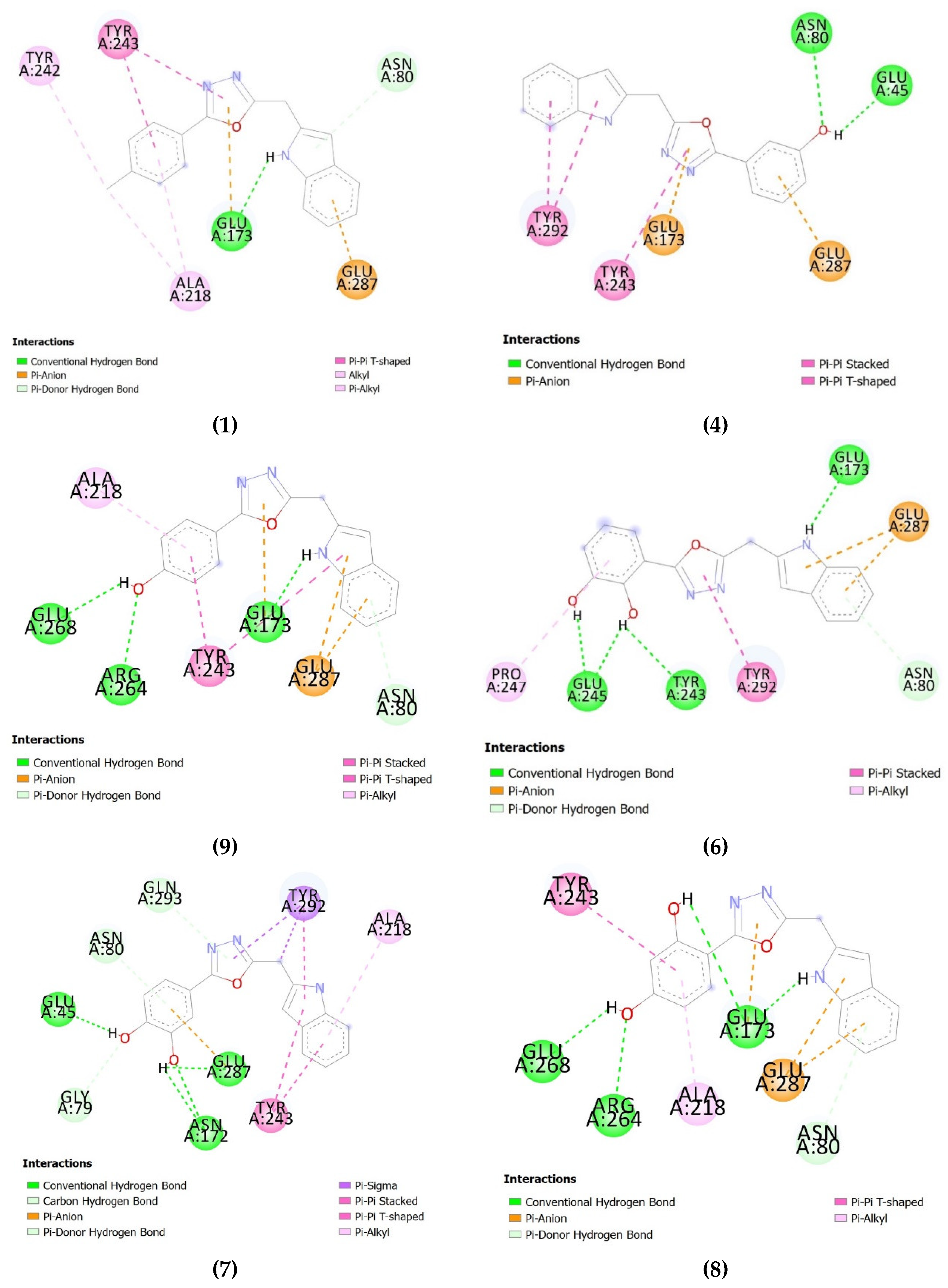
Synthesis, Molecular Docking and β-Glucuronidase Inhibitory Potential of Indole Base Oxadiazole Derivatives
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Biological Activity
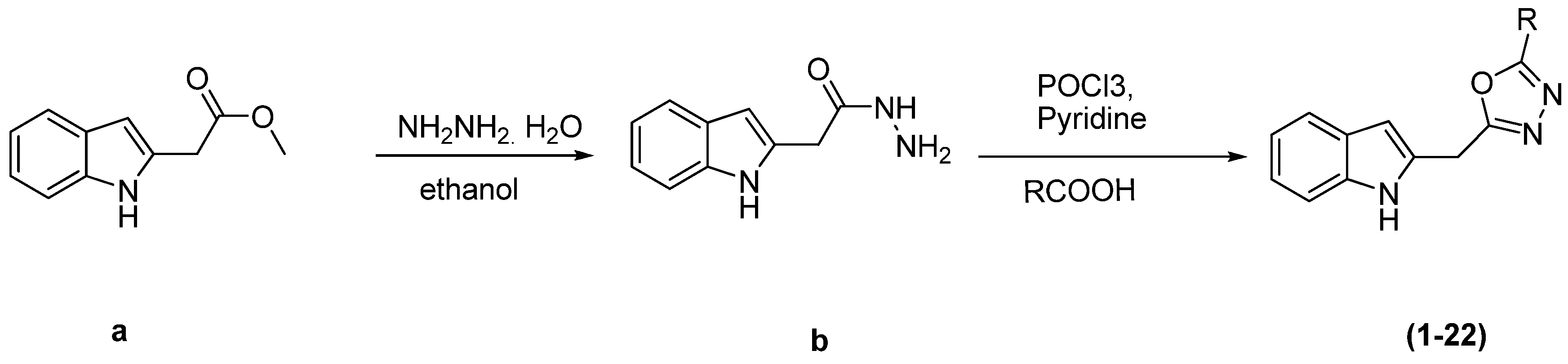
2.3. Molecular Docking Study
3. Materials and Methods
3.1. Instruments
3.2. Molecular Docking Details
3.3. Synthesis Methyl 2-(1H-indol-2-yl)acetate
3.4. General Procedure Indole Based 2,5-Disubstituted-1,3,4-Oxadiazoles (1–22)
3.4.1. 2-((1H-indol-2-yl) methyl)-5-(p-tolyl)-1,3,4-oxadiazole
3.4.2. 2-((1H-indol-2-yl) methyl)-5-(o-tolyl)-1,3,4-oxadiazole
3.4.3. 2-(5-((1H-indol-2-yl) methyl)-1,3,4-oxadiazol-2-yl) phenol
3.4.4. 3-(5-((1H-indol-2-yl) methyl)-1,3,4-oxadiazol-2-yl) phenol
3.4.5. 2-(5-((1H-indol-2-yl) methyl)-1,3,4-oxadiazol-2-yl) benzene-1,4-diol
3.4.6. 3-(5-((1H-indol-2-yl) methyl)-1,3,4-oxadiazol-2-yl) benzene-1,2-diol
3.4.7. 4-(5-((1H-indol-2-yl) methyl)-1,3,4-oxadiazol-2-yl) benzene-1,2-diol
3.4.8. 4-(5-((1H-indol-2-yl) methyl)-1,3,4-oxadiazol-2-yl) benzene-1,3-diol
3.4.9. 4-(5-((1H-indol-2-yl) methyl)-1,3,4-oxadiazol-2-yl) phenol
3.4.10. 2-((1H-indol-2-yl) methyl)-5-(2-nitrophenyl)-1,3,4-oxadiazole
3.4.11. 2-((1H-indol-2-yl) methyl)-5-(3-nitrophenyl)-1,3,4-oxadiazole
3.4.12. 2-((1H-indol-2-yl) methyl)-5-(4-nitrophenyl)-1,3,4-oxadiazole
3.4.13. 2-(5-((1H-indol-2-yl) methyl)-1,3,4-oxadiazol-2-yl)-4-methoxyphenol
3.4.14. 2-((1H-indol-2-yl) methyl)-5-(3-bromo-4-fluorophenyl)-1,3,4-oxadiazole
3.4.15. 2-((1H-indol-2-yl) methyl)-5-(3-chlorophenyl)-1,3,4-oxadiazole
3.4.16. 2-((1H-indol-2-yl) methyl)-5-(pyridin-3-yl)-1,3,4-oxadiazole
3.4.17. 2-((1H-indol-2-yl) methyl)-5-(pyridin-4-yl)-1,3,4-oxadiazole
3.4.18. 2-((1H-indol-2-yl) methyl)-5-(2-fluorophenyl)-1,3,4-oxadiazole
3.4.19. 2-((1H-indol-2-yl) methyl)-5-(3-fluorophenyl)-1,3,4-oxadiazole
3.4.20. 2-((1H-indol-2-yl) methyl)-5-(4-fluorophenyl)-1,3,4-oxadiazole
3.4.21. 2-((1H-indol-2-yl) methyl)-5-(2-chlorophenyl)-1,3,4-oxadiazole
3.4.22. 2-((1H-indol-2-yl) methyl)-5-(4-chlorophenyl)-1,3,4-oxadiazole
3.5. β-Glucuronidase Assay
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Khan, K.M.; Rahim, F.; Halim, S.A.; Taha, M.; Khan, M.; Perveen, S.; Mesaik, M.A.; Choudhary, M.I. Synthesis of novel inhibitors of β-glucuronidase based on benzothiazole skeleton and study of their binding affinity by molecular docking. Bioorg. Med. Chem. 2011, 19, 4286–4294. [Google Scholar] [CrossRef] [PubMed]
