A New Family of Homoleptic Copper Complexes of Curcuminoids: Synthesis, Characterization and Biological Properties
Abstract
1. Introduction
2. Results and Discussion
2.1. IR Spectra
2.2. NMR Spectra
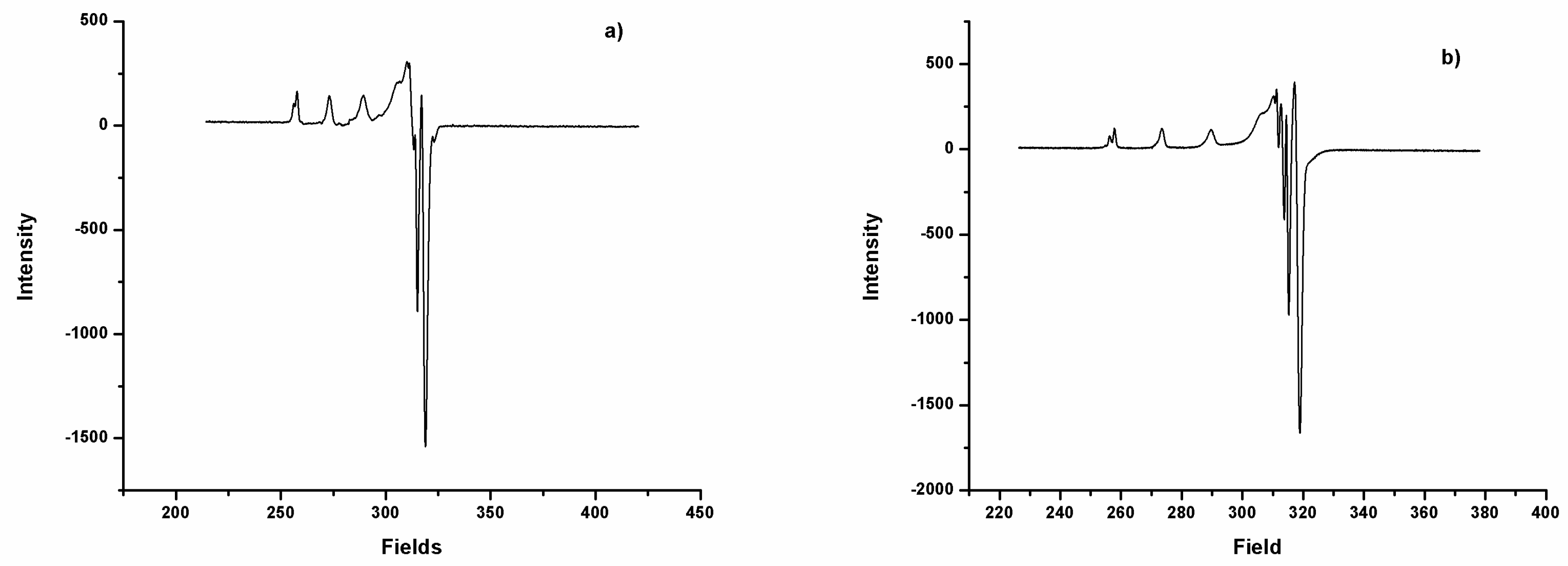
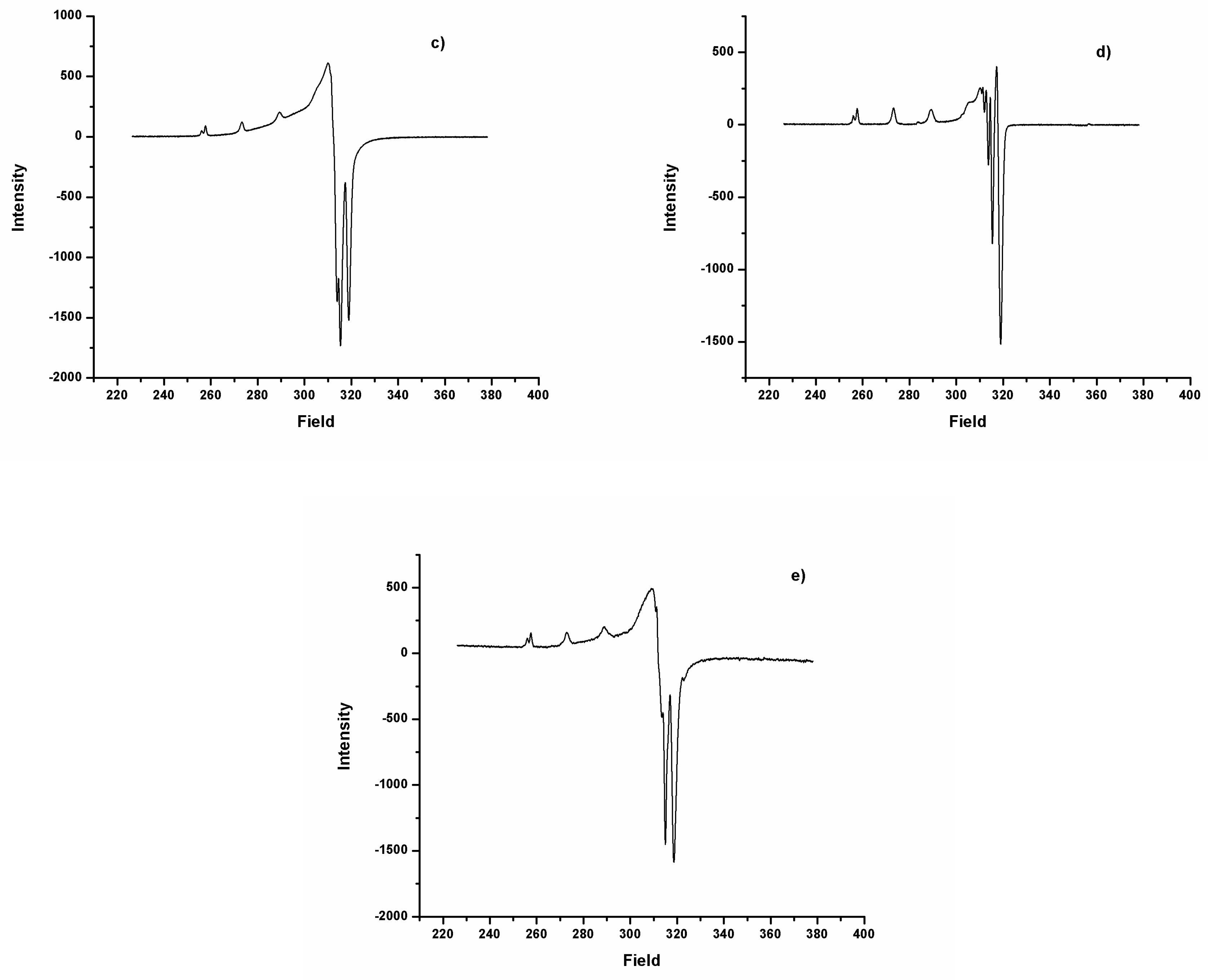
2.3. EPR Spectra
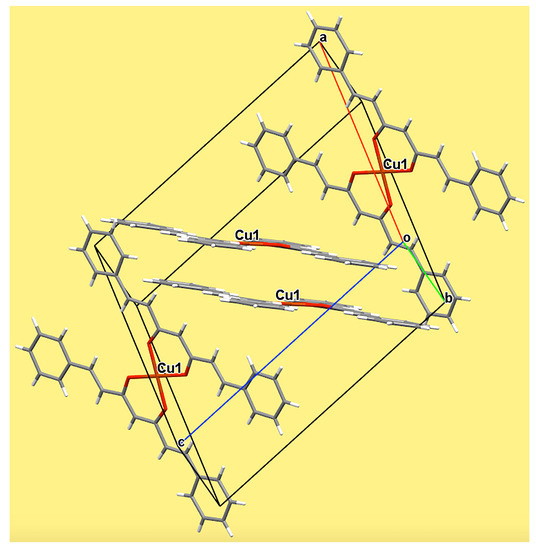
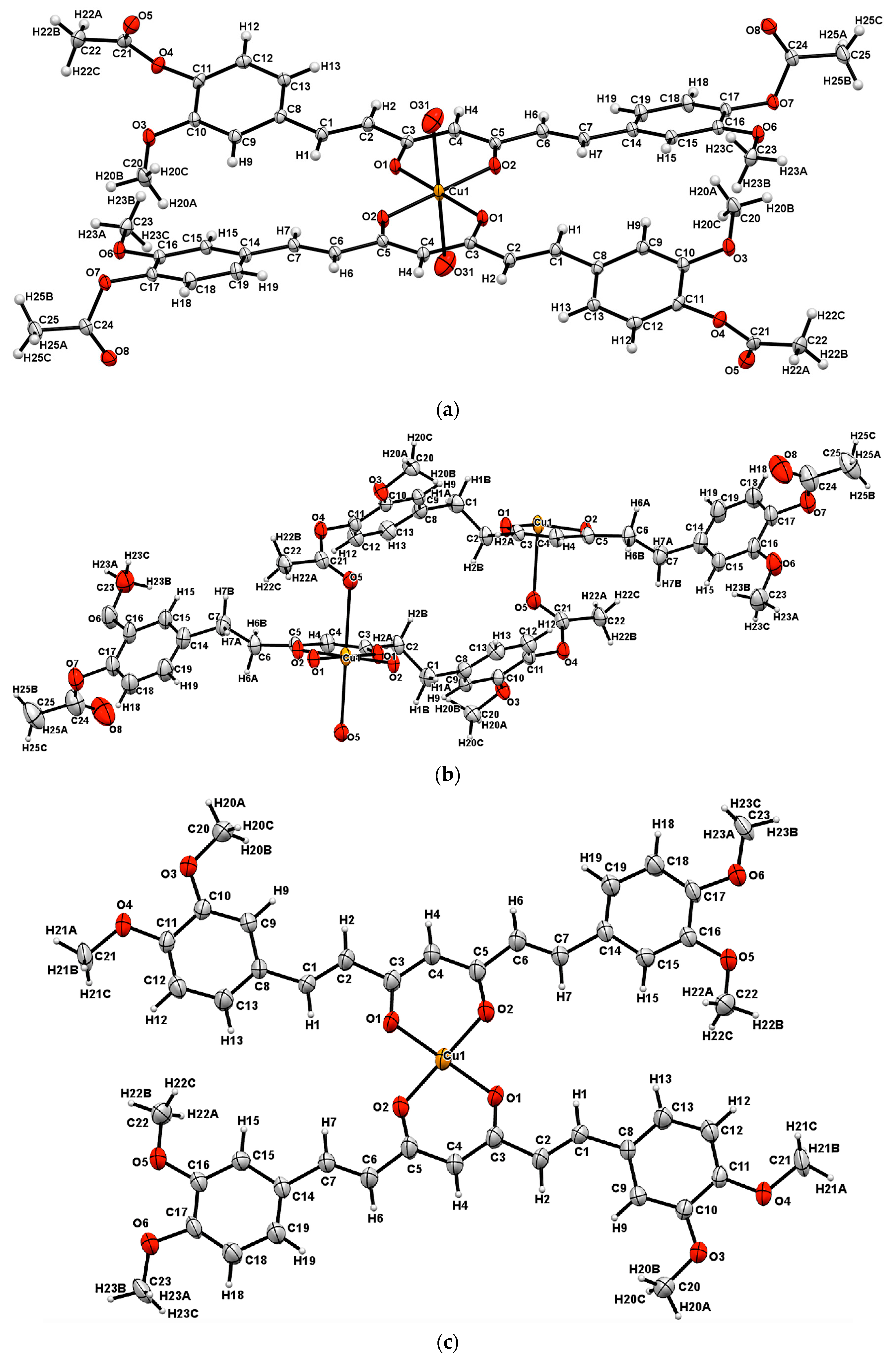
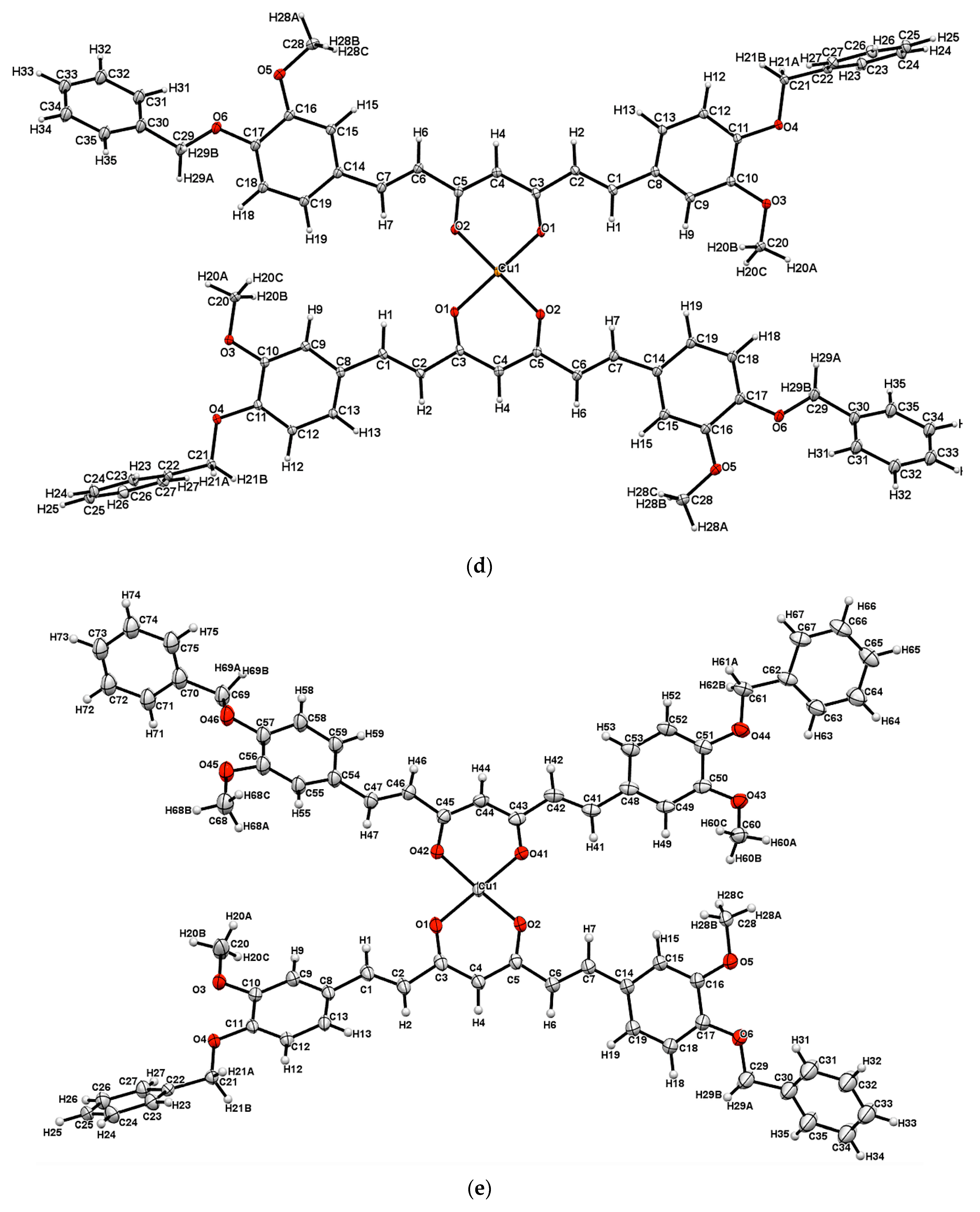
2.4. Single Crystal X-Ray Diffraction
2.5. Inhibition of Lipoperoxidation (LP) in Rat Brain Homogenate
2.6. Cytotoxic Activity
3. Materials and Methods
3.1. Physical Measurements
3.2. Spectroscopic Determinations
3.3. Inhibition of Lipid Peroxidation on Rat Brain
3.3.1. Animals
3.3.2. Rat Brain Homogenate Preparation
3.3.3. Induction of Lipid Peroxidation and Thiobarbituric Acid Reactive Substances (TBARS) Quantification
3.4. Citotoxic activity in Human Tumor Cells
3.5. Synthesis of Compounds
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wanninger, S.; Lorenz, V.; Subhan, A.; Edelmann, F.T. Metal complexes of curcumin-synthetic strategies, structures and medicinal applications. Chem. Soc. Rev. 2015, 44, 4986–5002. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Banerjee, S.; Sil, P.C. The beneficial role of curcumin on inflammation, diabetes and neurodegenerative disease: A recent update. Food Chem. Toxicol. 2015, 83, 111–124. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, B.B.; Surh, Y.-J.; Shishodia, S. The Molecular Target and Therapeutic Uses of Curcumin in Health an Disease. In Advances in Experimental Medicine and Biology; Cohen, I.R., Lajtha, I.R., Lambris, J.D., Paoletti, R., Rezaei, N., Eds.; Springer: New York, NY, USA, 2007; pp. 1–75. [Google Scholar] [CrossRef]
- Sanphui, P.; Bolla, D. Curcumin, a Biological Wonder Molecule: A Crystal Engineering Point of View. Cryst. Growth Des. 2018, 9, 5690–5711. [Google Scholar] [CrossRef]
