Abstract
Forward and backward stepwise regression (FR and BR, respectively) was applied for the structure–bioactivity prediction of angiotensin converting enzyme (ACE)-inhibitory/bitter-tasting dipeptides. The datasets used in this study consisted of 28 sequences and numerical variables reflecting dipeptides’ physicochemical nature. The data were acquired from the BIOPEP-UWM, Biological Magnetic Resonance Databank, ProtScale, and AAindex databases. The calculations were computed using STATISTICA®13.1. FR/BR models differed in R2 (0.91/0.76, respectively). The impact of C-atC(−) and N-Molw(+) on the dual function of dipeptides was observed. Positive (+) and negative (−) correlations with log IC50 are presented in parens. Moreover, C-Bur(+), N-atH(+), and N-Pol(−) were also found to be important in the FR model. The additional statistical significance of N-bul(−), N-Bur(−), and N-Hdr(+) was reported in the BR model. These attributes reflected the composition of the dipeptides. We report that the “ideal” bitter ACE inhibitor should be composed of P, Y, F (C-end) and G, V, I, L (N-end). Functions: log Rcaf. = f (observed log IC50) and log Rcaf. = f (predicted log IC50) revealed no direct relationships between ACE inhibition and the bitterness of the dipeptides. It probably resulted from some structural discrepancies between the ACE inhibitory/bitter peptides and/or the measure of activity describing one of the two bioactivities. Our protocol can be applicable for the structure–bioactivity prediction of other bioactivities peptides.
1. Introduction
It is well documented in the literature that peptides derived from food proteins exhibit diversity of bioactivities, such as antihypertensive, antioxidative, antithrombotic, antidiabetic, immunomodulating functions, etc. [1]. Peptides’ ability to inhibit angiotensin converting enzyme (ACE; EC 3.4.15.1) has been the most extensively described such bioactivity in papers compared with other biological functions [2]. Moreover, ACE-inhibiting peptides have been identified in proteins originating in practically all food sources [3].
Generally, ACE is involved in blood pressure regulation. Briefly, it transforms angiotensin I, a decapeptide with the sequence DRVYIHPFHL, into angiotensin II (an octapeptide: DRVYIHPF) and C-terminal dipeptide histidyl-leucine (HL). The transformation of angiotensin I into angiotensin II leads to vasoconstriction and finally to elevated blood pressure. In turn, ACE catalyzes the cleavage of vasodilating bradykinin (RPPGFSPFR). The products of this reaction are as follows: C-terminal dipeptide FR (phenyl-arginine) and RPPGFSP (bradykinin 1–7). Thus, the action of the such inhibiting peptides (e.g., derived from food proteins) affects blood pressure reduction [4,5].
Compared with the synthetic antihypertensive drugs, peptides acting as ACE inhibitors are considered to be milder, non-toxic, and safer [6]. Therefore, some ACE-inhibiting peptides have the potential to be used as diet-originating therapeutic agents in hypertension treatment [7]. This is consistent with the approach of [8] in considering that peptides—as functional food ingredients and nutraceuticals—may help avoid various undesirable side effects associated with organically synthesized chemical medicines and can reduce the costs of drug therapy. Moreover, many therapeutic peptides and proteins are classified as biodrugs and are studied by biotechnologists who seek the production of needle-free and more user-friendly drugs [9].
Apart from their biological functions, peptides may affect all five taste sensations, i.e., bitter, salty, sour, sweet, and umami [10,11]. Bitterness is the most common taste that is associated with the peptides generated during food protein hydrolysis. Thus, the formation of off-flavor bitter peptides during proteolysis is considered to be one of the limitations in the production of biologically, chemically, and functionally valuable food hydrolysates, which may negatively affect their practical application in the food and nutrition industries [12].
Although food-originating peptide ACE inhibitors have a beneficial impact on health, they are often carriers of unwanted bitter tastes, which can be an obstacle when considering the enhancement of foods with the abovementioned bioactivity [13]. Thus, scientists study biopeptides, including ACE inhibitors, using in silico, in vitro, and ex vivo/in vivo methods [14,15]. The first approach involves computer modeling (bio- and cheminformatic analyses), including the structure–function prediction of ACE inhibitors [16,17], the second includes, e.g., the identification of peptides in novel food sources and/or optimal technologies of their production [14], and the third one includes studies with humans and animals, in particular, spontaneously hypertensive rats (SHR) [18].
In silico methods have been found useful in predicting the properties that determine the bioactivity of peptides resulting from their chemical nature [19]. Such an approach is called quantitative structure–activity relationship (QSAR) and is applied to study “separately” ACE-inhibiting [17] and/or bitter peptides [20]. Our quick search using the words “bitter ACE inhibitors” as a query revealed only a few original papers concerning this subject. Three of them were found in the National Center for Biotechnology Information (NCBI) [21], and two were listed in Web of Science (WoS) [22]. Progress has been observed in computer technologies since the release of these papers. It has contributed to the increasing number of databases providing information about the peptides, programs for providing data on their physicochemical properties, and programs analyzing created datasets using chemometric methods, such as artificial neural networks (ANN), principal component analysis (PCA), multiple regression (MLR) techniques, etc. [19]. Such a variety of tools are suitable to create data matrices, which help to understand the impact of the chemical nature of peptides on their bioactivity/function [20].
Thus, the aim of our study was to predict which specific properties of amino acids forming dipeptide sequences determine their dual function (i.e., ACE inhibition and bitter taste) using MLR variants (i.e., forward and backward stepwise regression).