- Chojnowska, S.; Kępka, A.; Szajda, S.D.; Waszkiewicz, N.; Bierć, M.; Zwierz, K. Exoglycosidase markers of diseases. Biochem. Soc. Trans. 2011, 39, 406–409. [Google Scholar] [CrossRef] [PubMed]
- Szajda, S.D.; Jankowska, A.; Zwierz, K. Carbohydrate markers in colon carcinoma. Dis. Mark. 2008, 25, 233–242. [Google Scholar] [CrossRef]
- Kim, D.-H.; Jin, Y.-H. Intestinal bacterial β-glucuronidase activity of patients with colon cancer. Arch. Pharm. Res. 2001, 24, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.-C.; Roffler, S.R.; Tzou, S.-C.; Chuang, K.-H.; Su, Y.-C.; Chuang, C.-H.; Kao, C.-H.; Chen, C.-S.; Harn, I.-H.; Liu, K.-Y. An activity-based near-infrared glucuronide trapping probe for imaging β-glucuronidase expression in deep tissues. J. Am. Chem. Soc. 2012, 134, 3103–3110. [Google Scholar] [CrossRef] [PubMed]
- Szajda, S.D.; Waszkiewicz, S.D.; Zalewska, B.; Snarska, J.; Zwierz, K. Znaczenie diagnostyczne oznaczenia aktywności beta-glukuronidazy w surowicy krwi chorych na gruczolakoraka jelita grubego. In Proceedings of the XVI Zjazd Naukowy Polskiego Towarzystwa Diagnostyki Laboratoryjnej, Wrocław, Poland, 26–28 September 2007; Volume 43. [Google Scholar]
- Juan, T.-Y.; Roffler, S.R.; Hou, H.-S.; Huang, S.-M.; Chen, K.-C.; Leu, Y.-L.; Prijovich, Z.M.; Yu, C.-P.; Wu, C.-C.; Sun, G.-H. Antiangiogenesis targeting tumor microenvironment synergizes glucuronide prodrug antitumor activity. Clin. Cancer. Res. 2009, 15, 4600–4611. [Google Scholar] [CrossRef] [PubMed]
- Mürdter, T.E.; Friedel, G.; Backman, J.T.; McClellan, M.; Schick, M.; Gerken, M.; Bosslet, K.; Fritz, P.; Toomes, H.; Kroemer, H.K. Dose optimization of a doxorubicin prodrug (hmr 1826) in isolated perfused human lungs: Low tumor ph promotes prodrug activation by β-glucuronidase. J. Pharmacol. Exp. Ther. 2002, 301, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Falkenbach, A.; Wigand, R.; Unkelbach, U.; Jörgens, K.; Martinovic, A.; Scheuermann, E.; Seiffert, U.; Kaltwasser, J. Cyclosporin treatment in rheumatoid arthritis is associated with an increased serum activity of β-glucuronidase. Scand. J. Rheumatol. 1993, 22, 83–85. [Google Scholar] [CrossRef] [PubMed]