- Goel, A.; Kunnumakkara, A.B.; Aggarwal, B.B. Curcumin as “Curecumin”: From kitchen to clinic. Biochem. Pharmacol. 2008, 75, 787–809. [Google Scholar] [CrossRef] [PubMed]
- Naksuriya, O.; Okonogi, S.; Schiffelers, R.M.; Hennink, W.E. Curcumin nanoformulations: A review of pharmaceutical properties and preclinical studies and clinical data related to cancer treatment. Biomaterials 2014, 35, 3365–3383. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Shulin, Y.; Zhou, H.; Shao, L.; Huang, K.; Xiao, J.; Huang, Z.; Li, X. Synthesis, crystal structure and anti-inflammatory properties of curcumin analogs. Eur. J. Med. Chem. 2009, 44, 915–919. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wei, D.; Jiang, B.; Liu, T.; Ni, J.; Zhou, S. Two copper(II) complexes of curcumin derivatives: Synthesis, crystal structure and in vitro antitumor activity. Transit. Met. Chem. 2014, 39, 553–558. [Google Scholar] [CrossRef]
- Aliaga-Alcalde, N.; Marqués-Gallego, P.; Kraaijkamp, M.; Herranz-Lancho, C.; Dulk, H.D.; Görner, H.; Roubeau, O.; Teat, S.J.; Weyhermüller, T.; Reedijk, J. Copper Curcuminoids Containing Anthracene Groups: Fluorescent Molecules with Cytotoxic Activity. Inorg. Chem. 2010, 49, 9655–9663. [Google Scholar] [CrossRef] [PubMed]
- Aliaga-Alcalde, N.; Rodríquez, L.; Febinteanu, M.; Höfer, P.; Weyhermüller, T. Crystal Structure, Fluorescence, and Nanostructuration Studies of the First ZnII Anthracene-Based Curcuminoid. Inorg. Chem. 2012, 51, 864–873. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Xue, X.; Jiang, B.; Tian, Y. Metal complexes of a novel bis-β-diketone-type ligand and its copper(II) complexes of two-photon biological imaging. Sci. China Chem. 2012, 55, 334–340. [Google Scholar] [CrossRef]
- Sarkar, T.; Butcher, R.; Banerjee, S.; Mukherjee, S.; Hussain, A. Visible light-induced cytotoxicity of a dinuclear iron(III) complex of curcumin with low-micromolar IC50 value in cancer cells Dedicated to Professor Animesh Chakravorty on the occasion of his 80th birthday. Inorg. Chim. Acta 2016, 439, 8–17. [Google Scholar] [CrossRef]