2. Results
Two multilinear regression (MLR) models, namely forward regression (FR) and backward regression (BR), were carried out for two datasets composed of 28 peptidic bitter ACE inhibitors and 20 variables each. An F-test made for the analysis of significance of the estimated models led to rejecting the Hnull (briefly, independent variables do not affect the dependent variable, meaning log IC50). The reason for this was p < 0.01 and the relatively high values of the F-test. According to [23], the constructed regression model is considered to be suitable when an F-test or ANOVA analysis is carried out at least at p < 0.05. In the FR and BR models, an F-test is often applied as the default criterion to stop the stepwise regression procedure. Three values of α are applied when performing an F-test, namely 0.01, 0.02, and 0.05. However, some authors suggest using only the first two α-values rather than 0.05 to avoid the overfitting of the models [23]. Thus, in our study we applied the α-value of 0.01 to analyze the results of the F-test (see Section 4). The standard estimation error, being an indicator of differences between the observed and predicted log IC50, was 0.41 (FR model) and 0.58 (BR model) (Table 1). FR and BR models explained the variance (R2) in 91.0 and 76.0%, respectively. Normality Shapiro–Wilk’s (SW-W) test calculated for both regression models led to obtaining p > α (α = 0.01). The p-value was 0.254 (SW-W = 0.954) for FR and 0.136 for BR (SW-W = 0.943) (data not shown).

Table 1.
Statistical summary of forward and backward regression (FR and BR, respectively) made for the angiotensin converting enzyme (ACE)-inhibiting/bitter dipeptide dataset (α = 0.01).
Based on the results shown in Table 1, two equations explaining the impact of the individual variables on the log IC50 (Figure 1a,b) were generated. The first equation (Figure 1a) referred to the FR model, which was obtained in 10 steps, while the second one (Figure 1b), concerning the BR model, was calculated in 15 steps.
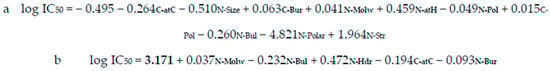
Figure 1.
FR (a) and BR (b) equations explaining the structure–function of the ACE-inhibiting/bitter dipeptides (statistically significant parameters are indicated in bold).
A total of five variables were statistically significant in both models (see also Table 1). Additionally, in the case of the BR model, the intercept was also found to be statistically significant. The importance of an individual variable in the bioactivity explanation was represented by a positive and/or negative value of a regression coefficient [17]. Thus, considering log IC50 parameter interpretation (the lower its value, the greater the bioactivity of a peptide), negative values of regression coefficients indicate that the higher value of a variable results in the lower final value of log IC50 [24]. For example, when looking at Figure 1a, if the molecular weight of the N-terminal residue raises, log IC50 will increase. To conclude, to get lower values of log IC50, it is better for peptidic bitter ACE inhibitors to be composed of N-terminal low molecular weight amino acids such as G (Molw = 75.07 Da).
The variables negatively correlated with log IC50 were as follows: C-atC, N-Size, N-Pol, and N-Polar (FR) and N-Bul, C-atC, and N-Bur (BR). The positively correlated ones were as follows: C-Bur, N-Molw, N-atH, C-Pol, and N-Str (FR) and N-Molw and N-Hdr (BR).
The ACE-inhibitory activity of the sequences derived from the BIOPEP-UWM database was expressed as IC50 [μM] [13] (see Section 4), whereas the bitter taste was defined as Rcaf., namely the bitterness of the peptide related to that of 1 mM caffeine solution [20]. Although our dipeptide data represented sequences with dual function expressed with two different parameters (IC50 and Rcaf.), the FR and BR equations were calculated taking into consideration log IC50 as a dependent variable. Then, the next step was to compare the dipeptide sequences with approximate observed versus predicted values of log IC50, with Rcaf. representing their potential as bitter tastants (see Table 2).

Table 2.
ACE-inhibiting and bitter-tasting dipeptides with approximate observed and predicted values of log IC50.
To achieve such data, based on the FR and BR, two linear regression curves were plotted (Figure 2a,b, respectively). They illustrate the distribution of the samples (dipeptides) in the area of a regression curve (solid line) or a confidence interval set by default at 95% (dashed line) considering the observed and predicted values of log IC50. The dipeptides possessing approximate observed vs. predicted log IC50 values were compared with Rcaf. estimating peptide bitterness. The FR results revealed 15 dipeptides with approximate observed versus predicted log IC50 values, whereas BR revealed 14 dipeptides (see Table 2). At total of nine common sequences possessing similar observed and predicted log IC50 values were observed in both models. They were as follows: PR, VY, VF, RF, RR, LF, GF, FG, and GV. Moreover, our dataset (see Section 4) was composed of 6 ACE-inhibiting sequences containing P, and four of them (KP, RP, YP, and PR) (Table 2; FR results) possessed approximate observed and predicted values of log IC50 values ranging from 0.61/0.69 (PR) to 2.86/2.83 (YP), respectively. Comparing their log IC50 with the log IC50 of other dipeptides, these values were observed to be relatively low, which indicated a high potential of P-containing dipeptides as ACE inhibitors. When looking at Rcaf. of the P-containing dipeptides (the higher the value, the more bitter the peptide) [20], they ranged from 0.05 (YP) to 1.25 (RP).