- Byass, P. The global burden of liver disease: A challenge for methods and for public health. BMC Med. 2014, 12, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.I.; Waheed, A.; Grubb, J.H.; Klei, H.E.; Korolev, S.; William, S.S. High Resolution Crystal Structure of Human β-Glucuronidase Reveals Structural Basis of Lysosome Targeting. PLoS ONE 2013, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Naz, H.; Islam, A.; Waheed, A.; Sly, W.S.; Ahmad, F.; Hassan, Md.I. Human β-Glucuronidase: Structure, Function, and application in enzyme replacement therapy. Rejuvenation Res. 2013, 16, 352–363. [Google Scholar] [CrossRef] [PubMed]
- Cacchi, S.; Fabrizi, G. Synthesis and functionalization of indoles through palladium-catalyzed reactions. Chem. Rev. 2005, 105, 2873–2920. [Google Scholar] [CrossRef] [PubMed]
- Imran, S.; Taha, M.; Hadiani Ismail, N. A review of bisindolylmethane as an important scaffold for drug discovery. Curr. Med. Chem. 2015, 22, 4412–4433. [Google Scholar] [CrossRef] [PubMed]
- Dadashpour, S.; Emami, S. Indole in the target-based design of anticancer agents: A versatile scaffold with diverse mechanisms. Eur. J. Med. Chem. 2018, 150, 9–29. [Google Scholar] [CrossRef] [PubMed]
- Eldehna, W.M.; Abo-Ashour, M.F.; Ibrahim, H.S.; Al-Ansary, G.H.; Ghabbour, H.A.; Elaasser, M.M.; Ahmed, H.Y.A.; Safwat, N.A. Novel [(3-indolylmethylene)hydrazono]indolin-2-ones as apoptotic anti-proliferative agents: Design, synthesis and in vitro biological evaluation. J. Enzyme Inhib. Med. Chem. 2018, 33, 686–700. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, H.S.; Abou-Seri, S.M.; Ismail, N.S.M.; Elaasser, M.M.; Aly, M.H.; Abdel-Aziz, H.A. Bis-isatinhydrazones with novel linkers: Synthesis and biological evaluation as cytotoxic agents. Eur. J. Med. Chem. 2016, 108, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Tantak, M.P.; Klingler, L.; Arun, V.; Kumar, A.; Sadana, R.; Kumar, D. Design and synthesis of bis(indolyl)ketohydrazide-hydrazones: Identification of potent and selective novel tubulin inhibitors. Eur. J. Med. Chem. 2017, 136, 184–194. [Google Scholar] [CrossRef] [PubMed]
