Structure–Activity Prediction of ACE Inhibitory/Bitter Dipeptides—A Chemometric Approach Based on Stepwise Regression
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Dataset Construction
4.1.1. Peptides
4.1.2. Variables
4.2. Protocol
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ACE | Angiotensin converting enzyme |
| α-value | Significance level |
| BR | Backward regression |
| FR | Forward regression |
| F-test | Fisher–Snedecor test |
| IC50 | Concentration of peptide corresponding to its half-maximal inhibitory activity [µM] |
| MLR | Multiple regression |
| p-value | Probability of a given statistical model |
| R | Correlation coefficient |
| Rcaf. | Bitterness of peptide related to that of 1 mM caffeine solution |
| R2 | Determination coefficient |
| SW-W | Normality Shapiro–Wilk’s test |
| t-test | Student’s t test |
References
- Albenzio, M.; Santillo, A.; Caroprese, M.; della Malva, A.; Marino, R. Bioactive peptides in animal food products. Foods 2017, 6, 35. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liao, W.; Udenigwe, C.C. Revisiting the mechanism of ACE inhibitory peptides from food proteins. Trends Food Sci. Food Technol. 2017, 69, 214–219. [Google Scholar] [CrossRef]
- Xie, C.-L.; Kim, J.-S.; Ha, J.-M.; Choung, S.-Y.; Choi, Y.-J. Angiotensin-I-converting enzyme inhibitor derived from cross-linked oyster protein. Biomed. Res. Int. 2015, 2015, 379234. [Google Scholar] [CrossRef] [PubMed]
- Shamloo, M.; Eck, P.; Beta, T. Angiotensin converting enzyme inhibitory peptides derived from cereals. J. Hum. Nutr. Food Sci. 2015, 3, 1057. [Google Scholar]
- Iwaniak, A.; Minkiewicz, P.; Darewicz, M. Food-originating ACE inhibitors, including antihypertensive peptides, as preventive food components in blood pressure reduction. Compr. Rev. Food Sci. Food Saf. 2014, 13, 114–134. [Google Scholar] [CrossRef]
- Kumar, R.; Kumar, A.; Sharma, R.; Baruwa, A. Pharmacological review on natural ACE inhibitors. Der Pharm. Lett. 2010, 2, 273–293. [Google Scholar]
- Tidona, F.; Criscione, A.; Guastella, A.M.; Zuccaro, A.; Bordonaro, S.; Marletta, D. Bioactive peptides in dairy products. Ital. J. Anim. Sci. 2009, 8, 315–340. [Google Scholar] [CrossRef]
- Mäkinen, S.; Johannson, T.; Vegarud, E.G.; Pihlava, J.M.; Pihlanto, A. Angiotensin I-converting enzyme inhibitory and antioxidant properties of rapeseed hydrolysates. J. Funct. Foods 2012, 4, 575–583. [Google Scholar] [CrossRef]
- Antosova, Z.; Mackova, M.; Kral, V.; Macek, T. Therapeutic application of peptides and proteins: Parenteral forever? Trends Biotechnol. 2009, 27, 629–635. [Google Scholar] [CrossRef]
- Iwaniak, A.; Minkiewicz, P.; Darewicz, M.; Hrynkiewicz, M. Food protein-originating peptides as tastants—Physiological, technological, sensory, and bioinformatic approaches. Food Res. Int. 2016, 89, 27–38. [Google Scholar] [CrossRef]
- Kęska, P.; Stadnik, J. Taste-active peptides and amino acids of pork meat as components of dry-cured meat products: An in-silico study. J. Sens. Stud. 2017, 32, e12301. [Google Scholar] [CrossRef]
