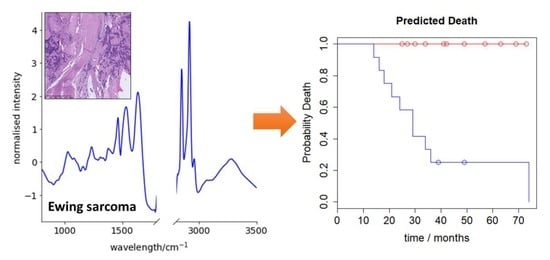
Predicting Ewing Sarcoma Treatment Outcome Using Infrared Spectroscopy and Machine Learning
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Sample Preparation
2.3. FTIR Spectroscopy
2.4. Statistical Analysis
3. Results
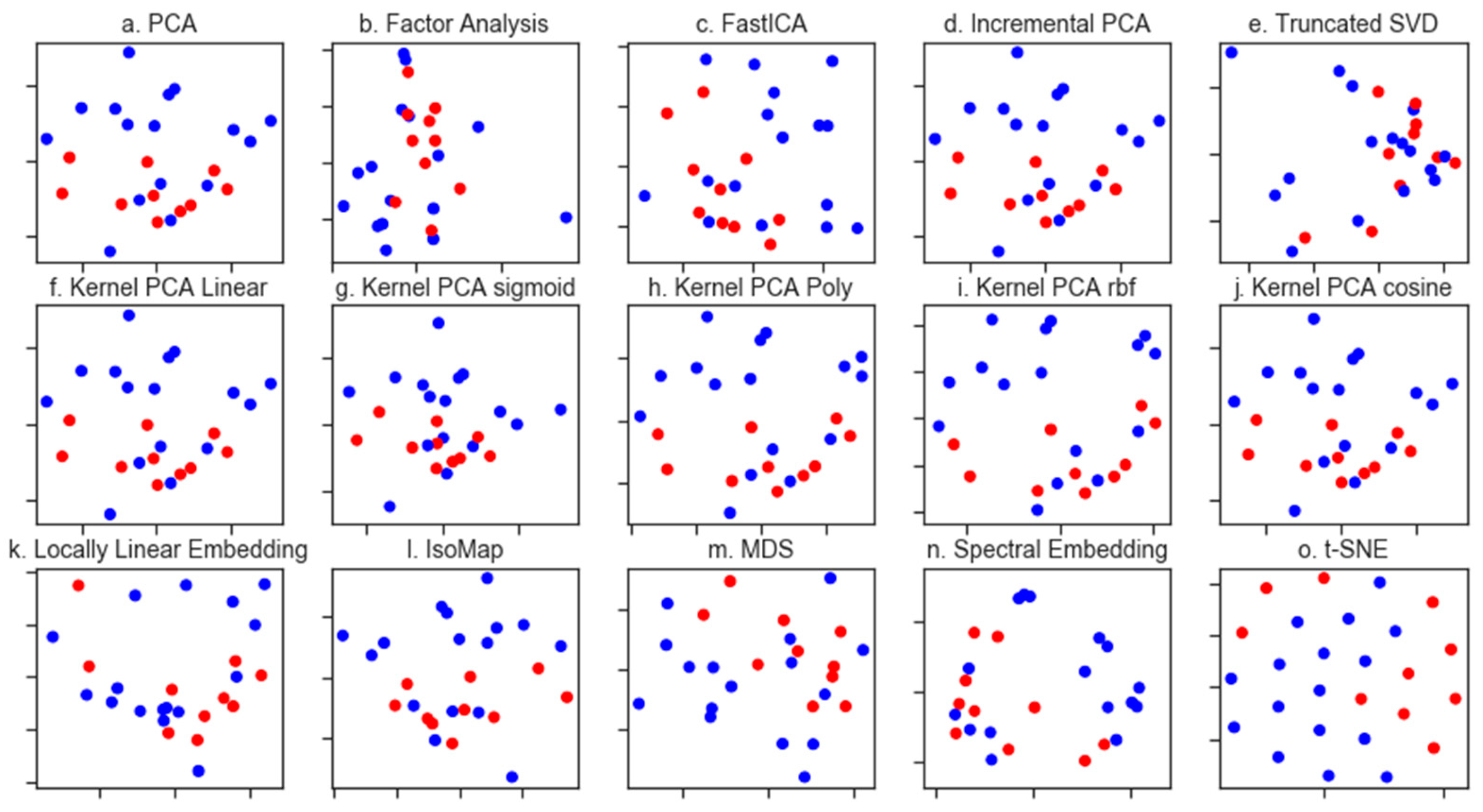
3.1. Exploratory Data Analysis
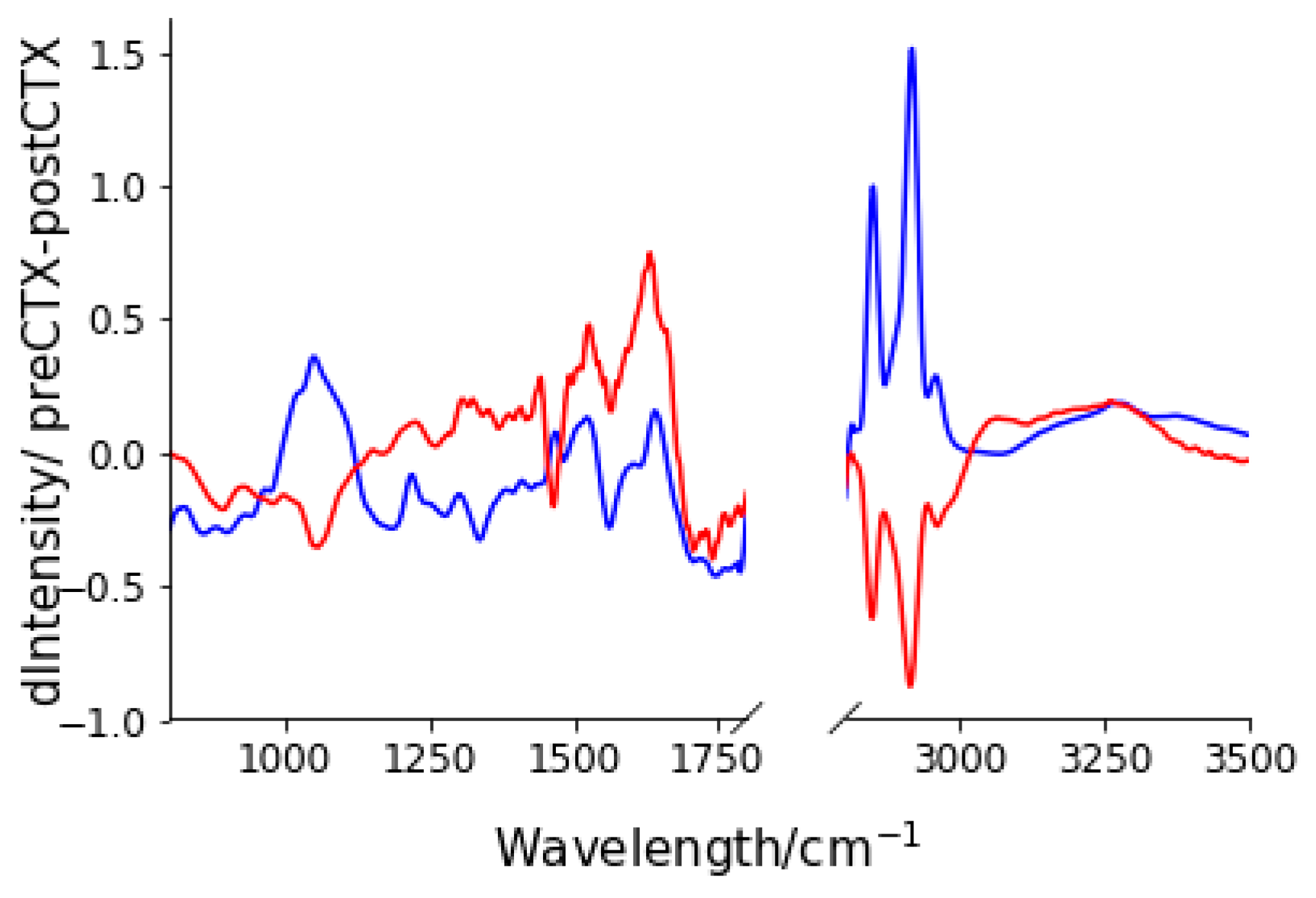
3.2. Spectral Changes after Neo-CTX
3.3. Generating a Predictive Model for Prognosis
3.4. Feature Generation
3.5. Supervised Learning
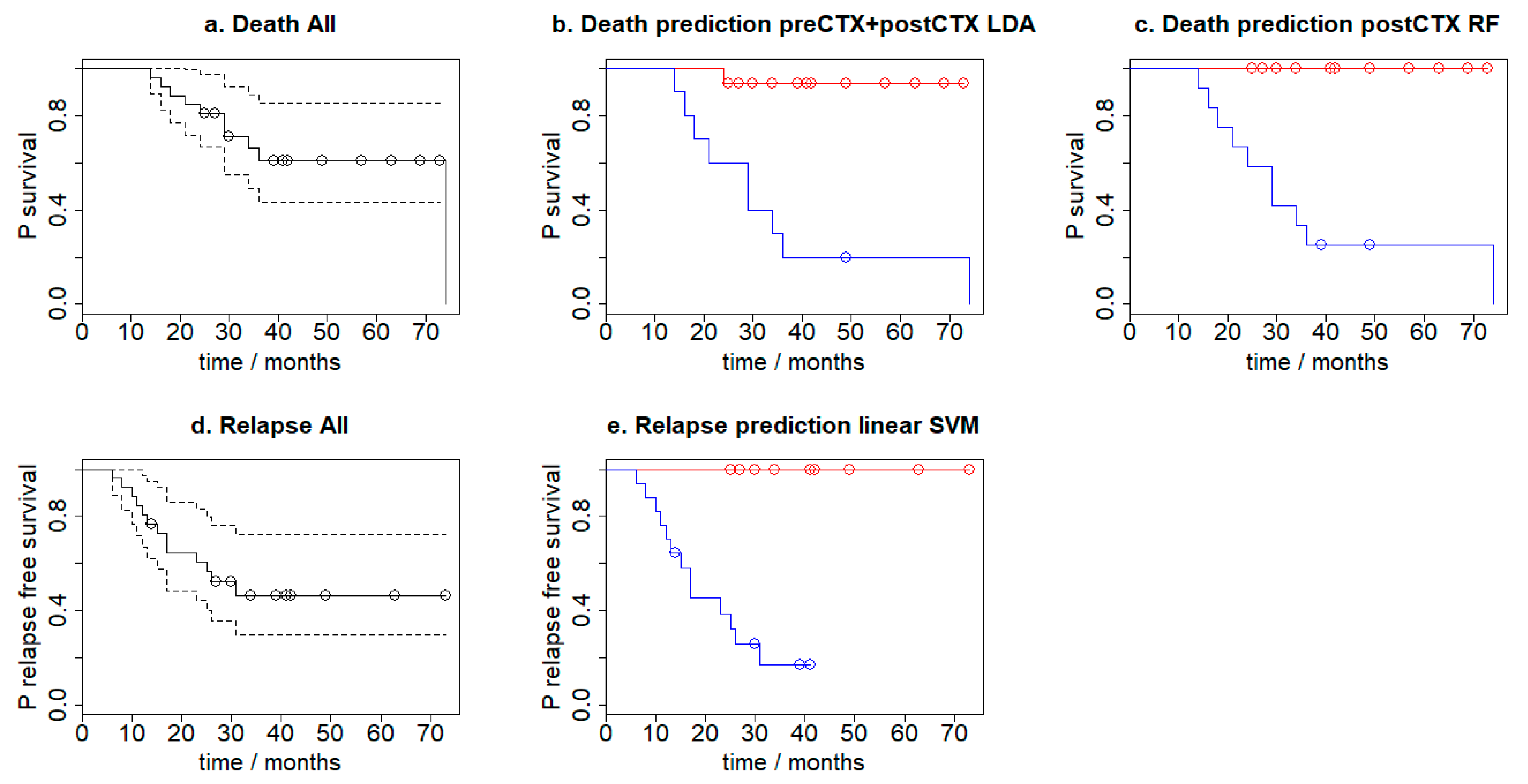
3.6. Survival Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Compliance with Ethical Standards
Abbreviations
| FTIR | Fourier Transform Infrared |
| ES | Ewing sarcoma |
| SVM | Support Vector Machine |
| neoCTX | neoadjuvant chemotherapy |
| preCTX | pre-neoadjuvant chemotherapy |
| postCTX | post-neoadjuvant chemotherapy |
| VIDE | vincristine, ifosfamide, doxorubicin, etoposide |
| FFPE | formalin-fixed paraffin-embedded |
| ATR | attenuated total reflection |
| PCA | principal component analysis |
| poly | polynomial |
| rbf | radial basis function |
| RF | Random Forest |
| LDA | Linear Discriminant Analysis |
| KNN | k-nearest neighbours |
| GDM | Gradient Boosted Classifier |
| CI | confidence interval |
| HR | hazard ratio |
References
- Gaspar, N.; Rey, A.; Bérard, P.M.; Michon, J.; Gentet, J.C.; Tabone, M.D.; Roché, H.; Defachelles, A.S.; Lejars, O.; Plouvier, E.; et al. Risk adapted chemotherapy for localised Ewing’s sarcoma of bone: The French EW93 study. Eur. J. Cancer 2012, 48, 1376–1385. [Google Scholar] [CrossRef] [PubMed]
- Belgaumi, A.F.; Al-Seraihy, A.; Siddiqui, K.S.; Ayas, M.; Bukhari, A.; Al-Musa, A.; Al-Ahmari, A.; El-Solh, H. Outcome of risk adapted therapy for relapsed/refractory acute lymphoblastic leukemia in children. Leuk. Lymphoma 2013, 54, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Sayed, H.; Farrag, A.; El-Sayed, M. Risk-based combined-modality therapy of pediatric Hodgkin’s lymphoma: A retrospective study. Leuk. Res. 2010, 34, 1447–1452. [Google Scholar] [CrossRef] [PubMed]