- Das Mukherjee, D.; Kumar, N.M.; Tantak, M.P.; Das, A.; Ganguli, A.; Datta, S.; Kumar, D.; Chakrabarti, G. Development of Novel Bis(indolyl)-hydrazide-Hydrazone Derivatives as Potent Microtubule-Targeting Cytotoxic Agents against A549 Lung Cancer Cells. Biochemistry 2016, 55, 3020–3035. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zhu, H.; Yang, Z.-M.; Zhu, H.-L. Synthesis, molecular modeling and biological evaluation of 2-aminomethyl-5-(quinolin-2-yl)-1, 3, 4-oxadiazole-2 (3h)-thione quinolone derivatives as novel anticancer agent. Eur. J. Med. Chem. 2013, 60, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Desai, N.; Bhatt, N.; Somani, H.; Trivedi, A. Synthesis, antimicrobial and cytotoxic activities of some novel thiazole clubbed 1, 3, 4-oxadiazoles. Eur. J. Med. Chem. 2013, 67, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Harish, K.P.; Mohana, K.N.; Mallesha, L. Synthesis of novel 1-[5-(4-methoxy-phenyl)-[1, 3, 4] oxadiazol-2-yl]-piperazine derivatives and evaluation of their in vivo anticonvulsant activity. Eur. J. Med. Chem. 2013, 65, 276–283. [Google Scholar] [CrossRef] [PubMed]
- Rajak, H.; Thakur, B.S.; Singh, A.; Raghuvanshi, K.; Sah, A.K.; Veerasamy, R.; Sharma, P.C.; Pawar, R.S.; Kharya, M.D. Novel limonene and citral based 2, 5-disubstituted-1, 3, 4-oxadiazoles: A natural product coupled approach to semicarbazones for antiepileptic activity. Bioorg. Med. Chem. Lett. 2013, 23, 864–868. [Google Scholar] [CrossRef] [PubMed]
- Ahsan, M.J.; Samy, J.G.; Jain, C.B.; Dutt, K.R.; Khalilullah, H.; Nomani, M.S. Discovery of novel antitubercular 1, 5-dimethyl-2-phenyl-4-([5-(arylamino)-1, 3, 4-oxadiazol-2-yl] methylamino)-1, 2-dihydro-3h-pyrazol-3-one analogues. Bioorg. Med. Chem. Lett. 2012, 22, 969–972. [Google Scholar] [CrossRef] [PubMed]
- Guda, D.R.; Park, S.-J.; Lee, M.-W.; Kim, T.-J.; Lee, M.E. Syntheses and anti-allergic activity of 2-((bis (trimethylsilyl) methylthio/methylsulfonyl) methyl)-5-aryl-1, 3, 4-oxadiazoles. Eur. J. Med. Chem. 2013, 62, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Farghaly, A. Synthesis, reactions and antimicrobial activity of some new indolyl-1, 3, 4-oxadiazole, triazole and pyrazole derivatives. J. Chin. Chem. Soc. 2004, 51, 147–156. [Google Scholar]
- Swain, C.; Baker, R.; Kneen, C.; Moseley, J.; Saunders, J.; Seward, E.; Stevenson, G.; Beer, M.; Stanton, J.; Watling, K. Novel 5-ht3 antagonists. Indole oxadiazoles. J. Med. Chem. 1991, 34, 140–151. [Google Scholar] [CrossRef] [PubMed]