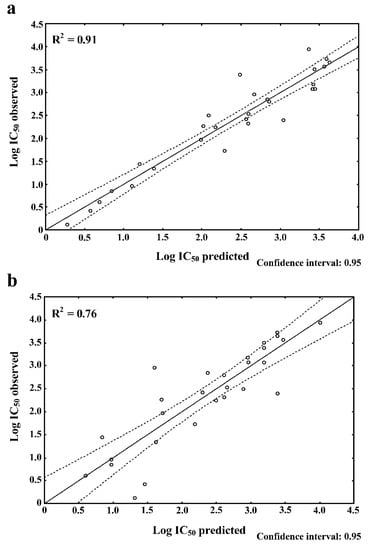
Figure 2.
Observed and predicted log IC50 of ACE-inhibiting/bitter dipeptides: (a) FR and (b) BR. The dashed line denotes the confidence interval.
Our study results show that the presence of P may determine the ACE-inhibitory activity and bitterness of a dipeptide. This may also be observed when searching databases of peptides, such as the BIOPEP-UWM database of sensory peptides and amino acids, in which the majority of bitter-tasting sequences with different chain length possess P and match the sequences with confirmed ACE-inhibitory bioactivity found in the BIOPEP-UWM database of bioactive peptide sequences [10,25,26].
So far, our abovementioned observations regarding the prediction of the dipeptide structure–ACE-inhibition/bitterness have concerned the association of the particular variables with the log IC50. No statistically significant results were found in both models (FR and BR) when directly comparing the ACE inhibition (predicted and observed log IC50) and the bitterness (log Rcaf.) of the dipeptides analyzed. The correlations between the abovementioned functions of dipeptides were relatively low (see Table 3).

Table 3.
Correlations between dipeptide ACE inhibition and bitterness obtained for the FR and BR models (α = 0.01).
The relatively low correlations were obtained in our studies (i.e., R2 = 0.32) between ACE inhibition and bitter taste of di- and tripeptides were higher than those obtained by other authors [12]. According to [12], this resulted from the discrepancies between the detailed structural requirements of peptides to act as ACE inhibitors (e.g., the presence of C-terminal bulky aromatic residue and positively charged amino acid in adjacent positions), as well as bitter tastants (e.g., the presence of hydrophobic side-chain C-terminal residue with aromatic amino acid in adjacent positions) [12].
3. Discussion
According to [13], the structure–bioactivity prediction of peptides showing more than one bioactivity is often difficult, because their biological function is measured for one effect. Thus, by using the BIOPEP-UWM database, a collection of bioactive [26] and sensory peptides [25], our strategy was to find the dipeptide sequences affecting both ACE inhibition and bitterness.
The main challenge for data analysts and scientists dealing with multivariate analyses, including multilinear regression, is to define which variables can be taken for model construction. Stepwise regression is one of the methods that allows the selecting of variables. Two stepwise regression methods are commonly used, namely forward and backward regression (FR and BR, respectively). Both of them enable building models to find the optimal solutions to a problem [27], such as predicting the impact of independent variables (e.g., numerically expressed properties) on a dependent variable (e.g., the measure of bioactivity) by describing the function of a sample (e.g., peptide) [19].
FR and BR are sequential models used to create a final model, which is built by adding or eliminating a predictor (a variable) [28]. Sequentiality relies on building successive models upon a prior model (unless the first model is a final one) when no more predictors based on specific criteria are included/excluded. Briefly, FR starts with no variables in the model. If the p-value of an individual variable is below the α-value, the one with the lowest value is included in the model, remaining in it forever (i.e., it affects the dependent variable). The procedure is continued until no variable reaches the entrance criteria. In BR, no predictors are found when starting. The predictors possessing the highest p-values among the pre-specified level are eliminated with no possibility of returning because their impact on the dependent variable is negligible. The resultant model is reduced and the procedure continues until the analysis of p-values for all remaining variables reaches the entrance criteria [29]. To conclude, the stepwise linear regression-driven approach enables determining the most influencing variables and including the most significant ones in the final analysis [30] and hence was taken into consideration when constructing the models to determine the impact of specific variables on the activity of dipeptides acting as ACE inhibitors and bitter tastants.
Our FR and BR models explained the variance (R2) in 91.0 and 76.0%, respectively (see Table 1). The latter value was comparable to the results reported by [31] concerning the application of partial least squares (PLS) in the analysis of structure–ACE-inhibition relations of 58 dipeptide sequences. In turn, our R2 = 0.91 approximated the PLS results of structure–bitterness studies of 48 dipeptides, in which R2 was 0.93 [31]. Our data confirmed a hypothesis about the normality of distribution [28], which indicated the suitability of FR and BR models for predicting the impact of the variables used for log IC50 prediction.
To summarize the statistical facts, although both models applied to the structural characteristics of dipeptide bitter ACE inhibitors possessed relatively low values of standard error of estimation and revealed the impact of five variables that were statistically significant, FR can be theoretically considered to be better than BR. The reason for such a conclusion is the higher values of R2 and adjusted R2 in the FR model compared with those of the BR model, meaning the model better fit to the data [28]. Thus, we decided to continue the procedure by determining the FR and BR equations describing the structure–activity relationships of the ACE-inhibiting/bitter-tasting dipeptides. As it was said above, the FR model was achieved in 10 steps, while the BR model was achieved in 15 steps. In both models, dependent variables illustrating the bioactivity of the dipeptides was represented by their log IC50 being the measure of ACE inhibition. The decision concerning the selection of such bioactivity measure resulted from an overview of scientific reports, according to which the QSAR models for ACE inhibitors were constructed with log IC50 [17]. Moreover, logarithms of IC50 (precisely, −log IC50) collected from the literature were applied for QSAR modeling of 48 bitter dipeptides to enable more efficient validation of new descriptors [32]. To conclude, we used log IC50 values as a “more leading” measure of the bioactivity of the analyzed peptides.