- Saletta, F.; Seng, M.S.; Lau, L.M.S. Advances in paediatric cancer treatment. Transl. Pediatr. 2014, 3, 156–182. [Google Scholar] [PubMed]
- Gurney, J.G.; Swensen, A.R.; Bulterys, M. Malignant bone tumors. In Cancer Incidence and Survival among Children and Adolescents: United States SEER Program 1975–1995; NIH Pub. No. 99–4649; Ries, L.A.G., Smith, M.A., Gumey, J.G., Eds.; National Cancer Institute, SEER Program: Bethesda, MD, USA, 1999. [Google Scholar]
- Toretsky, J.A.; Kim, A. Medscape, Ewing Sarcoma Epidemiology. Available online: http://emedicine.medscape.com/article/990378-overview#a6 (accessed on 11 July 2017).
- Raciborska, A.; Bilska, K.; Drabko, K.; Chaber, R.; Sobol, G.; Pogorzała, M.; Wyrobek, E.; Połczyńska, K.; Rogowska, E.; Rodriguez-Galindo, C.; et al. Validation of a multi-modal treatment protocol for Ewing sarcoma a report from the polish pediatric oncology group. Pediatr. Blood Cancer 2014, 61, 2170–2174. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.J.; Trevisan, J.; Bassan, P.; Bhargava, R.; Butler, H.J.; Dorling, K.M.; Fielden, P.R.; Fogarty, S.W.; Fullwood, N.J.; Heys, K.A.; et al. Using Fourier transform IR spectroscopy to analyze biological materials. Nature Protoc. 2014, 9, 1771–1791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bellisola, G.; Sorio, C. Infrared spectroscopy and microscopy in cancer research and diagnosis. Am. J. Cancer Res. 2012, 2, 1. [Google Scholar] [PubMed]
- Depciuch, J.; Kaznowska, E.; Zawlik, I.; Wojnarowska, R.; Cholewa, M.; Heraud, P.; Cebulski, J. Application of Raman Spectroscopy and Infrared Spectroscopy in the Identification of Breast Cancer. Appl. Spectrosc. 2016, 70, 251–263. [Google Scholar] [CrossRef]