- Wu, J.; Aluko, R.E. Quantitative structure-activity relationship study of bitter di- and tri-peptides including relationship with angiotensin I-converting enzyme inhibitory activity. J. Pept. Sci. 2007, 13, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Pripp, A.H.; Ardö, S. Modelling relationship between angiotensin-(I)-converting enzyme inhibition and the bitter taste of peptides. Food Chem. 2007, 102, 880–888. [Google Scholar] [CrossRef]
- Udenigwe, C.C. Bioinformatics approaches, prospects and challenges of food bioactive peptide research. Trends Food Sci. Technol. 2014, 36, 134–143. [Google Scholar] [CrossRef]
- Darewicz, M.; Borawska, J.; Vegarud, G.E.; Minkiewicz, P.; Iwaniak, A. Angiotensin I-converting enzyme (ACE) inhibitory activity and ACE inhibitory peptides of salmon (Salmo salar) protein hydrolysates obtained by human and porcine gastrointestinal enzymes. Int. J. Mol. Sci. 2014, 15, 14077–14101. [Google Scholar] [CrossRef] [PubMed]
- Pripp, A.H.; Isaksson, T.; Stepaniak, L.; Sørhaug, T. Quantitative structure-activity relationship modelling of ACE-inhibitory peptides derived from milk proteins. Eur. Food Res. Technol. 2004, 219, 579–583. [Google Scholar] [CrossRef]
- Wu, J.; Aluko, R.E.; Nakai, S. Structural requirements of angiotensin I-converting enzyme inhibitory peptides: Quantitative structure-activity relationship study of di- and tripeptides. J. Agric. Food Chem. 2006, 54, 732–738. [Google Scholar] [CrossRef]
- Koyama, M.; Hattori, S.; Amano, Y.; Watanabe, M.; Nakamura, K. Blood pressure-lowering peptides from neo-fermented buckwheat sprouts: A new approach to estimating ACE-inhibitory activity. PLoS ONE 2014, 9, e105802. [Google Scholar] [CrossRef]
- Iwaniak, A.; Minkiewicz, P.; Darewicz, M.; Protasiewicz, M.; Mogut, D. Chemometrics and cheminformatics in the analysis of biologically active peptides from food sources. J. Funct. Foods 2015, 16, 334–351. [Google Scholar] [CrossRef]
- Iwaniak, A.; Hrynkiewicz, M.; Bucholska, J.; Minkiewicz, P.; Darewicz, M. Understanding the nature of bitter-taste di- and tripeptides derived from food proteins based on chemometric analysis. J. Food Biochem. 2018, 42, e12500. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information. Available online: http://www.ncbi.nlm.nih.gov (accessed on 1 June 2018).
- Web of Science. Available online: https://webofknowledge.com (accessed on 1 June 2018).
- Granato, D.; de Araújo Calado, V.M.; Jarvis, B. Observations on the use of statistical methods in Food science and Technology. Food Res. Int. 2014, 55, 137–149. [Google Scholar] [CrossRef]
- Latha, D.P.P.; Sharmila, D.J.S. QSAR study for the prediction of IC50 and Log P for 5-N-acetyl-beta-D-neuraminic acid structurally similar compounds using stepwise (multivariate) linear regression. Int. J. Chem. Res. 2010, 2, 32–38. [Google Scholar]
- Iwaniak, A.; Minkiewicz, P.; Darewicz, M.; Sieniawski, K.; Starowicz, P. BIOPEP database of sensory peptides and amino acids. Food Res. Int. 2016, 85, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Minkiewicz, P.; Dziuba, J.; Iwaniak, A.; Dziuba, M.; Darewicz, M. BIOPEP and other programs for processing bioactive peptide sequences. J. Aoac Int. 2008, 91, 965–980. [Google Scholar] [PubMed]
- Bursac, Z.; Gauss, C.H.; Williams, D.K.; Hosmer, D.W. Purposeful selection of variables in logistic regression. Source Code Biol. Med. 2008, 3, 17. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wright, J.; Brownlee, A.; Buswell, R. A comparison of approaches to stepwise regression on variables sensitivities in building simulation and analysis. Energy Build. 2016, 127, 313–326. [Google Scholar] [CrossRef]
