Contributions of Mass Spectrometry to the Identification of Low Molecular Weight Molecules Able to Reduce the Toxicity of Amyloid-β Peptide to Cell Cultures and Transgenic Mouse Models of Alzheimer’s Disease
Abstract
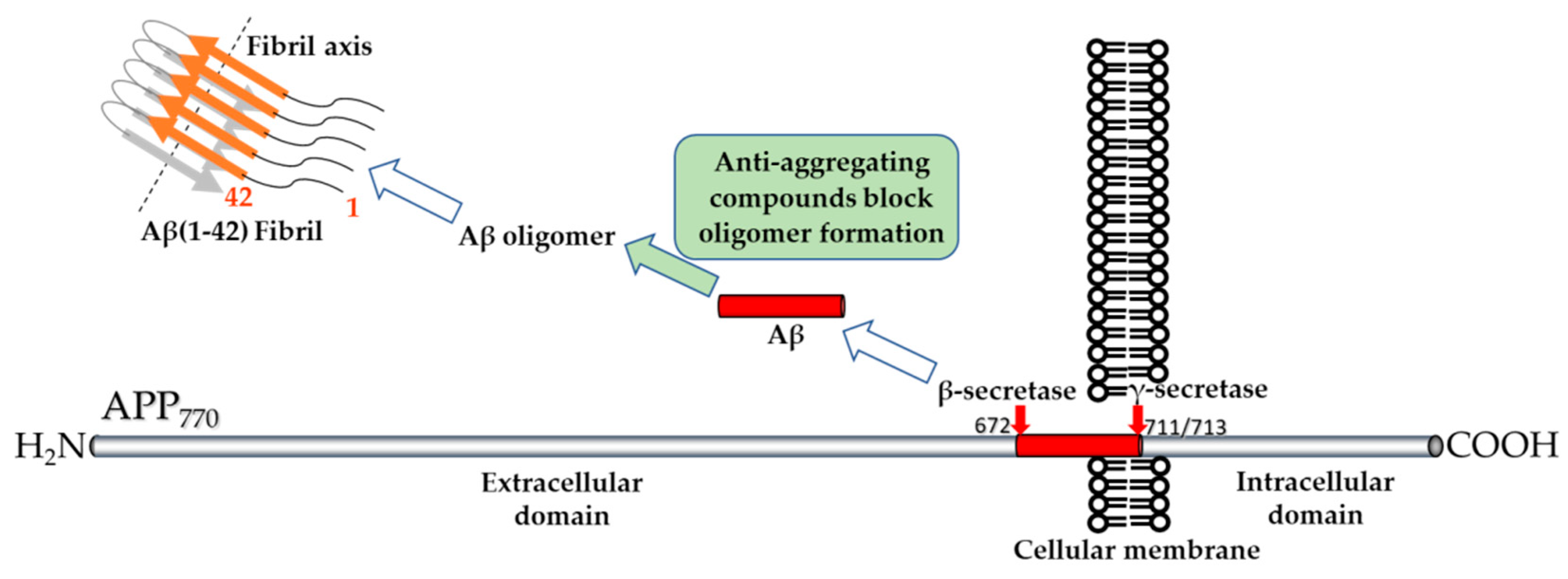
:1. Introduction
2. Low-Molecular Weight Compounds Tested for Decreasing Amyloid-Beta Peptide Oligomerization and Toxicity
2.1. Humanin Peptide (HN)
2.2. Tramiprosate
2.3. Melatonin
2.4. Oleuropein
2.5. Trehalose
2.6. β-cyclodextrin
2.7. GRKKRRQRRR-GGGG-DVEFRH Peptide
2.8. Ac-VVIA and VVIA-NH2 Peptides
2.9. RYYAAFFARR Peptide
2.10. RGTFEGKF Peptide
3. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- World Alzheimer Report 2018; Alzheimer’s Disease International: London, UK, 2018.