- Zawlik, I.; Kaznowska, E.; Cebulski, J.; Kolodziej, M.; Depciuch, J.; Vongsvivut, J.; Cholewa, M. FPA-FTIR Microspectroscopy for Monitoring Chemotherapy Efficacy in Triple-Negative Breast Cancer. Sci. Rep. 2016, 6, 37333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaznowska, E.; Depciuch, J.; Łach, K.; Kołodziej, M.; Koziorowska, A.; Vongsvivut, J.; Zawlik, I.; Cholewa, M.; Cebulski, J. The classification of lung cancers and their degree of malignancy by FTIR, PCA-LDA analysis, and a physics-based computational model. Talanta 2018, 186, 337–345. [Google Scholar] [CrossRef]
- Kaznowska, E.; Łach, K.; Depciuch, J.; Chaber, R.; Koziorowska, A.; Slobodian, S.; Kiper, K.; Chlebus, A.; Cebulski, J. Application of infrared spectroscopy for the identification of squamous cell carcinoma (lung cancer). Preliminary study. Infrared Phys. Technol. 2018, 89, 282–290. [Google Scholar] [CrossRef]
- Mehrotra, R.; Tyagi, G.; Jangir, D.K.; Dawar, R.; Gupta, N. Analysis of ovarian tumor pathology by Fourier Transform Infrared Spectroscopy. J. Ovarian Res. 2010, 3, 27. [Google Scholar] [CrossRef]
- Hands, J.R.; Clemens, G.; Stables, R.; Ashton, K.; Brodbelt, A.; Davis, C.; Dawson, T.P.; Jenkinson, M.D.; Lea, R.W.; Walker, C.; et al. Brain tumour differentiation: Rapid stratified serum diagnostics via attenuated total reflection Fourier-transform infrared spectroscopy. J. Neurooncol. 2016, 127, 463–472. [Google Scholar] [CrossRef]
- Lyng, F.M.; Faoláin, E.O.; Conroy, J.; Meade, A.D.; Knief, P.; Duffy, B.; Hunter, M.B.; Byrne, J.M.; Kelehan, P.; Byrne, H.J. Vibrational spectroscopy for cervical cancer pathology, from biochemical analysis to diagnostic tool. Exp. Mol. Pathol. 2007, 82, 121–129. [Google Scholar] [CrossRef]