Both models (i.e., FR and BR) revealed five variables that were statistically significant in explaining the structure–bioactivity relationship of the dipeptides analyzed (see Table 1 and Figure 1a,b). The comparison of the two equations revealed the existence of two common, statistically significant variables influencing the double function of the dipeptides, namely C-atC (negative correlations) and N-Molw (positive correlations). Some common structural features of the ACE-inhibiting peptides that can be associated with bitterness were reported by other authors in their quantitative structural descriptions [33]. According to [33], the bitterness of the peptides was associated with relatively low values of a Balaban index related to the number of atoms in a molecule (the more atoms in a molecule, the lower the Balaban index). This observation matches the impact of C-Molw and C-atC on the dual bioactivity of the analyzed dipeptides. This may result from the presence of relatively high molecular weight residues, such as F and/or Y in a dipeptide sequence. According to [34], peptide bitterness was also determined by the presence of the abovementioned amino acids. Moreover, bitterness may be intensified by the presence of G [34], which was found to be statistically significant in our regression equations represented by positive correlations with log IC50 of N-Molw (FR and BR models) and N-atH (BR model). Finally, our results concerning the interpretation of variables in the context of their association with the specific amino acids present in bitter ACE-inhibiting dipeptides are also consistent with studies by [35] who found a correlation between the ACE inhibition and bitterness (R2 = 0.87) of enzymatic hydrolysates produced from shrimp (Pandalopsis dispar) protein byproducts. They observed that ACE inhibition and bitter-tasting hydrolysates were related to the presence of Y, F, L, I, V, and K [35].
It is well known that the N- and C-terminal locations of a residue in a sequence determines peptide bioactivity [36]. According to the scientific reports, the presence of the N-terminal G, I, L, and V in a peptide chain is preferable for ACE inhibition, while P, Y, and W are the favored C-terminal amino acids [37,38]. Such observations are consistent with the results obtained in our study. According to the regression models (Figure 1a,b), our potent ACE-inhibiting bitter peptides should be composed of C-terminal amino acids with a relatively high molecular weight (C-Molw), which is affected by an increasing number of carbon atoms (C-atC), as well as polarity (C-Pol). This regularity was represented by the presence of C-terminal Y, R, and K of a dipeptide dataset. According to [36], the enhancement of the ACE-inhibitory potential of peptides composed of 2–6 amino acids depended on the increased side-chain hydrophobicity of C-terminal amino acid and the decreased side-chain size of a residue in an adjacent position. It may refer to dipeptides possessing C-terminal F, Y, and/or P. Taking into consideration the N-terminal residues of the dipeptides analyzed, they were composed of amino acids with a relatively low molecular weight (G, I, L) and rather hydrophobic side chains, which was reflected by the positive correlations in our stepwise regression models. According to [17], highly potent dipeptides with an ACE-inhibiting effect should generally be composed of bulky and hydrophobic side chain residues. Hydrophobicity and the total length of a peptide were also indicated by other authors [39] as important parameters affecting the bitterness of single amino acids, as well as di- and tripeptides.
When looking at both equations, it can be observed that the buriability of the N- and C-terminal residue (N-Bur and C-Bur, respectively) had a statistically significant impact on log IC50. This parameter explains how the burial of hydrophobic or hydrophilic amino acids inside a protein sequence contributes to its stability. It is suggested that hydrophilic residues have a negative or minor impact on the stability of proteins compared with the hydrophobic amino acids present in a protein interior [40]. To conclude, the buriability scale proposed by [40] describes the residue–residue and residue–solvent interactions and was significantly and positively correlated with the hydrophobicity of a residue. Referring these findings to our results in the FR model, we observed that “Bur” of a C-terminal amino acid in a dipeptide sequence had an impact on log IC50 (positive correlation). No impact on the dipeptide log IC50 was observed for C-Hdr. Although, “Bur” is the parameter used to estimate protein stability [40], the lack of correlations between C-Hdr and C-Bur in the FR model could legitimate the application of buriability for the analysis of dipeptide structure–ACE-inhibition/bitterness. On the other hand, our results from the BR model indicated that N-Hdr of a residue (positive correlations) was in opposition to N-bur (negative correlations). Considering the fact that the buriability scale is useful for “residue-type” interactions (see above) [40], it can also be used to describe the interactions of short-chain peptides, such as ACE inhibitors with proteins (e.g., bitter taste receptors).
As it was said above, peptide bioactivity is determined by its amino acid composition [41]. When looking at both sets of ACE-inhibiting/bitter dipeptides found in Table 2, the dominant amino acids forming the peptide sequences were as follows: F, G, and R located at both the N- and C-end of a dipeptide chain. Some authors [42] have summarized the structural features affecting peptide bitterness. Apart from net hydrophobicity of a whole sequence, it was found that the presence of specific residues and their locations in a peptide chain were important as well. For example, peptides containing P within the sequence with the additional presence of R, V, and L (both N- and C-terminal) were indispensable for peptides to be bitter [42]. Such peptide sequences (i.e., containing P) could be also observed among the dipeptides possessing approximate observed versus predicted values of log IC50 with Rcaf. (see Table 2).