- Selkoe, D.J. Alzheimer’s Disease: Genes, Proteins, and Therapy. Physiol. Rev. 2001, 81, 741–766. [Google Scholar] [CrossRef] [PubMed]
- Kuperstein, I.; Broersen, K.; Benilova, I.; Rozenski, J.; Jonckheere, W.; Debulpaep, M.; Vandersteen, A.; Segers-Nolten, I.; Van Der Werf, K.; Subramaniam, V.; et al. Neurotoxicity of Alzheimer’s disease Aβ peptides is induced by small changes in the Aβ42 to Aβ40 ratio. EMBO J. 2010, 29, 3408–3420. [Google Scholar] [CrossRef]
- Scheuner, D.; Eckman, C.; Jensen, M.; Song, X.; Citron, M.; Suzuki, N.; Bird, T.D.; Hardy, J.; Hutton, M.; Kukull, W.; et al. Secreted amyloid β-protein similar to that in the senile plaques of Alzheimer’s disease is increased in vivo by the presenilin 1 and 2 and APP mutations linked to familial Alzheimer’s disease. Nat. Med. 1996, 2, 864–870. [Google Scholar] [CrossRef]
- Tanzi, R.E. The genetics of Alzheimer disease. Cold Spring Harb. Perspect. Med. 2012, 2, a006296. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, S.L.; Dupuis, N.F.; Lazo, N.D.; Wyttenbach, T.; Condron, M.M.; Bitan, G.; Teplow, D.B.; Shea, J.E.; Ruotolo, B.T.; Robinson, C.V.; et al. Amyloid-β protein oligomerization and the importance of tetramers and dodecamers in the aetiology of Alzheimer’s disease. Nat. Chem. 2009, 1, 326–331. [Google Scholar] [CrossRef] [PubMed]
- Ryan, P.; Patel, B.; Makwana, V.; Jadhav, H.R.; Kiefel, M.; Davey, A.; Reekie, T.A.; Rudrawar, S.; Kassiou, M. Peptides, Peptidomimetics, and Carbohydrate-Peptide Conjugates as Amyloidogenic Aggregation Inhibitors for Alzheimer’s Disease. ACS Chem. Neurosci. 2018, 9, 1530–1551. [Google Scholar] [CrossRef]
- Opuni, K.F.M.; Al-Majdoub, M.; Yefremova, Y.; El-Kased, R.F.; Koy, C.; Glocker, M.O. Mass spectrometric epitope mapping. Mass Spectrom. Rev. 2018, 37, 229–241. [Google Scholar] [CrossRef] [PubMed]
- Shipley, M.M.; Mangold, C.A.; Szpara, M.L. Differentiation of the SH-SY5Y Human Neuroblastoma Cell Line. J. Vis. Exp. 2016, e53193. [Google Scholar] [CrossRef] [PubMed]
- Kovalevich, J.; Langford, D. Considerations for the Use of SH-SY5Y Neuroblastoma Cells in Neurobiology BT—Neuronal Cell Culture: Methods and Protocols. In Neuronal Cell Culture; Amini, S., White, M.K., Eds.; Humana Press: Totowa, NJ, USA, 2013; Volume 1078, pp. 9–21. [Google Scholar]
- Ross, R.A.; Spengler, B.A.; Biedler, J.L. Coordinate morphological and biochemical interconversion of human neuroblastoma cells. J. Natl. Cancer Inst. 1983, 71, 741–747. [Google Scholar] [PubMed]
- Eves, E.M.; Tucker, M.S.; Roback, J.D.; Downen, M.; Rosner, M.R.; Wainer, B.H. Immortal rat hippocampal cell lines exhibit neuronal and glial lineages and neurotrophin gene expression. Proc. Natl. Acad. Sci. USA 1992, 89, 4373–4377. [Google Scholar] [CrossRef] [PubMed]