- Liu, H.; Su, Q.; Sheng, D.; Zheng, W.; Wang, X. Comparison of red blood cells from gastric cancer patients and healthy persons using FTIR spectroscopy. J. Mol. Struct. 2017, 1130, 33–37. [Google Scholar] [CrossRef]
- Khanmohammadi, M.; Garmarudi, A.B.; Ghasemi, K.; Jaliseh, H.K.; Kaviani, A. Diagnosis of colon cancer by attenuated total reflectance-fourier transform infrared microspectroscopy and soft independent modeling of class analogy. Med. Oncol. 2009, 26, 292–297. [Google Scholar] [CrossRef]
- Baker, M.J.; Gazi, E.; Brown, M.D.; Shanks, J.H.; Gardner, P.; Clarke, N.W. FTIR-based spectroscopic analysis in the identification of clinically aggressive prostate cancer. Br. J. Cancer 2008, 99, 1859–1866. [Google Scholar] [CrossRef] [Green Version]
- Wald, N.; Le Corre, Y.; Martin, L.; Mathieu, V.; Goormaghtigh, E. Infrared spectra of primary melanomas can predict response to chemotherapy: The example of dacarbazine. Biochim. Biophys. Acta 2016, 1862, 174–181. [Google Scholar] [CrossRef]
- Chaber, R.; Łach, K.; Szmuc, K.; Michalak, E.; Raciborska, A.; Mazur, D.; Machaczka, M.; Cebulski, J. Application of infrared spectroscopy in the identification of Ewing sarcoma: A preliminary report. Infrared Phys. Technol. 2017, 83, 200–205. [Google Scholar] [CrossRef]
- Chaber, R.; Łach, K.; Arthur, C.J.; Raciborska, A.; Michalak, E.; Ciebiera, K.; Bilska, K.; Drabko, K.; Cebulski, J. Prediction of Ewing Sarcoma treatment outcome using attenuated tissue reflection FTIR tissue spectroscopy. Sci. Rep. 2018, 8, 12299. [Google Scholar] [CrossRef]
- Schoelkopf, B.; Smola, A.J.; Mueller, K.-R. Kernel principal component analysis. In Advances in Kernel Methods; MIT Press: Cambridge, MA, USA, 1999; pp. 327–352. [Google Scholar]