- Rajesh, J.; Gubendran, A.; Rajagopal, G.; Athappan, P. Synthesis, spectra and DNA interactions of certain mononuclear transition metal(II) complexes of macrocyclic tetra-aza diacetyl curcumin ligand. J. Mol. Struct. 2012, 1010, 169–178. [Google Scholar] [CrossRef]
- Pi, Z.; Wang, J.; Jiang, B.; Cheng, G.; Zhou, S. A curcumin-based TPA four-branched copper(II) complex probe for in vivo early tumor detection. Mater. Sci. Eng. C Mater. Biol. Appl. 2015, 46, 565–571. [Google Scholar] [CrossRef] [PubMed]
- Thomachan, S.; Sindhu, S.; John, V.D. Synthesis, Characterization, Antibacterial, Antifungal and Cytotoxic Activity of Curcuminoid Analogues with Trisubstituted Phenyl and Anthracenyl ring and their Zinc (II), Copper (II) and Vanadyl (IV) Chelates. Int. J. Pharm. Chem. 2016, 6, 78–86. [Google Scholar] [CrossRef]
- Pucci, D.; Bellini, T.; Crispini, A.; D’Agnano, I.; Liguori, P.; Garcia-Orduña, P.; Pirillo, S.; Valentini, A.; Zanchetta, G. DNA binding and cytotoxicity of fluorescent curcumin-based Zn(II) complexes. Med. Chem. Commun. 2012, 3, 462–468. [Google Scholar] [CrossRef]
- Zhang, W.; Chen, C.; Shi, H.; Yang, M.; Liu, Y.; Ji, P.; Chen, H.; Tan, R.X.; Li, E. Curcumin is a biologically active copper chelator with antitumor activity. Phytomedicine 2016, 23, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Sumanont, Y.; Murakami, Y.; Tohda, M.; Vajragupta, O.; Watanabe, H.; Matsumoto, K. Effects of Manganese Complexes of Curcumin and Diacetylcurcumin on Kainic Acid-Induced Neurotoxic Responses in the Rat Hippocampus. Biol. Pharm. Bull. 2007, 30, 1732–1739. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.Z.; Jiang, T.; Wang, L.; Yang, H.; Zhang, S.; Zhou, P.J. Interaction of curcumin with Zn(II) and Cu(II) ions based on experiment and theoretical calculation. J. Mol. Struct. 2010, 984, 316–325. [Google Scholar] [CrossRef]