- Ross, C.A.; Akimov, S.S. Human-induced pluripotent stem cells: Potential for neurodegenerative diseases. Hum. Mol. Genet. 2014, 23, R17–R26. [Google Scholar] [CrossRef]
- Perlman, R.L. Mouse Models of Human Disease: An Evolutionary Perspective. Evol. Med. Public Heal. 2016, eow014. [Google Scholar] [CrossRef]
- Elder, G.A.; Sosa, M.A.G.; Gasperi, R. De Transgenic Mouse Models of Alzheimer ’ s Disease. Mt. Sinai Sch. Med. 2010, 77, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Kitazawa, M.; Medeiros, R.; Laferla, F.M. Transgenic Mouse Models of Alzheimer Disease: Developing a Better Model as a Tool for Therapeutic Interventions. Curr. Pharm. Des. 2012, 18, 1131–1147. [Google Scholar] [CrossRef]
- Gong, Z.; Tas, E.; Muzumdar, R. Humanin and age-related diseases: A new link? Front. Endocrinol. (Lausanne) 2014, 5, 210. [Google Scholar] [CrossRef]
- Maftei, M.; Tian, X.; Manea, M.; Exner, T.E.; Schwanzar, D.; von Arnim, C.A.F.; Przybylski, M. Interaction structure of the complex between neuroprotective factor humanin and Alzheimer’s β-amyloid peptide revealed by affinity mass spectrometry and molecular modeling. J. Pept. Sci. 2012, 18, 373–382. [Google Scholar] [CrossRef]
- Jin, H.; Liu, T.; Wang, W.-X.; Xu, J.-H.; Yang, P.-B.; Lu, H.-X.; Sun, Q.-R.; Hu, H.-T. Protective effects of [Gly14]-Humanin on β-amyloid-induced PC12 cell death by preventing mitochondrial dysfunction. Neurochem. Int. 2010, 56, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Yen, K.; Wan, J.; Mehta, H.H.; Miller, B.; Christensen, A.; Levine, M.E.; Salomon, M.P.; Brandhorst, S.; Xiao, J.; Kim, S.; et al. Humanin Prevents Age-Related Cognitive Decline in Mice and is Associated with Improved Cognitive Age in Humans. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Tajima, H.; Kawasumi, M.; Chiba, T.; Yamada, M.; Yamashita, K.; Nawa, M.; Kita, Y.; Kouyama, K.; Aiso, S.; Matsuoka, M.; et al. A humanin derivative, S14G-HN, prevents amyloid-β-induced memory impairment in mice. J. Neurosci. Res. 2005, 79, 714–723. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, W.; Li, Z.; Hao, J.; Zhang, Z.; Liu, L.; Mao, N.; Miao, J.; Zhang, L. S14G-Humanin improves cognitive deficits and reduces amyloid pathology in the middle-aged APPswe/PS1dE9 mice. Pharmacol. Biochem. Behav. 2012, 100, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Niikura, T.; Sidahmed, E.; Hirata-Fukae, C.; Aisen, P.S.; Matsuoka, Y. A humanin derivative reduces amyloid beta accumulation and ameliorates memory deficit in triple transgenic mice. PLoS ONE 2011, 6, e16259. [Google Scholar] [CrossRef] [PubMed]
- Park, T.Y.; Kim, S.H.; Shin, Y.C.; Lee, N.H.; Lee, R.K.C.; Shim, J.H.; Glimcher, L.H.; Mook-Jung, I.; Cheong, E.; Kim, W.K.; et al. Amelioration of neurodegenerative diseases by cell death-induced cytoplasmic delivery of humanin. J. Control. Release 2013, 166, 307–315. [Google Scholar] [CrossRef]