Although we could observe some similarities in the amino acid composition of bitter ACE inhibitors, which was reflected by the variables indicated in the FR and BR models, there were also some dipeptides with more diversified structures that did not match the above-described regularities. These results were reflected by relatively low correlations between the dipeptide ACE inhibition and bitterness obtained for the FR and BR models (see Table 3). According to other authors, although it is possible to find some correlations between ACE-inhibitory activity and the bitterness of the peptides, further direct comparison of these two properties is rather difficult due to one measure of activity applied. Usually, it is the measure describing ACE inhibition or bitterness [13].
To summarize the above discussion, our protocol, consisting of the application of curated databases and cheminformatic websites for the construction of data matrices, can be useful in chemometric analyses of the structure–function prediction of peptides with double bioactivity. Although we found FR to be statistically better than BR, considering that only the first one cannot be considered to be unequivocal. Both models indicated some common features, which influence the bitterness and ACE-inhibitory potential of dipeptides; however, looking at FR and BR in detail, they differed in the number of other descriptors important in the data interpretation. This is due to the fact that FR and BR are methods of variable selection, enabling the construction of models “eliminating” those whose impact on the dependent variable is negligible. However, FR and BR can be recommended for result interpretation. Such an approach allows for more consistent inferences and explanations of the relationship between the structure and ACE-inhibitory potential of bitter dipeptides. Our procedure and results obtained can be useful for structure–function analysis of peptides with other bioactivities and can be applicable when developing technologies for the production and/or elimination of peptides with desired/undesired properties.
4. Materials and Methods
4.1. Dataset Construction
4.1.1. Peptides
The samples (i.e., objects and cases) taken to create a dataset were 28 dipeptide sequences acting simultaneously as ACE inhibitors and bitter tastants, i.e., FL(0.12), EY(0.43), PR(0.61), VY(0.85), VF(0.96), KP(1.34), KF(1.45), IL(1.74), RF(1.97), RP(2.26), AF(2.28), GY(2.32), GP(2.40), RR(2.43), FP(2.50), LF(2.54), GF(2.80), YP(2.86), IF(2.97), IG(3.08), GI(3.08), YG(3.18), GL(3.40), GR(3.51), FG(3.57), GV(3.66), GE(3.73), and LG(3.94). Their ACE-inhibiting measure was an IC50 value defined as the concentration [µM] of peptides corresponding to its half-maximal inhibition [13]. The abovementioned values assigned to each peptide were derived from the literature, which was cited in the “References” toolbar of the BIOPEP-UWM database of bioactive peptides [26,43]. Moreover, the specific value of IC50 can be found in the window called “EC50 [µM]”. It results from the universal graphical layout of the BIOPEP-UWM database of bioactive peptides containing sequences with over 40 bioactivities, which can be expressed as EC50, i.e., substance concentration giving half-maximal effect [44]. Thus, in the case of ACE-inhibiting peptides, their “EC50” found in the BIOPEP-UWM database toolbar should be understood as “inhibition” meaning IC50 [45].
Finally, the IC50 parameter of each dipeptide was transformed into a logarithm to enable multilinear regression analyses (see above in parens).
4.1.2. Variables
Values of log IC50 were dependent variables (see the chapter above). The independent variables were the descriptors (i.e., attributes and descriptors) associated with the physicochemical properties of N- and C-terminal amino acids forming ACE-inhibiting/bitter dipeptides. They were as follows: molecular weight (Molw), bulkiness (Bul) [46], polarity (Pol) [47], hydrophobicity (Hdr) [48], the number of carbon atoms (atC), the number of hydrogen atoms (atH), steric parameter (Str) [49], polarizability parameter (Polar) [50], size (Size) [51], and buriability (Bur) [40]. The abbreviations of variables are provided in parens. Moreover, all the abbreviations possessed an “N-” and “C-” prefix to associate an appropriate variable with the dipeptide N- and C-terminal amino acid residue, respectively. For example, a variable “C-atC” should be read as the number of carbon atoms in a C-terminal amino acid of a dipeptide sequence.
The abovementioned attributes were implemented from the following databases: ProtScale (Molw, Bul, Pol, and Hdr) [52,53], Biological Magnetic Resonance Data Bank (atC and atH) [54,55], and AAindex (Str, Polar, Size, and Bur) [56,57]. All the databases were accessed between April and June 2018.
4.2. Protocol
Multiple regression (MLR) for the ACE-inhibiting/bitter dipeptide dataset was made using STATISTICA®13.1 software (StatSoft, Cracow, Poland). The calculations were run by opening the “Statistics” menu and selecting the function “Multiple Regression”. After the selection of dependent and independent variables (see chapters above) and opening the toolbar “Advanced”, the following options were ticked: “Advanced options (stepwise or ridge regression)” and “Extended precision computations”. It enabled opening the window called “Model Definition”, selecting the function entitled “Stepwise” and then “Forward stepwise” and ticking “At each step” from the window called “Display results”. Choosing the option entitled “At each step” allowed for the observation of consecutive changes in the models during the procedure of particular model estimation (i.e., backward/forward stepwise regression models). An identical protocol was applied to make a backward stepwise regression by ticking the function called “Backward stepwise” instead of “Forward stepwise” in the “Model Definition” window.