- Rodríguez-Galindo, C.; Navid, F.; Liu, T.; Billups, C.A.; Rao, B.N.; Krasin, M.J. Prognostic factors for local and distant control in Ewing sarcoma family of tumors. Ann. Oncol. 2008, 19, 814–820. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Galindo, C.; Liu, T.; Krasin, M.J.; Wu, J.; Billups, C.A.; Daw, N.C.; Spunt, S.L.; Rao, B.N.; Santana, V.M.; Navid, F. Analysis of prognostic factors in Ewing sarcoma family of tumors: Review of St. Jude Children’s Research Hospital studies. Cancer 2007, 110, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Picci, P.; Böhling, T.; Bacci, G.; Ferrari, S.; Sangiorgi, L.; Mercuri, M.; Ruggieri, P.; Manfrini, M.; Ferraro, A.; Casadei, R.; et al. Chemotherapy-induced tumor necrosis as a prognostic factor in localized Ewing’s sarcoma of the extremities. J. Clin. Oncol. 1997, 15, 1553–1559. [Google Scholar] [CrossRef]
- Albergo, J.I.; Gaston, C.L.; Laitinen, M.; Darbyshire, A.; Jeys, L.M.; Sumathi, V.; Parry, M.; Peake, D.; Carter, S.R.; Tillman, R.; et al. Ewing’s sarcoma only patients with 100% of necrosis after chemotherapy should be classified as having a good response. Bone Jt. J. 2016, 98-B, 1138–1144. [Google Scholar] [CrossRef] [PubMed]
- Hattinger, C.M.; Vella, S.; Tavanti, E.; Fanelli, M.; Picci, P.; Serra, M. Pharmacogenomics of second-line drugs used for treatment of unresponsive or relapsed osteosarcoma patients. Pharmacogenomics 2016, 17, 2097–2114. [Google Scholar] [CrossRef] [Green Version]
- Zambelli, D.; Zuntini, M.; Nardi, F.; Manara, M.C.; Serra, M.; Landuzzi, L.; Lollini, P.; Ferrari, S.; Alberghini, M.; Llombart-Bosch, A.; et al. Biological indicators of prognosis in Ewing’s sarcoma: An emerging role for lectin galactoside-binding soluble 3 binding protein (LGALS3BP). Int. J. Cancer 2010, 126, 41–52. [Google Scholar] [CrossRef]
- Kikuta, K.; Tochigi, N.; Shimoda, T.; Yabe, H.; Morioka, H.; Toyama, Y.; Hosono, A.; Beppu, Y.; Kawai, A.; Hirohashi, S.; et al. Nucleophosmin as a Candidate Prognostic Biomarker of Ewing’s Sarcoma Revealed by Proteomics. Clin. Cancer Res. 2009, 15, 2885–2894. [Google Scholar] [CrossRef]