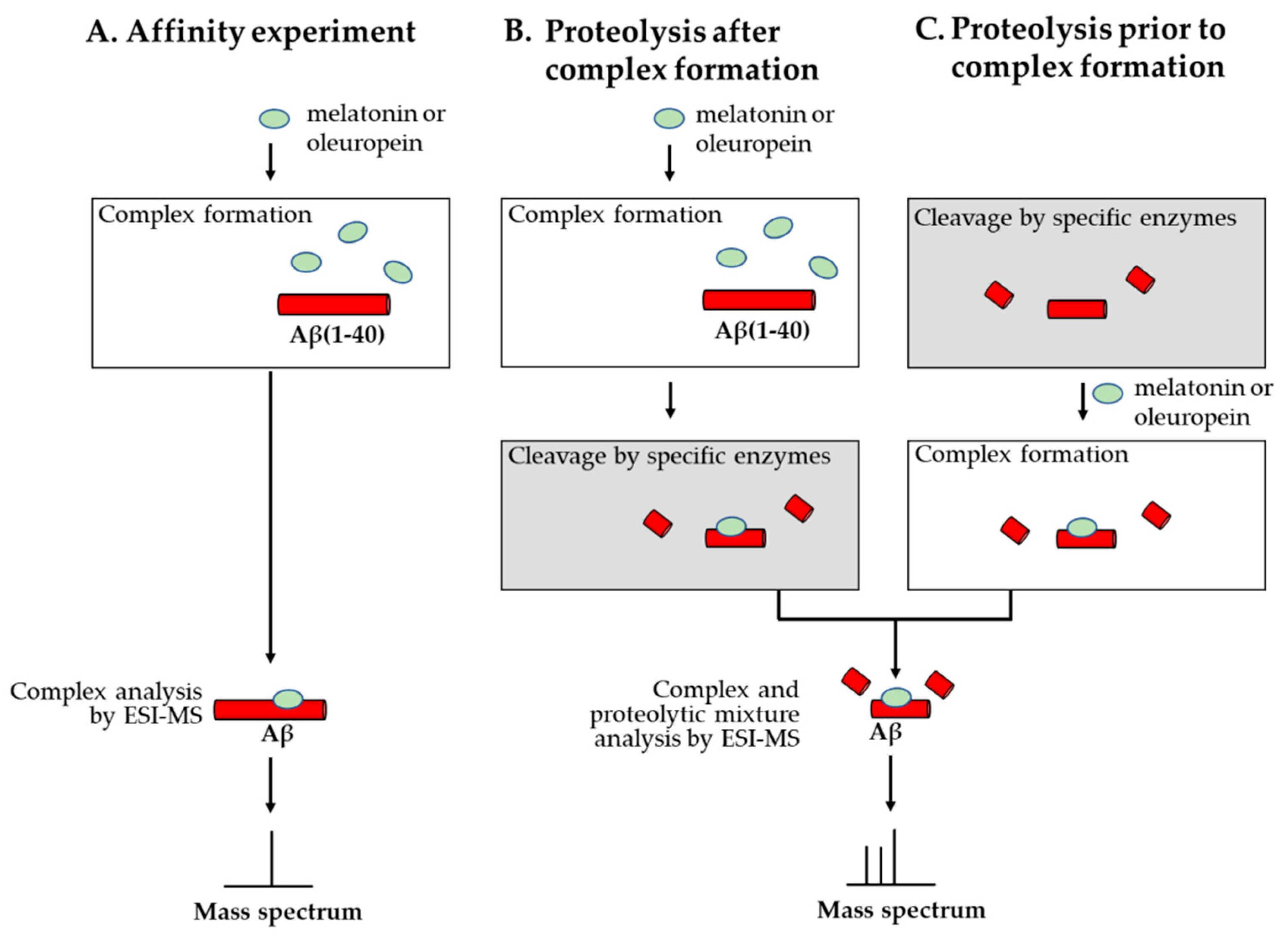
- Martineau, E.; de Guzman, J.M.; Rodionova, L.; Kong, X.; Mayer, P.M.; Aman, A.M. Investigation of the noncovalent interactions between anti-amyloid agents and amyloid β peptides by ESI-MS. J. Am. Soc. Mass Spectrom. 2010, 21, 1506–1514. [Google Scholar] [CrossRef]
- Gervais, F.; Paquette, J.; Morissette, C.; Krzywkowski, P.; Yu, M.; Azzi, M.; Lacombe, D.; Kong, X.; Aman, A.; Laurin, J.; et al. Targeting soluble Aβ peptide with Tramiprosate for the treatment of brain amyloidosis. Neurobiol. Aging 2007, 28, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Kocis, P.; Tolar, M.; Yu, J.; Sinko, W.; Ray, S.; Blennow, K.; Fillit, H.; Hey, J.A. Elucidating the Aβ42 Anti-Aggregation Mechanism of Action of Tramiprosate in Alzheimer’s Disease: Integrating Molecular Analytical Methods, Pharmacokinetic and Clinical Data. CNS Drugs 2017, 31, 495–509. [Google Scholar] [CrossRef] [PubMed]
- Bazoti, F.M.; Tsarbopoulos, A.; Markides, K.E.; Bergquist, J. Study of the non-covalent interaction between amyloid-β-peptide and melatonin using electrospray ionization mass spectrometry. J. Mass Spectrom. 2005, 40, 182–192. [Google Scholar] [CrossRef]
- Pappolla, M.A.; Sos, M.; Omar, R.A.; Bick, R.J.; Hickson-Bick, D.L.; Reiter, R.J.; Efthimiopoulos, S.; Robakis, N.K. Melatonin prevents death of neuroblastoma cells exposed to the Alzheimer amyloid peptide. J. Neurosci. Off. J. Soc. Neurosci. 1997, 17, 1683–1690. [Google Scholar] [CrossRef]
- Matsubara, E.; Bryant-Thomas, T.; Quinto, J.P.; Henry, T.L.; Poeggeler, B.; Herbert, D.; Cruz-Sanchez, F.; Chyan, Y.J.; Smith, M.A.; Perry, G.; et al. Melatonin increases survival and inhibits oxidative and amyloid pathology in a transgenic model of Alzheimer’s disease. J. Neurochem. 2003, 85, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.; Kulhanek, D.; Nowlin, J.; Jones, R.; Praticò, D.; Rokach, J.; Stackman, R. Chronic melatonin therapy fails to alter amyloid burden or oxidative damage in old Tg2576 mice: Implications for clinical trials. Brain Res. 2005, 1037, 209–213. [Google Scholar] [CrossRef]
- Gong, Y.H.; Hua, N.; Zang, X.; Huang, T.; He, L. Melatonin ameliorates Aβ1–42-induced Alzheimer’s cognitive deficits in mouse model. J. Pharm. Pharmacol. 2018, 70, 70–80. [Google Scholar] [CrossRef]
- Matsubara, E.; Vidal, R.; Pacheco-Quinto, J.; Pappolla, M.A.; Matsubara, E.; Vidal, R.; Pacheco-Quinto, J.; Poeggeler, B.; Zagorski, M.; Sambamurti, K. Melatonin Treatment Enhances Aβ Lymphatic Clearance in a Transgenic Mouse Model of Amyloidosis Send Orders for Reprints to reprints@benthamscience.ae Melatonin Treatment Enhances Aβ Lymphatic Clearance in a Transgenic Mouse Model of Amyloidosis. Curr. Alzheimer Res. 2018, 15, 1–6. [Google Scholar]