Statistical procedures run to check if the regression analyses were applied appropriately when considering the representativeness of the samples and variables were carried out according to the recommendations in [23]. For example, the statistical significance of the estimated regression equations was determined using the global Fisher–Snedecor test (F-test) [23] at α = 0.01 and verifying the null versus the alternative hypotheses (Hnull, Halt, respectively). Hnull meant that the independent variables have no impact on the dependent variable. Halt was in opposition to Hnull. Rejection of Hnull indicated that at least 1 regression coefficient was non-zero. The statistical significance of particular predictors (i.e., regression coefficients; b) was analyzed by t-test at α = 0.01 [28].
5. Conclusions
The FR and BR models differed in R2, which suggests that the FR model is statistically better than the BR model in the structure–activity prediction of the ACE-inhibiting/bitter-tasting dipeptides. Both models showed some common properties, which determine the dual function of the dipeptides. The FR model revealed more descriptors important in data interpretation than did the BR model. The FR procedure led to finding the following statistically significant regularities, which determine the ACE-inhibitory/bitterness of dipeptides expressed by their log IC50: C-atC(−), C-Bur(+), N-Molw(+), N-atH(+), N-Pol(−). In turn, the BR model revealed the importance of additional variables, such as N-bul(−), N-Bur(−), and N-Hdr(+). The values in parens represent positive and negative correlations with log IC50, respectively, while the bold font shows statistically significant variables indicated in both the FR and BR models. The properties of the amino acids represented by individual variables affected the composition of the sequences analyzed. It was found that bitter ACE inhibitors preferred P, Y, F (C-end) and G, V, I, L (N-end).
Based on the results obtained, FR and BR can be recommended as chemometric techniques for the prediction of relationships between the structure and activity of peptides, such as ACE inhibitors with bitter taste. Our protocol, including curated databases and programs containing chemometric data, can be also applicable for the structure–function analysis of peptides with other biological functions.
Author Contributions
M.H. and A.I. designed the protocol, constructed the datasets, and discussed the results. J.B. was engaged in the analysis of the results and the drafting the manuscript. P.M. and M.D. prepared the literature overview used in the Introduction, and they prepared the tables and figures used in the manuscript. All the authors accepted the final version of manuscript.
Funding
The project was financially supported by the Minister of Science and Higher Education in the range of the program entitled “Regional Initiative of Excellence” for the years 2019–2022, Project No. 010/RID/2018/19, amount of funding 12,000,000 PLN and UNIVERSITY OF WARMIA AND MAZURY, grant number 17.610.014-300.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
| ACE | Angiotensin converting enzyme |
| α-value | Significance level |
| BR | Backward regression |
| FR | Forward regression |
| F-test | Fisher–Snedecor test |
| IC50 | Concentration of peptide corresponding to its half-maximal inhibitory activity [µM] |
| MLR | Multiple regression |
| p-value | Probability of a given statistical model |
| R | Correlation coefficient |
| Rcaf. | Bitterness of peptide related to that of 1 mM caffeine solution |
| R2 | Determination coefficient |
| SW-W | Normality Shapiro–Wilk’s test |
| t-test | Student’s t test |
References
- Albenzio, M.; Santillo, A.; Caroprese, M.; della Malva, A.; Marino, R. Bioactive peptides in animal food products. Foods 2017, 6, 35. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liao, W.; Udenigwe, C.C. Revisiting the mechanism of ACE inhibitory peptides from food proteins. Trends Food Sci. Food Technol. 2017, 69, 214–219. [Google Scholar] [CrossRef]
- Xie, C.-L.; Kim, J.-S.; Ha, J.-M.; Choung, S.-Y.; Choi, Y.-J. Angiotensin-I-converting enzyme inhibitor derived from cross-linked oyster protein. Biomed. Res. Int. 2015, 2015, 379234. [Google Scholar] [CrossRef] [PubMed]
- Shamloo, M.; Eck, P.; Beta, T. Angiotensin converting enzyme inhibitory peptides derived from cereals. J. Hum. Nutr. Food Sci. 2015, 3, 1057. [Google Scholar]
- Iwaniak, A.; Minkiewicz, P.; Darewicz, M. Food-originating ACE inhibitors, including antihypertensive peptides, as preventive food components in blood pressure reduction. Compr. Rev. Food Sci. Food Saf. 2014, 13, 114–134. [Google Scholar] [CrossRef]
- Kumar, R.; Kumar, A.; Sharma, R.; Baruwa, A. Pharmacological review on natural ACE inhibitors. Der Pharm. Lett. 2010, 2, 273–293. [Google Scholar]
- Tidona, F.; Criscione, A.; Guastella, A.M.; Zuccaro, A.; Bordonaro, S.; Marletta, D. Bioactive peptides in dairy products. Ital. J. Anim. Sci. 2009, 8, 315–340. [Google Scholar] [CrossRef]