- Wiegand, R.E. Performance of using multiple stepwise algorithms for variable selection. Stat. Med. 2010, 29, 1647–1659. [Google Scholar] [CrossRef]
- Latha, D.P.P.; Sharmila, D.J.S. QSAR study for the prediction of half maximal inhibitory concentration of compounds structurally similar to glicerol. Turk. J. Biochem. 2010, 35, 289–292. [Google Scholar]
- Collantes, E.R.; Dunn, W.J. Amino acid side chain descriptors for quantitative structure-activity relationship studies of peptide analogues. J. Med. Chem. 1995, 38, 2705–2713. [Google Scholar] [CrossRef]
- Tong, J.-B.; Chang, J.; Liu, S.-L.; Bai, M. A quantitative structure-activity relationship (QSAR) study of drugs based on a new descriptor of amino acid. J. Serb. Chem. Soc. 2015, 80, 343–353. [Google Scholar] [CrossRef]
- Soltani, S.; Haghaei, H.; Shayanfar, A.; Vallipour, J.; Zeynali, K.A.; Jouyban, A. QSBR study of bitter peptides: Application of GA-PLS in combination with MLR, SVM, and ANN approaches. Biomed. Res. Int. 2013, 2013, 501310. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Li-Chan, E.C.Y. Application of Fourier transform Raman spectroscopy for prediction of bitterness of peptides. Appl. Spectrosc. 2006, 60, 1297–1306. [Google Scholar] [CrossRef] [PubMed]
- Cheung, I.W.Y.; Li-Chan, E.C.Y. Angiotensin-I converting enzyme inhibitory activity and bitterness of enzymatically-produced hydrolysates of shrimp (Pandalopsis dispar) processing byproducts investigated by Taguchi design. Food Chem. 2010, 122, 1003–1012. [Google Scholar] [CrossRef]
- Pripp, A.H.; Isaksson, T.; Stepaniak, L.; Sørhaug, T.; Ardö, Y. Quantitative structure activity relationship modelling of peptides and proteins as a tool food science. Trends Food Sci. Technol. 2005, 16, 484–494. [Google Scholar] [CrossRef]
- FitzGerald, R.J.; Murray, B.A.; Walsh, D.J. Hypotensive peptides from milk proteins. J. Nutr. 2004, 134, 980S–988S. [Google Scholar] [CrossRef] [PubMed]
- Vermeirssen, V.; van der Bent, A.; Van Camp, J.; van Amerongen, A.; Verstraete, W. A quantitative in silico analysis calculates angiotensin I converting enzyme (ACE) inhibitory activity in pea and whey protein digests. Biochimie 2004, 86, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Asao, M.; Iwamura, H.; Akamatsu, M.; Fujita, T. Quantitative structure-activity relationships of the bitter thresholds of amino acids, peptides, and their derivatives. J. Med. Chem. 1987, 30, 1873–1879. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Zhou, Y. Quantifying the effect of burial of amino acid residues on protein stability. Proteins 2004, 54, 315–322. [Google Scholar] [CrossRef]
- Shahidi, F.; Zhong, Y. Bioactive peptides. J. AOAC Int. 2008, 91, 914–932. [Google Scholar]
- Aluko, R.E. Structural characteristics of food protein-derived bitter peptides. In Bitterness. Perception, Chemistry and Food Processing, 1st ed.; Aliani, M., Eskin, M.N.A., Eds.; Jon Wiley & Sons, Inc.: Hoboken, NJ, USA, 2017; pp. 105–129. ISBN 978-1118590284. [Google Scholar]
- BIOPEP-UWM Database. Available online: http://www.uwm.edu.pl/biochemia (accessed on 1 April–30 June 2018).