- Bazoti, F.N.; Bergquist, J.; Markides, K.E.; Tsarbopoulos, A. Noncovalent interaction between amyloid-beta-peptide (1–40) and oleuropein studied by electrospray ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 2006, 17, 568–575. [Google Scholar] [CrossRef]
- Bazoti, F.N.; Bergquist, J.; Markides, K.; Tsarbopoulos, A. Localization of the noncovalent binding site between amyloid-beta-peptide and oleuropein using electrospray ionization FT-ICR mass spectrometry. J. Am. Soc. Mass Spectrom. 2008, 19, 1078–1085. [Google Scholar] [CrossRef]
- Omar, S.H.; Scott, C.J.; Hamlin, A.S.; Obied, H.K. Olive biophenols reduces alzheimer’s pathology in SH-SY5Y cells and APPswe mice. Int. J. Mol. Sci. 2019, 20. [Google Scholar] [CrossRef]
- Grossi, C.; Rigacci, S.; Ambrosini, S.; Ed Dami, T.; Luccarini, I.; Traini, C.; Failli, P.; Berti, A.; Casamenti, F.; Stefani, M. The Polyphenol Oleuropein Aglycone Protects TgCRND8 Mice against Aß Plaque Pathology. PLoS ONE 2013, 8, e71702. [Google Scholar] [CrossRef]
- Pantano, D.; Luccarini, I.; Nardiello, P.; Servili, M.; Stefani, M.; Casamenti, F. Oleuropein aglycone and polyphenols from olive mill waste water ameliorate cognitive deficits and neuropathology. Br. J. Clin. Pharmacol. 2017, 83, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Li, Y.; Zheng, M.; Xi, X.; Zhang, X.; Han, C. Structure Properties, Acquisition Protocols, and Biological Activities of Oleuropein Aglycone. Front. Chem. 2018, 6, 239. [Google Scholar] [CrossRef]
- Qi, W.; Zhang, A.; Good, T.A.; Fernandez, E.J. Two disaccharides and trimethylamine N-oxide affect Aβ aggregation differently, but all attenuate oligomer-induced membrane permeability. Biochemistry 2009, 48, 8908–8919. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Barkhordarian, H.; Emadi, S.; Park, C.B.; Sierks, M.R. Trehalose differentially inhibits aggregation and neurotoxicity of beta-amyloid 40 and 42. Neurobiol. Dis. 2005, 20, 74–81. [Google Scholar] [CrossRef]
- Portbury, S.D.; Hare, D.J.; Sgambelloni, C.; Perronnes, K.; Portbury, A.J.; Finkelstein, D.I.; Adlard, P.A. Trehalose Improves Cognition in the Transgenic Tg2576 Mouse Model of Alzheimer’s Disease. J. Alzheimer’s Dis. 2017, 60, 549–560. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Liang, Y.; Xu, F.; Sun, B.; Wang, Z. Trehalose rescues Alzheimer’s disease phenotypes in APP/PS1 transgenic mice. J. Pharm. Pharmacol. 2013, 65, 1753–1756. [Google Scholar] [CrossRef] [PubMed]
- Van Bulck, M.; Sierra-Magro, A.; Alarcon-Gil, J.; Perez-Castillo, A.; Morales-Garcia, J. Novel Approaches for the Treatment of Alzheimer’s and Parkinson’s Disease. Int. J. Mol. Sci. 2019, 20, 719. [Google Scholar] [CrossRef] [PubMed]
- Camilleri, P.; Haskins, N.J.; Hewlett, D.R. β-Cyclodextrin interacts with the Alzheimer amyloid β-A4 peptide. FEBS Lett. 1994, 341, 256–258. [Google Scholar] [CrossRef]