- Taha, M.; Ismail, N.H.; Imran, S.; Selvaraj, M.; Rashwan, H.; Farhanah, F.U.; Rahim, F.; Kesavanarayanan, K.S.; Ali, M. Synthesis of benzimidazole derivatives as potent β-glucuronidase inhibitors. Bioorg. Chem. 2015, 61, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Taha, M.; Ismail, N.H.; Imran, S.; Selvaraj, M.; Rahim, F. Synthesis of novel inhibitors of β-glucuronidase based on the benzothiazole skeleton and their molecular docking studies. RSC Adv. 2016, 6, 3003–3012. [Google Scholar] [CrossRef]
- Taha, M.; Ismail, N.H.; Imran, S.; Selvaraj, M.; Rahim, A.; Ali, M.; Siddiqui, S.; Rahim, F.; Khan, K.M. Synthesis of novel benzohydrazone–oxadiazole hybrids as β-glucuronidase inhibitors and molecular modeling studies. Bioorg. Med. Chem. 2015, 23, 7394–7404. [Google Scholar] [CrossRef] [PubMed]
- Baharudin, M.S.; Taha, M.; Imran, S.; Ismail, N.H.; Rahim, F.; Javid, M.T.; Khan, K.M.; Ali, M. Synthesis of indole analogs as potent β-glucuronidase inhibitors. Bioorg. Chem. 2017, 72, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Taha, M.; Ismail, N.H.; Imran, S.; Rashwan, H.; Jamil, W.; Ali, S.; Kashif, S.M.; Rahim, F.; Salar, U.; Khan, K.M. Synthesis of 6-chloro-2-aryl-1h-imidazo [4, 5-b] pyridine derivatives: Antidiabetic, antioxidant, β-glucuronidase inhibiton and their molecular docking studies. Bioorg. Chem. 2016, 65, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Taha, M.; Sultan, S.; Nuzar, H.A.; Rahim, F.; Imran, S.; Ismail, N.H.; Naz, H.; Ullah, H. Synthesis and biological evaluation of novel n-arylidenequinoline-3-carbohydrazides as potent β-glucuronidase inhibitors. Bioor. Med. Chem. 2016, 24, 3696–3704. [Google Scholar] [CrossRef] [PubMed]
- Taha, M.; Ismail, N.H.; Imran, S.; Rahim, F.; Wadood, A.; Khan, H.; Ullah, H.; Salar, U.; Khan, K.M. Synthesis, β-glucuronidase inhibition and molecular docking studies of hybrid bisindole-thiosemicarbazides analogs. Bioorg. Chem. 2016, 68, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Taha, M.; Ismail, N.H.; Jamil, W.; Khan, K.M.; Salar, U.; Kashif, S.M.; Rahim, F.; Latif, Y. Synthesis and evaluation of unsymmetrical heterocyclic thioureas as potent β-glucuronidase inhibitors. Med. Chem. Res. 2015, 24, 3166–3173. [Google Scholar] [CrossRef]