- John, V.D.; Krishnankutty, K. Antitumour activity of synthetic curcuminoid analogs (1,7-diaryl-1,6-heptadiene-3,5-diones) and their copper complexes. Appl. Organomet. Chem. 2006, 20, 477–482. [Google Scholar] [CrossRef]
- Asti, M.; Ferrari, E.; Croci, S.; Atti, G.; Rubagotti, S.; Iori, M.; Cappon, P.C.; Zerbini, A.; Saladini, M.; Versari, A. Synthesis and Characterization of 68Ga-Labeled Curcumin and Curcuminoid Complexes as Potential Radiotracers for Imaging of Cancer and Alzheimer’s Disease. Inorg. Chem. 2014, 53, 4922–4933. [Google Scholar] [CrossRef] [PubMed]
- Attanasio, D.; Collamati, I.; Ercolani, C. Ligand arrangement in tetragonally CuO4N and CuO4N2 chromophores formed from copper(II) α-nitroketonates and sterically hindered N-bases. Dalton Trans. 1974, 2442–2448. [Google Scholar] [CrossRef]
- Garribba, E.; Micera, G. The Determination of the Geometry of Cu(II) Complexes an EPR Spectroscopy Experiment. J. Chem. Educ. 2006, 83, 1229–1232. [Google Scholar] [CrossRef]
- Rajagopal, G.; Prasanna, N.; Athappan, P. Copper(II) and Ruthenium(II)/(III) Schiff base complexes. Trans. Met. Chem. 1999, 24, 251–257. [Google Scholar] [CrossRef]
- Barik, A.; Mishra, B.; Shen, L.; Mohan, H.; Kadam, R.M.; Dutla, S.; Zhang, H.; Priyadarsini, K.L. Evaluation of a new copper(II) curcumin complex as superoxide dismutase mimic and its free radical reactions. Free Radic. Biol. Med. 2005, 39, 811–822. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Chakravarty, A.R. Metal complexes of curcumin for cellular imaging, targeting, and photoinduced anticancer activity. Acc. Chem. Res. 2012, 48, 2075–2083. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.R.; Decker, E.A. The role of oxygen in lipid oxidation reactions: A review. Annu. Rev. Food Sci. Technol. 2015, 6, 171–194. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Gu, Z.; Zhang, C.; Li, S.; Zhang, L.; Zhou, G.; Wang, S.; Zhang, J. Synthesis, characterization and ROS-mediated antitumor effects of palladium(II) complexes of curcuminoids. Eur. J. Med. Chem. 2018, 144, 662–671. [Google Scholar] [CrossRef] [PubMed]