- Ren, B.; Jiang, B.; Hu, R.; Zhang, M.; Chen, H.; Ma, J.; Sun, Y.; Jia, L.; Zheng, J. HP-β-cyclodextrin as an inhibitor of amyloid-β aggregation and toxicity. Phys. Chem. Chem. Phys. 2016, 18, 20476–20485. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Ho, D.; Calingasan, N.Y.; Pipalia, N.H.; Lin, M.T.; Beal, M.F. Neuroprotection by cyclodextrin in cell and mouse models of Alzheimer disease. J. Exp. Med. 2012, 209, 2501–2513. [Google Scholar] [CrossRef] [PubMed]
- Maulik, M.; Westaway, D.; Jhamandas, J.H.; Kar, S. Role of cholesterol in APP metabolism and its significance in Alzheimer’s disease pathogenesis. Mol. Neurobiol. 2013, 47, 37–63. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.S.; Stavrides, P.; Kumar, A.; Jiang, Y.; Mohan, P.S.; Ohno, M.; Dobrenis, K.; Davidson, C.D.; Saito, M.; Pawlik, M.; et al. Cyclodextrin has conflicting actions on autophagy flux in vivo in brains of normal and Alzheimer model mice. Hum. Mol. Genet. 2017, 26, 843–859. [Google Scholar] [CrossRef] [PubMed]
- Cimini, S.; Sclip, A.; Mancini, S.; Colombo, L.; Messa, M.; Cagnotto, A.; Di Fede, G.; Tagliavini, F.; Salmona, M.; Borsello, T. The cell-permeable Aβ1–6A2VTAT(D) peptide reverts synaptopathy induced by Aβ1–42wt. Neurobiol. Dis. 2016, 89, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Di Fede, G.; Catania, M.; Maderna, E.; Morbin, M.; Moda, F.; Colombo, L.; Rossi, A.; Cagnotto, A.; Virgilio, T.; Palamara, L.; et al. Tackling amyloidogenesis in Alzheimer’s disease with A2V variants of Amyloid-β. Sci. Rep. 2016, 6, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Giunta, B.; Hou, H.; Zhu, Y.; Rrapo, E.; Tian, J.; Takashi, M.; Commins, D.; Singer, E.; He, J.; Fernandez, F.; et al. HIV-1 tat contributes to Alzheimer’s disease-like pathology in PSAPP mice. Int. J. Clin. Exp. Pathol. 2009, 2, 433–443. [Google Scholar]
- Zheng, X.; Wu, C.; Liu, D.; Li, H.; Bitan, G.; Shea, J.E.; Bowers, M.T. Mechanism of C-Terminal Fragments of Amyloid β-Protein as Aβ Inhibitors: Do C-Terminal Interactions Play a Key Role in Their Inhibitory Activity? J. Phys. Chem. B 2016, 120, 1615–1623. [Google Scholar] [CrossRef]
- Li, H.; Du, Z.; Lopes, D.H.J.; Fradinger, E.A.; Wang, C.; Bitan, G. C-terminal tetrapeptides inhibit aβ42-induced neurotoxicity primarily through specific interaction at the N-Terminus of Aβ42. J. Med. Chem. 2011, 54, 8451–8460. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, W.; Zhang, Q.; Zhang, S.; Yuan, Z. Study on the efficiency and interaction mechanism of a decapeptide inhibitor of β-amyloid aggregation. Biomacromolecules 2014, 15, 931–939. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, Q.; Hu, X.; Wang, W.; Zhu, X.; Yuan, Z. Pharmacodynamics in Alzheimer’s disease model rats of a bifunctional peptide with the potential to accelerate the degradation and reduce the toxicity of amyloid β-Cu fibrils. Acta Biomater. 2018, 65, 327–338. [Google Scholar] [CrossRef]