- Neubig, R.E.; Spedding, M.; Kenakin, T.; Christopoulos, A. International Union of Pharmacology Committee on Receptor Nomenclature and Drug Classification. XXXVIII. Update on terms and symbols in quantitative pharmacology. Pharmacol. Rev. 2003, 55, 597–606. [Google Scholar] [CrossRef]
- Iwaniak, A.; Dziuba, J. Animal and plant origin proteins as the precursors of peptides with ACE inhibitory activity. Proteins evaluation by means of in silico methods. Food Technol. Biotechnol. 2009, 47, 441–449. [Google Scholar]
- Zimmerman, J.M.; Eliezer, N.; Simha, R.J. The characterization of amino acid sequences in proteins by statistical methods. Theor. Biol. 1968, 21, 170–201. [Google Scholar] [CrossRef]
- Grantham, R. Amino acid difference formula to explain protein evolution. Science 1974, 185, 862–864. [Google Scholar] [CrossRef] [PubMed]
- Kyte, J.; Doolitle, R.F. A simple method for displaying the hydrophatic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Charton, M. Protein folding and the genetic code: An alternative quantitative model. J. Theor. Biol. 1981, 91, 115–123. [Google Scholar] [CrossRef]
- Charton, M.; Charton, B.I. The structural dependence of amino acid hydrophobicity parameters. J. Theor. Biol. 1982, 99, 629–644. [Google Scholar] [CrossRef]
- Dawson, D.M. The Biochemical Genetics of Man; Brock, D.J.H., Mayo, O., Eds.; Academic Press: New York, NY, USA, 1972; pp. 1–38. [Google Scholar]
- ProtScale. Available online: http://web.expasy.org/protscale (accessed on 1 April–30 June 2018).
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the expasy server. In The Proteomics Protocols Handbook; Walkes, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. ISBN 978-1-58829-343-5. [Google Scholar]