- Wilson, J.J.; Lippard, S.J. Synthetic methods for the preparation of platinum anticancer complexes. Chem. Rev. 2014, 114, 4470–4495. [Google Scholar] [CrossRef] [PubMed]
- Armarego, W.L.F.; Perrin, D.D. Purification of Laboratory Chemicals, 6th ed.; Butterworth Heinemann: Oxford, UK, 2009; pp. 138–159. [Google Scholar]
- Bruker. APEX2, and SAINT-Plus; Bruker AXS Inc.: Madison, WI, USA, 2004. [Google Scholar]
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. Sec. A 2008, 64, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Domínguez, M.; Nieto, A.; Marín, J.C.; Keck, A.S.; Jeffery, E.; Céspedes, C.L. Antioxidant activities of extracts from Barkleyanthus salicifollius (Asteraceae) and Penstemon gentianoides (Scrophulariaceae). J. Agric. Food Chem. 2005, 53, 5889–5895. [Google Scholar] [CrossRef] [PubMed]
- Rossato, J.I.; Ketzer, L.A.; Centuriao, F.B.; Silva, S.J.; Lüdtke, D.S.; Zeni, G.; Braga, A.L.; Rubin, M.A.; Rocha, B.T. Antioxidant properties of new chalcogenides against lipid peroxidation in rat brain. Neurochem. Res. 2002, 27, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [PubMed]
- Ng, T.B.; Liu, F.; Wang, Z.T. Antioxidative activity of natural products from plants. Life Sci. 2000, 66, 709–723. [Google Scholar] [CrossRef]
- Ohkawa, H.; Ohishi, N.; Yagi, K. Assay for lipid peroxides in animal tissues by thiobarbituric acid reaction. Anal. Biochem. 1979, 95, 351–358. [Google Scholar] [CrossRef]
- Monks, A.; Scudiero, D.; Skehan, P.; Shoemaker, R.; Paul, K.; Vistica, D.; Hose, C.; Langley, J.; Cronise, P.; Vaigro-Wolff, A.; et al. Feasibility of a high-flux anticancer drug screen using a diverse panel of cultured human tumor cell lines. J. Natl. Cancer Inst. 1991, 38, 757–766. [Google Scholar] [CrossRef]