- Shukla, N.; Schiffman, J.; Reed, D.; Davis, I.J.; Womer, R.B.; Lessnick, S.L.; Lawlor, E.R. COG Ewing Sarcoma Biology Committee, Biomarkers in Ewing Sarcoma: The Promise and Challenge of Personalized Medicine. A Report from the Children’s Oncology Group. Front. Oncol. 2013, 3, 141. [Google Scholar] [CrossRef]
- Vo, K.T.; Edwards, J.V.; Epling, C.L.; Sinclair, E.; Hawkins, D.S.; Grier, H.E.; Janeway, K.A.; Barnette, P.; McIlvaine, E.; Krailo, M.D.; et al. Impact of Two Measures of Micrometastatic Disease on Clinical Outcomes in Patients with Newly Diagnosed Ewing Sarcoma: A Report from the Children’s Oncology Group. Clin. Cancer Res. 2016, 22, 3643–3650. [Google Scholar] [CrossRef] [Green Version]
- Meyers, P.A. Systemic therapy for osteosarcoma and Ewing sarcoma. Am. Soc. Clin. Oncol. Educ. Book 2015, e644–e647. [Google Scholar] [CrossRef]
- Gaspar, N.; Hawkins, D.S.; Dirksen, U.; Lewis, I.J.; Ferrari, S.; Le Deley, M.C.; Kovar, H.; Grimer, R.; Whelan, J.; Claude, L.; et al. Ewing Sarcoma: Current Management and Future Approaches Through Collaboration. J. Clin. Oncol. 2015, 33, 3036–3046. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: The histological samples of tumours are partially available from the authors. |

| Gender Male/Female | 12/15 |
|---|---|
| Age (years) range, median | 5–20 years 14 years |
| Localised/disseminated | 11/16 |
| Tumour resection complete/incomplete | 1/26 |
| Necrosis ≥ 90% vs. <90% | 20/7 |
| Local radiotherapy | 17 |
| Auto HSCT | 9 |
| Relapses (progressions)/deaths | 14/10 |
| Follow up time (months) range (median) | 14–74 (34) |
| Death | ||||
| preCTX | postCTX | preCTX-postCTX | preCTX+postCTX | |
| KNN | 0.692 | 0.692 | 0.808 | 0.692 |
| Linear SVM | 0.615 | 0.885 | 0.538 | 0.846 |
| Random Forest | 0.769 | 0.923 | 0.577 | 0.808 |
