; Martin, L.; Spitale, R.C.; Do, B.T.; Sagan, S.M.; Zarnegar, B.; Qu, K.; Khavari, P.A.; Quake, S.R.; Sarnow, P.; et al. Dissecting noncoding and pathogen RNA-protein interactomes. RNA 2015, 21, 135–143. [Google Scholar] [CrossRef]
- Martinez-Salas, E.; Francisco-Velilla, R.; Fernandez-Chamorro, J.; Lozano, G.; Diaz-Toledano, R. Picornavirus ires elements: RNA structure and host protein interactions. Virus Res. 2015, 206, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Fujimura, K.; Kano, F.; Murata, M. Identification of pcbp2, a facilitator of ires-mediated translation, as a novel constituent of stress granules and processing bodies. RNA 2008, 14, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Perera, R.; Daijogo, S.; Walter, B.L.; Nguyen, J.H.C.; Semler, B.L. Cellular protein modification by poliovirus: The two faces of poly(rc)-binding protein. J. Virol. 2007, 81, 8919–8932. [Google Scholar] [CrossRef] [PubMed]
- Lozano, G.; Fernandez, N.; Martinez-Salas, E. Modeling three-dimensional structural motifs of viral ires. J. Mol. Biol. 2016, 428, 767–776. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Chamorro, J.; Francisco-Velilla, R.; Ramajo, J.; Martinez-Salas, E. Rab1b and arf5 are novel RNA-binding proteins involved in fmdv ires-driven RNA localization. Life Sci. Alliance 2019, 2. [Google Scholar] [CrossRef] [PubMed]
- Venables, J.P.; Koh, C.S.; Froehlich, U.; Lapointe, E.; Couture, S.; Inkel, L.; Bramard, A.; Paquet, E.R.; Watier, V.; Durand, M.; et al. Multiple and specific mrna processing targets for the major human hnrnp proteins. Mol. Cell. Biol. 2008, 28, 6033–6043. [Google Scholar] [CrossRef] [PubMed]
- Makeyev, A.V.; Liebhaber, S.A. The poly(c)-binding proteins: A multiplicity of functions and a search for mechanisms. RNA 2002, 8, 265–278. [Google Scholar] [CrossRef]
- Li, F.; Bullough, K.Z.; Vashisht, A.A.; Wohlschlegel, J.A.; Philpott, C.C. Poly(rc)-binding protein 2 regulates hippo signaling to control growth in breast epithelial cells. Mol. Cell. Biol. 2016, 36, 2121–2131. [Google Scholar] [CrossRef]
- Adinolfi, S.; Bagni, C.; Castiglione Morelli, M.A.; Fraternali, F.; Musco, G.; Pastore, A. Novel RNA-binding motif: The kh module. Biopolymers 1999, 51, 153–164. [Google Scholar] [CrossRef]
- Yeap, B.B.; Voon, D.C.; Vivian, J.P.; McCulloch, R.K.; Thomson, A.M.; Giles, K.M.; Czyzyk-Krzeska, M.F.; Furneaux, H.; Wilce, M.C.J.; Wilce, J.A.; et al. Novel binding of hur and poly(c)-binding protein to a conserved uc-rich motif within the 3′-untranslated region of the androgen receptor messenger RNA. J. Biol. Chem. 2002, 277, 27183–27192. [Google Scholar] [CrossRef] [PubMed]
- Nabavinia, M.S.; Charbgoo, F.; Alibolandi, M.; Mosaffa, F.; Gholoobi, A.; Ramezani, M.; Abnous, K. Comparison of flow cytometry and elasa for screening of proper candidate aptamer in cell-selex pool. Appl. Biochem. Biotechnol. 2018, 184, 444–452. [Google Scholar] [CrossRef] [PubMed]
- Chang, A.L.; McKeague, M.; Smolke, C.D. Facile characterization of aptamer kinetic and equilibrium binding properties using surface plasmon resonance. Methods Enzymol. 2014, 549, 451–466. [Google Scholar]
- Potty, A.S.; Kourentzi, K.; Fang, H.; Jackson, G.W.; Zhang, X.; Legge, G.B.; Willson, R.C. Biophysical characterization of DNA aptamer interactions with vascular endothelial growth factor. Biopolymers 2009, 91, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Luque, F.J.; Stich, M.; Manrubia, S.; Briones, C.; Berzal-Herranz, A. Efficient hiv-1 inhibition by a 16 nt-long RNA aptamer designed by combining in vitro selection and in silico optimisation strategies. Sci. Rep. 2014, 4, 6242. [Google Scholar] [CrossRef] [PubMed]