- Mäkinen, S.; Johannson, T.; Vegarud, E.G.; Pihlava, J.M.; Pihlanto, A. Angiotensin I-converting enzyme inhibitory and antioxidant properties of rapeseed hydrolysates. J. Funct. Foods 2012, 4, 575–583. [Google Scholar] [CrossRef]
- Antosova, Z.; Mackova, M.; Kral, V.; Macek, T. Therapeutic application of peptides and proteins: Parenteral forever? Trends Biotechnol. 2009, 27, 629–635. [Google Scholar] [CrossRef]
- Iwaniak, A.; Minkiewicz, P.; Darewicz, M.; Hrynkiewicz, M. Food protein-originating peptides as tastants—Physiological, technological, sensory, and bioinformatic approaches. Food Res. Int. 2016, 89, 27–38. [Google Scholar] [CrossRef]
- Kęska, P.; Stadnik, J. Taste-active peptides and amino acids of pork meat as components of dry-cured meat products: An in-silico study. J. Sens. Stud. 2017, 32, e12301. [Google Scholar] [CrossRef]
- Wu, J.; Aluko, R.E. Quantitative structure-activity relationship study of bitter di- and tri-peptides including relationship with angiotensin I-converting enzyme inhibitory activity. J. Pept. Sci. 2007, 13, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Pripp, A.H.; Ardö, S. Modelling relationship between angiotensin-(I)-converting enzyme inhibition and the bitter taste of peptides. Food Chem. 2007, 102, 880–888. [Google Scholar] [CrossRef]
- Udenigwe, C.C. Bioinformatics approaches, prospects and challenges of food bioactive peptide research. Trends Food Sci. Technol. 2014, 36, 134–143. [Google Scholar] [CrossRef]
- Darewicz, M.; Borawska, J.; Vegarud, G.E.; Minkiewicz, P.; Iwaniak, A. Angiotensin I-converting enzyme (ACE) inhibitory activity and ACE inhibitory peptides of salmon (Salmo salar) protein hydrolysates obtained by human and porcine gastrointestinal enzymes. Int. J. Mol. Sci. 2014, 15, 14077–14101. [Google Scholar] [CrossRef] [PubMed]
- Pripp, A.H.; Isaksson, T.; Stepaniak, L.; Sørhaug, T. Quantitative structure-activity relationship modelling of ACE-inhibitory peptides derived from milk proteins. Eur. Food Res. Technol. 2004, 219, 579–583. [Google Scholar] [CrossRef]
- Wu, J.; Aluko, R.E.; Nakai, S. Structural requirements of angiotensin I-converting enzyme inhibitory peptides: Quantitative structure-activity relationship study of di- and tripeptides. J. Agric. Food Chem. 2006, 54, 732–738. [Google Scholar] [CrossRef]
- Koyama, M.; Hattori, S.; Amano, Y.; Watanabe, M.; Nakamura, K. Blood pressure-lowering peptides from neo-fermented buckwheat sprouts: A new approach to estimating ACE-inhibitory activity. PLoS ONE 2014, 9, e105802. [Google Scholar] [CrossRef]
- Iwaniak, A.; Minkiewicz, P.; Darewicz, M.; Protasiewicz, M.; Mogut, D. Chemometrics and cheminformatics in the analysis of biologically active peptides from food sources. J. Funct. Foods 2015, 16, 334–351. [Google Scholar] [CrossRef]
- Iwaniak, A.; Hrynkiewicz, M.; Bucholska, J.; Minkiewicz, P.; Darewicz, M. Understanding the nature of bitter-taste di- and tripeptides derived from food proteins based on chemometric analysis. J. Food Biochem. 2018, 42, e12500. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information. Available online: http://www.ncbi.nlm.nih.gov (accessed on 1 June 2018).
- Web of Science. Available online: https://webofknowledge.com (accessed on 1 June 2018).
- Granato, D.; de Araújo Calado, V.M.; Jarvis, B. Observations on the use of statistical methods in Food science and Technology. Food Res. Int. 2014, 55, 137–149. [Google Scholar] [CrossRef]
- Latha, D.P.P.; Sharmila, D.J.S. QSAR study for the prediction of IC50 and Log P for 5-N-acetyl-beta-D-neuraminic acid structurally similar compounds using stepwise (multivariate) linear regression. Int. J. Chem. Res. 2010, 2, 32–38. [Google Scholar]
- Iwaniak, A.; Minkiewicz, P.; Darewicz, M.; Sieniawski, K.; Starowicz, P. BIOPEP database of sensory peptides and amino acids. Food Res. Int. 2016, 85, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Minkiewicz, P.; Dziuba, J.; Iwaniak, A.; Dziuba, M.; Darewicz, M. BIOPEP and other programs for processing bioactive peptide sequences. J. Aoac Int. 2008, 91, 965–980. [Google Scholar] [PubMed]
- Bursac, Z.; Gauss, C.H.; Williams, D.K.; Hosmer, D.W. Purposeful selection of variables in logistic regression. Source Code Biol. Med. 2008, 3, 17. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wright, J.; Brownlee, A.; Buswell, R. A comparison of approaches to stepwise regression on variables sensitivities in building simulation and analysis. Energy Build. 2016, 127, 313–326. [Google Scholar] [CrossRef]
- Wiegand, R.E. Performance of using multiple stepwise algorithms for variable selection. Stat. Med. 2010, 29, 1647–1659. [Google Scholar] [CrossRef]
- Latha, D.P.P.; Sharmila, D.J.S. QSAR study for the prediction of half maximal inhibitory concentration of compounds structurally similar to glicerol. Turk. J. Biochem. 2010, 35, 289–292. [Google Scholar]
- Collantes, E.R.; Dunn, W.J. Amino acid side chain descriptors for quantitative structure-activity relationship studies of peptide analogues. J. Med. Chem. 1995, 38, 2705–2713. [Google Scholar] [CrossRef]