| LDA | 0.808 | 0.538 | 0.692 | 0.923 |
| GaussianBoosted | 0.769 | 0.769 | 0.654 | 0.692 |
| Relapse | ||||
| KNN | 0.615 | 0.5 | 0.769 | 0.615 |
| Linear SVM | 0.923 | 0.808 | 0.577 | 0.769 |
| Random Forest | 0.692 | 0.769 | 0.462 | 0.808 |
| LDA | 0.692 | 0.654 | 0.577 | 0.692 |
| GaussianBoosted | 0.615 | 0.769 | 0.5 | 0.654 |
| Necrosis > 90% | ||||
| KNN | 0.769 | 0.769 | 0.769 | 0.731 |
| Linear SVM | 0.769 | 0.538 | 0.846 | 0.692 |
| Random Forest | 0.654 | 0.769 | 0.692 | 0.654 |
| LDA | 0.654 | 0.808 | 0.615 | 0.769 |
| GaussianBoosted | 0.577 | 0.769 | 0.538 | 0.538 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chaber, R.; Arthur, C.J.; Łach, K.; Raciborska, A.; Michalak, E.; Bilska, K.; Drabko, K.; Depciuch, J.; Kaznowska, E.; Cebulski, J. Predicting Ewing Sarcoma Treatment Outcome Using Infrared Spectroscopy and Machine Learning. Molecules 2019, 24, 1075. https://doi.org/10.3390/molecules24061075
Chaber R, Arthur CJ, Łach K, Raciborska A, Michalak E, Bilska K, Drabko K, Depciuch J, Kaznowska E, Cebulski J. Predicting Ewing Sarcoma Treatment Outcome Using Infrared Spectroscopy and Machine Learning. Molecules. 2019; 24(6):1075. https://doi.org/10.3390/molecules24061075
Chicago/Turabian StyleChaber, Radosław, Christopher J. Arthur, Kornelia Łach, Anna Raciborska, Elżbieta Michalak, Katarzyna Bilska, Katarzyna Drabko, Joanna Depciuch, Ewa Kaznowska, and Józef Cebulski. 2019. "Predicting Ewing Sarcoma Treatment Outcome Using Infrared Spectroscopy and Machine Learning" Molecules 24, no. 6: 1075. https://doi.org/10.3390/molecules24061075
APA StyleChaber, R., Arthur, C. J., Łach, K., Raciborska, A., Michalak, E., Bilska, K., Drabko, K., Depciuch, J., Kaznowska, E., & Cebulski, J. (2019). Predicting Ewing Sarcoma Treatment Outcome Using Infrared Spectroscopy and Machine Learning. Molecules, 24(6), 1075. https://doi.org/10.3390/molecules24061075