- Sumantran, V.N. Cellular chemosensitivity assays: An overview. In Cancer Cell Culture: Methods and Protocols, 2nd ed.; Cree, I.A., Ed.; Humana Press: New York, NY, USA, 2011; Chapter 19; pp. 219–236. [Google Scholar]
- Lozada, C.; Soria-Arteche, O.; Ramírez-Apan, M.T.; Nieto-Camacho, A.; Enríquez, R.G.; Izquierdo, T.; Jiménez-Corona, A. Synthesis, cytotoxic and antioxidant evaluations of amino derivatives. Bioorg. Med. Chem. 2012, 20, 5077–5084. [Google Scholar] [CrossRef] [PubMed]
- Akram Khan, M.; El-Khatib, R.; Rainsford, K.D.; Whitehouse, M.W. Synthesis and anti-inflammatory properties of some aromatic and heterocyclic curcuminoids. Bioorg. Chem. 2012, 40, 30–38. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |



| Compounds | Keto-enol (cm−1) | -CH=C- (cm−1) | C-H Stretch (cm−1) | C-H Bending (cm−1) | Vibrations M-O (cm−1) |
|---|---|---|---|---|---|
| DAC | 1755 and 1795 | 966 | - | - | - |
| DACH4 | 1757 and 1797 | - | 2966–2841 | - | - |
| DiMeOC | 1620 and 1663 | 964 | - | - | - |
| DiBcOC | 1625 and 1730 | 970 | - | 849–694 | - |
| PhCurcu | 1619 and 1670 | 968 | - | - | - |
| DAC-Cu | - | - | - | - | 1514 and 484 |
| DACH4-Cu | - | - | - | - | 1508 and 467 |
| DiMeOC-Cu | - | - | - | - | 1506 and 463 |
| DiBcOC-Cu | - | - | - | - | 1501 and 465 |
| PhCurcu-Cu | - | - | - | - | 1511 and 420 |
| Compounds | OH (δ) | Methine (δ) | α to the Diketone (δ) | β to the Diketone (δ) | Methoxyl (δ) | Acetyl (δ) | Benzyl (δ) | Aromatic (δ) |
|---|---|---|---|---|---|---|---|---|
| DAC | 16.12 | 6.20 | 6.99 | 7.66 | 3.85 | 2.28 | - | 7.16–7.52 |
| DACH4 | 15.53 | 5.78 | 2.77 | 2.85 | 3.74 | 2.23 | - | 5.78–7.00 |
| DiMeOC | 16.27 | 5.80 | 6.47 | 7.58 | 3.91–3.89 | - | - | 6.85–7.05 |
| DiBcOC | 16.30 | 6.11 | 6.48 | 7.59 | 3.84 | - | 5.14 | 7.34–7.59 |
| PhCurcu | 16.11 | 6.21 | 6.96 | 7.67 | - | - | - | 7.73 |