- Biological Magnetic Resonance Data Bank. Available online: http://www.bmrb.wisc.edu/ref_info/aadata.dat (accessed on 1 April–30 June 2018).
- Ulrich, E.L.; Akutsu, H.; Doreleijers, J.F.; Harano, Y.; Ioannidis, Y.E.; Lin, J.; Markley, J.L. BioMagResBank. Nucl. Acids Res. 2008, 36, D402–D408. [Google Scholar] [CrossRef]
- AAindex Database. Available online: http://www.genome.jp (accessed on 1 April 2018).
- Kawashima, S.; Pokarowski, P.; Pokarowska, M.; Kolinski, A.; Katayama, T.; Kanehisa, M. AAindex: Amino acid index database, progress report 2008. Nucleic Nucl. Acids Res. 2008, 36, D202–D205. [Google Scholar] [CrossRef]
Sample Availability: Not available. |
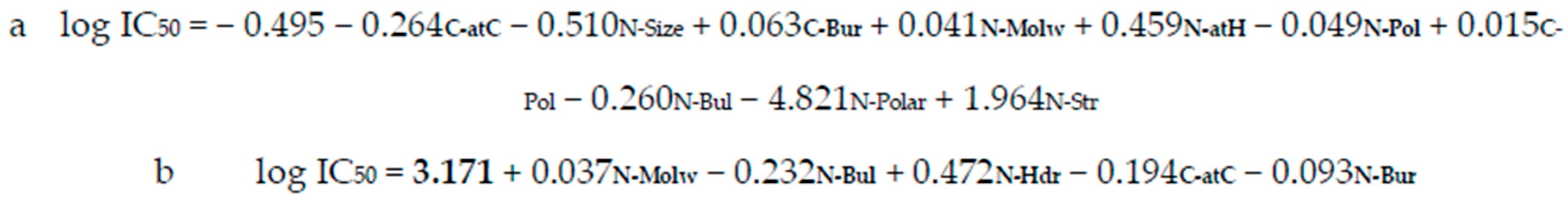
| Statistical Data | Forward Regression (FR) | Backward Regression (BR) |
|---|---|---|
| F | (10,17) 1 17.1 | (5,22) 1 14.1 |
| R | 0.95 | 0.87 |
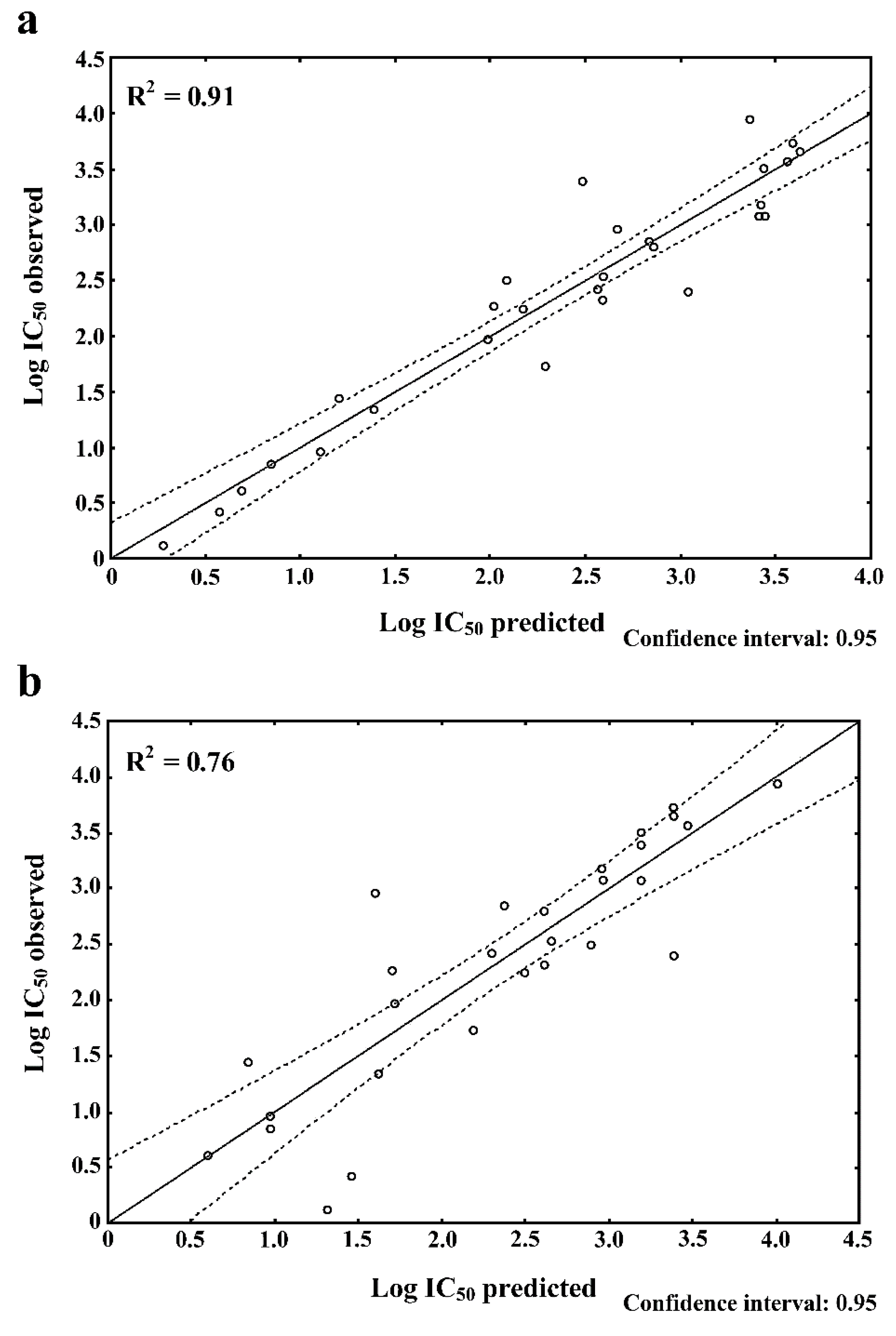
| R2 | 0.91 | 0.76 |
| Adjusted R2 | 0.86 | 0.71 |
| p | 0.000001 | 0.000003 |
| Standard estimation error | 0.41 | 0.58 |
| Statistically significant variables 2 | C-atC, C-Bur, N-Molw, N-atH, N-Pol | N-Molw, N-Bul, N-Hdr, C-atC, N-Bur |
| FR | BR | ||||||
|---|---|---|---|---|---|---|---|
| Dipeptides | Log IC50 | Rcaf. 1 | Peptides | Log IC50 | Rcaf. | ||
| Observed | Predicted | Observed | Predicted | ||||
| GR | 3.51 | 3.43 | 0.01 | GL | 3.40 | 3.19 | 0.04 |
| YP | 2.86 | 2.83 | 0.05 | LG | 3.94 | 4.00 | 0.05 |
| RR | 2.43 | 2.56 | 0.13 | RR | 2.43 | 2.30 | 0.13 |
| FG2 | 3.57 | 3.55 | 0.17 | FG | 3.57 | 3.47 | 0.17 |
| GV | 3.66 | 3.62 | 0.22 | GV | 3.66 | 3.38 | 0.22 |
| EY | 0.43 | 0.57 | 0.25 | IG | 3.08 | 2.96 | 0.22 |
| KP | 1.34 | 1.38 | 0.33 | YG | 3.18 | 2.95 | 0.33 |
| PR | 0.61 | 0.69 | 0.33 | PR2 | 0.61 | 0.59 | 0.33 |
| VY | 0.85 | 0.84 | 0.33 | VY | 0.85 | 0.97 | 0.33 |
| VF | 0.96 | 1.10 | 0.33 | VF | 0.96 | 0.97 | 0.33 |