- Zawawi, N.K.N.A.; Taha, M.; Ahmat, N.; Wadood, A.; Ismail, N.H.; Rahim, F.; Ali, M.; Abdullah, N.; Khan, K.M. Novel 2, 5-disubtituted-1, 3, 4-oxadiazoles with benzimidazole backbone: A new class of β-glucuronidase inhibitors and in silico studies. Bioorg. Med. Chem. 2015, 23, 3119–3125. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Autodock4 and autodocktools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Jiang, J.; Jin, Y.; Kallemeijn, W.W.; Kuo, C.L.; Artola, M.; Dai, W.; van Elk, C.; van Eijk, M.; van der Marel, G.A.; et al. Activity-based probes for functional interrogation of retaining beta-glucuronidases. Nat. Chem. Biol. 2017, 13, 867–873. [Google Scholar] [CrossRef] [PubMed]
- Taha, M.; Ismail, N.H.; Imran, S.; Wadood, A.; Rahim, F.; Al Muqarrabin, L.M.R.; Zaki, H.M.; Ahmat, N.; Nasir, A.; Khan, F. Synthesis of novel disulfide and sulfone hybrid scaffolds as potent β-glucuronidase inhibitor. Bioorg. Chem. 2016, 68, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Drendel, W.B.; Chen, Z.W.; Mathews, F.S.; Sly, W.S.; Grubb, J.H. Structure of human beta-glucuronidase reveals candidate lysosomal targeting and active-site motifs. Nat. Struct. Biol. 1996, 3, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Jamil, W.; Perveen, S.; Shah, S.A.A.; Taha, M.; Ismail, N.H.; Perveen, S.; Ambreen, N.; Khan, K.M.; Choudhary, M.I. Phenoxyacetohydrazide Schiff bases: β-Glucuronidase inhibitors. Molecules 2014, 19, 8788–8802. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are available from the authors. |
| S. No. | Yield (%) | IC50 (μM ± SEM a) | S. No. | Yield (%) | IC50 (μM ± SEM a) | ||
|---|---|---|---|---|---|---|---|
| 1 | 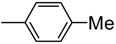 | 36.4 ± 0.6 | 12 | 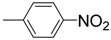 | 90 | 37.3 ± 0.7 | |
| 2 | 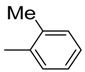 | 26.2 ± 0.8 | 13 | 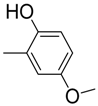 | 82 | 20.6 ± 0.5 | |
| 3 | 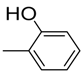 | 6.2 ± 0.2 | 14 | 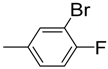 | 84 | NA b | |
| 4 | 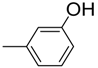 | 84 | 17.90 ± 0.4 | 15 |  | 83 | 34.6 ± 0.7 |
| 5 | 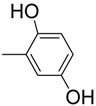 | 85 | 11.4 ± 0.30 | 16 | 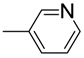 | 85 | 46.4 ± 0.9 |
| 6 |  | 86 | 0.9 ± 0.01 | 17 |  | 87 | 43.1 ± 0.8 |
| 7 | 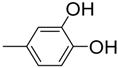 | 81 | 1.2 ± 0.01 | 18 | 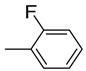 | 88 | 5.0 ± 0.1 |
| 8 | 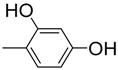 | 83 | 7.2 ± 0.10 | 19 | 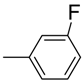 | 90 | 10.5 ± 0.2 |