| DAC-Cu | - | - | - | - | - | 2.27 | 3.85 | - |
| DACH4-Cu | - | - | - | - | - | 4.01 | - | - |
| DiMeOC-Cu | - | - | - | - | - | 4.01 | - | - |
| DiBcOC-Cu | - | - | - | - | - | 3.86 | - | - |
| PhCurcu-Cu | - | - | 6.91 | 7.66 | - | - | - | - |
| Complexes | g‖ | g┴ | A‖ (10−4 cm−1) | A┴ (10−4 cm−1) | g‖/A‖ (cm−1) | µeffec |
|---|---|---|---|---|---|---|
| DAC-Cu | 2.29 | 2.06 | 162.3 | 11.3 | 141.1 | 1.69 |
| DACH4-Cu | 2.29 | 2.06 | 161.8 | 10.5 | 141.5 | 2.01 |
| DiMeOC-Cu | 2.30 | 2.07 | 160.8 | 9.5 | 143.0 | 1.99 |
| DiBcOC-Cu | 2.30 | 2.06 | 159.9 | 12.4 | 143.8 | 2.00 |
| PhCurcu-Cu | 2.30 | 2.07 | 157.3 | 18.11 | 146.3 | 2.09 |
| Products | Concentration (uM) | D. O. 540 nm | nmol/mg prot. | Inhibition (%) |
|---|---|---|---|---|
| Basal | - | 0.004 | 0.169 | - |
| FeSO4 | - | 0.851 | 11.379 | - |
| Curcu | 10 | 0.000 | 0.116 | 98.67 |
| 100 | 0.000 | 0.116 | 98.67 | |
| DAC | 10 | 0.030 | 0.516 | 94.66 |
| 100 | 0.012 | 0.275 | 97.16 | |
| DAC-Cu | 10 | 0.022 | 0.410 | 95.75 |
| 100 | 0.018 | 0.348 | 96.40 | |
| DACH4 | 10 | 0.313 | 4.257 | 55.95 |
| 100 | 0.015 | 0.308 | 96.81 | |
| DACH4-Cu | 10 | 0.020 | 0.377 | 96.16 |
| 100 | 0.024 | 0.427 | 95.65 | |
| DiMeOC | 10 | 0.733 | 9.808 | 1.47 |
| 100 | 0.169 | 2.355 | 75.63 | |
| DiMeOC-Cu | 10 | 0.321 | 4.360 | 55.61 |
| 100 | 0.225 | 3.086 | 68.57 | |
| DiBncOC | 10 | 0.785 | 10.496 | 7.76 |
| 100 | 0.612 | 8.217 | 27.79 | |
| DiBncOC-Cu | 10 | 0.795 | 10.628 | 6.60 |
| 100 | 0.769 | 10.287 | 9.59 | |
| PhCurcu | 10 | 0.712 | 9.530 | 8.22 |
| 100 | 0.208 | 2.865 | 72.41 | |
| PhCurcu-Cu | 10 | 0.474 | 6.388 | 38.48 |
| 100 | 0.217 | 2.990 | 71.20 |
| Products | Concentration (μM) | TBARS (nmol/mg prot.) | Inhibition (%) | IC50 (μM) |
|---|---|---|---|---|
| Basal | - | 0.24 ± 0.09 | - | - |
| FeSO4 100 µM | - | 9.15 ± 0.32 | - | - |