- Stuart, C.H.; Riley, K.R.; Boyacioglu, O.; Herpai, D.M.; Debinski, W.; Qasem, S.; Marini, F.C.; Colyer, C.L.; Gmeiner, W.H. Selection of a novel aptamer against vitronectin using capillary electrophoresis and next generation sequencing. Mol. Ther. Nucleic Acids 2016, 5, e386. [Google Scholar] [CrossRef]
- Pinto, A.; Bermudo Redondo, M.C.; Ozalp, V.C.; O’Sullivan, C.K. Real-time apta-pcr for 20 000-fold improvement in detection limit. Mol. BioSyst. 2009, 5, 548–553. [Google Scholar] [CrossRef]
- Civit, L.; Pinto, A.; Rodrigues-Correia, A.; Heckel, A.; O’Sullivan, C.K.; Mayer, G. Sensitive detection of cancer cells using light-mediated apta-pcr. Methods 2016, 97, 104–109. [Google Scholar] [CrossRef]
- Pinto, A.; Polo, P.; Henry, O.; Redondo, M.C.; Svobodova, M.; O’Sullivan, C. Label-free detection of gliadin food allergen mediated by real-time apta-pcr. Anal. Bioanal. Chem. 2014, 406, 515–524. [Google Scholar] [CrossRef]
- Bueno, R.; Marciello, M.; Moreno, M.; Sánchez-Sánchez, C.; Martinez, J.I.; Martinez, L.; Prats-Alfonso, E.; Guimerà-Brunet, A.; Garrido, J.A.; Villa, R.; et al. Versatile graphene-based platform for robust nanobiohybrid interfaces. ACS Omega 2019, 4, 3287–3297. [Google Scholar] [CrossRef]
- Sean, P.; Nguyen, J.H.; Semler, B.L. Altered interactions between stem-loop iv within the 5′ noncoding region of coxsackievirus RNA and poly(rc) binding protein 2: Effects on ires-mediated translation and viral infectivity. Virology 2009, 389, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Francisco-Velilla, R.; Fernandez-Chamorro, J.; Lozano, G.; Diaz-Toledano, R.; Martinez-Salas, E. RNA-protein interaction methods to study viral ires elements. Methods 2015, 91, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Hall, B.; Micheletti, J.M.; Satya, P.; Ogle, K.; Pollard, J.; Ellington, A.D. Design, Synthesis, and Amplification of DNA Pools for in Vitro Selection; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2001. [Google Scholar]
- Fernandez, N.; Garcia-Sacristan, A.; Ramajo, J.; Briones, C.; Martinez-Salas, E. Structural analysis provides insights into the modular organization of picornavirus ires. Virology 2011, 409, 251–261. [Google Scholar] [CrossRef]
- Fernandez-Chamorro, J.; Pineiro, D.; Gordon, J.M.; Ramajo, J.; Francisco-Velilla, R.; Macias, M.J.; Martinez-Salas, E. Identification of novel non-canonical RNA-binding sites in gemin5 involved in internal initiation of translation. Nucleic Acids Res. 2014, 42, 5742–5754. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moreno, M.; Fernández-Algar, M.; Fernández-Chamorro, J.; Ramajo, J.; Martínez-Salas, E.; Briones, C. A Combined ELONA-(RT)qPCR Approach for Characterizing DNA and RNA Aptamers Selected against PCBP-2. Molecules 2019, 24, 1213. https://doi.org/10.3390/molecules24071213
Moreno M, Fernández-Algar M, Fernández-Chamorro J, Ramajo J, Martínez-Salas E, Briones C. A Combined ELONA-(RT)qPCR Approach for Characterizing DNA and RNA Aptamers Selected against PCBP-2. Molecules. 2019; 24(7):1213. https://doi.org/10.3390/molecules24071213
Chicago/Turabian StyleMoreno, Miguel, María Fernández-Algar, Javier Fernández-Chamorro, Jorge Ramajo, Encarnación Martínez-Salas, and Carlos Briones. 2019. "A Combined ELONA-(RT)qPCR Approach for Characterizing DNA and RNA Aptamers Selected against PCBP-2" Molecules 24, no. 7: 1213. https://doi.org/10.3390/molecules24071213
APA StyleMoreno, M., Fernández-Algar, M., Fernández-Chamorro, J., Ramajo, J., Martínez-Salas, E., & Briones, C. (2019). A Combined ELONA-(RT)qPCR Approach for Characterizing DNA and RNA Aptamers Selected against PCBP-2. Molecules, 24(7), 1213. https://doi.org/10.3390/molecules24071213