| RF | 1.97 | 1.98 | 0.4 | RF | 1.97 | 1.72 | 0.4 |
| GE | 3.73 | 3.58 | 0.67 | GI | 3.08 | 3.19 | 0.44 |
| LF | 2.54 | 2.59 | 0.77 | LF | 2.54 | 2.65 | 0.77 |
| GF | 2.80 | 2.85 | 0.83 | GF | 2.80 | 2.61 | 0.83 |
| RP | 2.26 | 2.17 | 1.25 | ↑ Total: 14 (BR) | |||
| ↑ Total: 15 (FR) | Common dipeptides (9): PR, VY, VF, RF, RR, LF, GF, FG, GV | ||||||
| Model | Log Rcaf. = f (Observed log IC50) | Log Rcaf. = f (Predicted log IC50) | ||
|---|---|---|---|---|
| FR | R2 = 0.10 | R = −0.32 | R2 = 0.10 | R = −0.22 |
| BR | R2 = 0.05 | R = −0.32 | R2 = 0.05 | R = −0.23 |
| p | 0.09 | 0.25 | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hrynkiewicz, M.; Iwaniak, A.; Bucholska, J.; Minkiewicz, P.; Darewicz, M. Structure–Activity Prediction of ACE Inhibitory/Bitter Dipeptides—A Chemometric Approach Based on Stepwise Regression. Molecules 2019, 24, 950. https://doi.org/10.3390/molecules24050950
Hrynkiewicz M, Iwaniak A, Bucholska J, Minkiewicz P, Darewicz M. Structure–Activity Prediction of ACE Inhibitory/Bitter Dipeptides—A Chemometric Approach Based on Stepwise Regression. Molecules. 2019; 24(5):950. https://doi.org/10.3390/molecules24050950
Chicago/Turabian StyleHrynkiewicz, Monika, Anna Iwaniak, Justyna Bucholska, Piotr Minkiewicz, and Małgorzata Darewicz. 2019. "Structure–Activity Prediction of ACE Inhibitory/Bitter Dipeptides—A Chemometric Approach Based on Stepwise Regression" Molecules 24, no. 5: 950. https://doi.org/10.3390/molecules24050950
APA StyleHrynkiewicz, M., Iwaniak, A., Bucholska, J., Minkiewicz, P., & Darewicz, M. (2019). Structure–Activity Prediction of ACE Inhibitory/Bitter Dipeptides—A Chemometric Approach Based on Stepwise Regression. Molecules, 24(5), 950. https://doi.org/10.3390/molecules24050950