- Zhang, Q.; Hu, X.; Wang, W.; Yuan, Z. Study of a Bifunctional Aβ Aggregation Inhibitor with the Abilities of Antiamyloid-β and Copper Chelation. Biomacromolecules 2016, 17, 661–668. [Google Scholar] [CrossRef] [PubMed]
- Barucker, C.; Bittner, H.J.; Chang, P.K.Y.; Cameron, S.; Hancock, M.A.; Liebsch, F.; Hossain, S.; Harmeier, A.; Shaw, H.; Charron, F.M.; et al. Aβ42-oligomer Interacting Peptide (AIP) neutralizes toxic amyloid-β42 species and protects synaptic structure and function. Sci. Rep. 2015, 5, 15410. [Google Scholar] [CrossRef] [PubMed]
- Yefremova, Y.; Opuni, K.F.M.; Danquah, B.D.; Thiesen, H.J.; Glocker, M.O. Intact Transition Epitope Mapping (ITEM). J. Am. Soc. Mass Spectrom. 2017, 28, 1612–1622. [Google Scholar] [CrossRef] [PubMed]
- Ștefănescu, R.; Lupu, L.; Manea, M.; Iacob, R.E.; Przybylski, M. Molecular characterization of the β-amyloid(4-10) epitope of plaque specific Aβ antibodies by affinity-mass spectrometry using alanine site mutation. J. Pept. Sci. 2018, 24, e3047. [Google Scholar] [CrossRef] [PubMed]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ştefănescu, R.; Stanciu, G.D.; Luca, A.; Caba, I.C.; Tamba, B.I.; Mihai, C.T. Contributions of Mass Spectrometry to the Identification of Low Molecular Weight Molecules Able to Reduce the Toxicity of Amyloid-β Peptide to Cell Cultures and Transgenic Mouse Models of Alzheimer’s Disease. Molecules 2019, 24, 1167. https://doi.org/10.3390/molecules24061167
Ştefănescu R, Stanciu GD, Luca A, Caba IC, Tamba BI, Mihai CT. Contributions of Mass Spectrometry to the Identification of Low Molecular Weight Molecules Able to Reduce the Toxicity of Amyloid-β Peptide to Cell Cultures and Transgenic Mouse Models of Alzheimer’s Disease. Molecules. 2019; 24(6):1167. https://doi.org/10.3390/molecules24061167
Chicago/Turabian StyleŞtefănescu, Raluca, Gabriela Dumitriṭa Stanciu, Andrei Luca, Ioana Cezara Caba, Bogdan Ionel Tamba, and Cosmin Teodor Mihai. 2019. "Contributions of Mass Spectrometry to the Identification of Low Molecular Weight Molecules Able to Reduce the Toxicity of Amyloid-β Peptide to Cell Cultures and Transgenic Mouse Models of Alzheimer’s Disease" Molecules 24, no. 6: 1167. https://doi.org/10.3390/molecules24061167
APA StyleŞtefănescu, R., Stanciu, G. D., Luca, A., Caba, I. C., Tamba, B. I., & Mihai, C. T. (2019). Contributions of Mass Spectrometry to the Identification of Low Molecular Weight Molecules Able to Reduce the Toxicity of Amyloid-β Peptide to Cell Cultures and Transgenic Mouse Models of Alzheimer’s Disease. Molecules, 24(6), 1167. https://doi.org/10.3390/molecules24061167