| 9 | 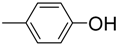 | 88 | 11.0 ± 0.4 | 20 | 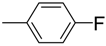 | 92 | 17.0 ± 0.4 |
| 10 | 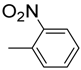 | 90 | 40.0 ± 0.7 | 21 | 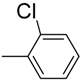 | 90 | 6.0 ± 0.2 |
| 11 | 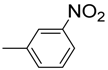 | 87 | NA b | 22 | 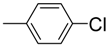 | 82 | 22.2 ± 0.5 |
| d-saccharic acid 1,4 lactone c | 48.1 ± 1.2 | ||||||
| Indole Based Oxadiazole Derivatives | Free Binding Energy (kcal/mol) | H-Bonds | Number of Closest Residues to the Docked Ligand in the Active Site | IC50 ± SEM |
|---|---|---|---|---|
| 1 | −8.74 | 1 | 5 | 36.4 ± 0.6 |
| 3 | −8.17 | 1 | 4 | 6.2 ± 0.2 |
| 4 | −8.53 | 2 | 6 | 17.9 ± 0.4 |
| 5 | −9.16 | 3 | 7 | 11.4 ± 0.3 |
| 6 | −8.77 | 4 | 7 | 0.9 ± 0.01 |
| 7 | −8.88 | 4 | 9 | 1.2 ± 0.01 |
| 8 | −8.56 | 4 | 7 | 7.2 ± 0.10 |
| 9 | −8.65 | 3 | 7 | 11.04 ± 0.4 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anouar, E.H.; Moustapha, M.E.; Taha, M.; Geesi, M.H.; Farag, Z.R.; Rahim, F.; Almandil, N.B.; Farooq, R.K.; Nawaz, M.; Mosaddik, A. Synthesis, Molecular Docking and β-Glucuronidase Inhibitory Potential of Indole Base Oxadiazole Derivatives. Molecules 2019, 24, 963. https://doi.org/10.3390/molecules24050963
Anouar EH, Moustapha ME, Taha M, Geesi MH, Farag ZR, Rahim F, Almandil NB, Farooq RK, Nawaz M, Mosaddik A. Synthesis, Molecular Docking and β-Glucuronidase Inhibitory Potential of Indole Base Oxadiazole Derivatives. Molecules. 2019; 24(5):963. https://doi.org/10.3390/molecules24050963
Chicago/Turabian StyleAnouar, El Hassane, Moustapha Eid Moustapha, Muhammad Taha, Mohammed H. Geesi, Zeinab R. Farag, Fazal Rahim, Noor Barak Almandil, Rai Khalid Farooq, Muhammad Nawaz, and Ashik Mosaddik. 2019. "Synthesis, Molecular Docking and β-Glucuronidase Inhibitory Potential of Indole Base Oxadiazole Derivatives" Molecules 24, no. 5: 963. https://doi.org/10.3390/molecules24050963
APA StyleAnouar, E. H., Moustapha, M. E., Taha, M., Geesi, M. H., Farag, Z. R., Rahim, F., Almandil, N. B., Farooq, R. K., Nawaz, M., & Mosaddik, A. (2019). Synthesis, Molecular Docking and β-Glucuronidase Inhibitory Potential of Indole Base Oxadiazole Derivatives. Molecules, 24(5), 963. https://doi.org/10.3390/molecules24050963