- Tong, J.-B.; Chang, J.; Liu, S.-L.; Bai, M. A quantitative structure-activity relationship (QSAR) study of drugs based on a new descriptor of amino acid. J. Serb. Chem. Soc. 2015, 80, 343–353. [Google Scholar] [CrossRef]
- Soltani, S.; Haghaei, H.; Shayanfar, A.; Vallipour, J.; Zeynali, K.A.; Jouyban, A. QSBR study of bitter peptides: Application of GA-PLS in combination with MLR, SVM, and ANN approaches. Biomed. Res. Int. 2013, 2013, 501310. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Li-Chan, E.C.Y. Application of Fourier transform Raman spectroscopy for prediction of bitterness of peptides. Appl. Spectrosc. 2006, 60, 1297–1306. [Google Scholar] [CrossRef] [PubMed]
- Cheung, I.W.Y.; Li-Chan, E.C.Y. Angiotensin-I converting enzyme inhibitory activity and bitterness of enzymatically-produced hydrolysates of shrimp (Pandalopsis dispar) processing byproducts investigated by Taguchi design. Food Chem. 2010, 122, 1003–1012. [Google Scholar] [CrossRef]
- Pripp, A.H.; Isaksson, T.; Stepaniak, L.; Sørhaug, T.; Ardö, Y. Quantitative structure activity relationship modelling of peptides and proteins as a tool food science. Trends Food Sci. Technol. 2005, 16, 484–494. [Google Scholar] [CrossRef]
- FitzGerald, R.J.; Murray, B.A.; Walsh, D.J. Hypotensive peptides from milk proteins. J. Nutr. 2004, 134, 980S–988S. [Google Scholar] [CrossRef] [PubMed]
- Vermeirssen, V.; van der Bent, A.; Van Camp, J.; van Amerongen, A.; Verstraete, W. A quantitative in silico analysis calculates angiotensin I converting enzyme (ACE) inhibitory activity in pea and whey protein digests. Biochimie 2004, 86, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Asao, M.; Iwamura, H.; Akamatsu, M.; Fujita, T. Quantitative structure-activity relationships of the bitter thresholds of amino acids, peptides, and their derivatives. J. Med. Chem. 1987, 30, 1873–1879. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Zhou, Y. Quantifying the effect of burial of amino acid residues on protein stability. Proteins 2004, 54, 315–322. [Google Scholar] [CrossRef]
- Shahidi, F.; Zhong, Y. Bioactive peptides. J. AOAC Int. 2008, 91, 914–932. [Google Scholar]
- Aluko, R.E. Structural characteristics of food protein-derived bitter peptides. In Bitterness. Perception, Chemistry and Food Processing, 1st ed.; Aliani, M., Eskin, M.N.A., Eds.; Jon Wiley & Sons, Inc.: Hoboken, NJ, USA, 2017; pp. 105–129. ISBN 978-1118590284. [Google Scholar]
- BIOPEP-UWM Database. Available online: http://www.uwm.edu.pl/biochemia (accessed on 1 April–30 June 2018).
- Neubig, R.E.; Spedding, M.; Kenakin, T.; Christopoulos, A. International Union of Pharmacology Committee on Receptor Nomenclature and Drug Classification. XXXVIII. Update on terms and symbols in quantitative pharmacology. Pharmacol. Rev. 2003, 55, 597–606. [Google Scholar] [CrossRef]
- Iwaniak, A.; Dziuba, J. Animal and plant origin proteins as the precursors of peptides with ACE inhibitory activity. Proteins evaluation by means of in silico methods. Food Technol. Biotechnol. 2009, 47, 441–449. [Google Scholar]
- Zimmerman, J.M.; Eliezer, N.; Simha, R.J. The characterization of amino acid sequences in proteins by statistical methods. Theor. Biol. 1968, 21, 170–201. [Google Scholar] [CrossRef]
- Grantham, R. Amino acid difference formula to explain protein evolution. Science 1974, 185, 862–864. [Google Scholar] [CrossRef] [PubMed]
- Kyte, J.; Doolitle, R.F. A simple method for displaying the hydrophatic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Charton, M. Protein folding and the genetic code: An alternative quantitative model. J. Theor. Biol. 1981, 91, 115–123. [Google Scholar] [CrossRef]
- Charton, M.; Charton, B.I. The structural dependence of amino acid hydrophobicity parameters. J. Theor. Biol. 1982, 99, 629–644. [Google Scholar] [CrossRef]
- Dawson, D.M. The Biochemical Genetics of Man; Brock, D.J.H., Mayo, O., Eds.; Academic Press: New York, NY, USA, 1972; pp. 1–38. [Google Scholar]
- ProtScale. Available online: http://web.expasy.org/protscale (accessed on 1 April–30 June 2018).
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the expasy server. In The Proteomics Protocols Handbook; Walkes, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. ISBN 978-1-58829-343-5. [Google Scholar]
- Biological Magnetic Resonance Data Bank. Available online: http://www.bmrb.wisc.edu/ref_info/aadata.dat (accessed on 1 April–30 June 2018).
- Ulrich, E.L.; Akutsu, H.; Doreleijers, J.F.; Harano, Y.; Ioannidis, Y.E.; Lin, J.; Markley, J.L. BioMagResBank. Nucl. Acids Res. 2008, 36, D402–D408. [Google Scholar] [CrossRef]
- AAindex Database. Available online: http://www.genome.jp (accessed on 1 April 2018).
- Kawashima, S.; Pokarowski, P.; Pokarowska, M.; Kolinski, A.; Katayama, T.; Kanehisa, M. AAindex: Amino acid index database, progress report 2008. Nucleic Nucl. Acids Res. 2008, 36, D202–D205. [Google Scholar] [CrossRef]
Sample Availability: Not available. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).