| α-Tocopherol (n = 4) | 0.1 0.32 1 3.16 10 31.62 100 | 6.28 ± 0.18 6.04 ± 0.24 5.21 ± 0.33 * 3.67 ± 0.56 ** 2.72 ± 0.33 ** 1.84 ± 0.31 ** 1.40 ± 0.36 ** | 4.62 ± 0.57 8.26 ± 1.31 21.13 ± 2.56 * 44.84 ± 6.74 ** 59.00 ± 3.71 ** 72.3 ± 3.87 ** 79.09 ± 4.79 ** | 6.78 ± 2.16 |
| DAC | 0.1 0.32 1 3.16 10 | 9.68 ± 0.43 9.29 ± 0.67 7.78 ± 0.53 * 5.22 ± 0.35 ** 1.43 ± 0.08 ** | 4.16 ± 1.97 8.32 ± 1.49 23.19 ± 0.92 * 48.46 ± 1.53 ** 85.82 ± 0.60 ** | 3.21 ± 0.16 |
| DAC-Cu | 0.1 0.32 1 3.16 10 | 9.08 ± 0.66 8.04 ± 0.70 6.24 ± 0.71 ** 3.08 ± 0.21 ** 1.32 ± 0.48 ** | 10.34 ± 1.48 20.78 ± 2.44 38.70 ± 3.82 ** 69.57 ± 1.56 ** 86.58 ± 5.21 ** | 1.55 ± 0.15 |
| DACH4 | 1.78 3.16 5.62 10 17.78 31.62 56.23 | 8.32 ± 0.31 7.85 ± 0.41 * 7.25 ± 0.36 ** 6.32 ± 0.18 ** 4.45 ± 0.22 ** 0.36 ± 0.06 ** 0.14 ± 0.01 ** | 9.05 ± 1.03 14.28 ± 1.59 * 20.86 ± 1.48 ** 30.84 ± 0.57 ** 51.41 ± 0.92 ** 96.02 ± 0.57 ** 98.39 ± 0.18 ** | 16.46 ± 0.30 |
| DACH4-Cu | 1.78 3.16 5.62 10 17.78 31.62 56.23 | 7.32 ± 0.30 ** 6.67 ± 0.34 ** 5.89 ± 0.33 ** 3.28 ± 0.37 ** 0.28 ± 0.05 ** 0.20 ± 0.02 ** 0.14 ± 0.007 ** | 19.93 ± 2.01 ** 27.14 ± 1.30 ** 35.74 ± 1.88 ** 64.27 ± 2.74 ** 96.90 ± 0.63 ** 97.73 ± 0.29 ** 98.39 ± 0.03 ** | 7.93 ± 0.41 |
| DiMeOC | 17.78 31.62 56.23 100 177.83 | 5.51± 0.13 ** 3.40 ± 0.007 ** 2.67 ± 0.06 ** 2.27 ± 0.09 ** 2.08 ± 0.10 ** | 38.66 ± 3.26 ** 62.16 ± 1.18 ** 70.33 ± 0.63 ** 74.75 ± 0.92 ** 76.91 ± 0.88 ** | 23.01 ± 1.37 |
| DiMeOC-Cu | 1 3.16 10 31.62 100 316.23 | 8.58 ± 0.28 7.42 ± 0.20 ** 4.28 ± 0.06 ** 3.10 ± 0.12 ** 2.89 ± 0.16 ** 2.8 0± 0.23 ** | 4.55 ± 4.35 17.51 ± 2.63 ** 52.41 ± 1.20 ** 65.56 ± 1.49 ** 67.90 ± 1.17 ** 68.97 ± 1.99 ** | 9.35 ± 0.34 |
| % of Inhibition | ||||||
|---|---|---|---|---|---|---|
| Products (25 μM) | U251 | PC-3 | K562 | HCT-15 | MCF-7 | SKLU-1 |
| DAC | 46.2 | 77.54 | 67.8 | 46.88 | 41.74 | 41.74 |
| DAC-Cu | 8.6 | 13.19 | 19.7 | 4.16 | NC | NC |
| DACH4 | 22.8 | 26.08 | 39.4 | 31.12 | 7.16 | 7.16 |
| DACH4-Cu | NC | 31.5 | 15.4 | 3.9 | 3.0 | 11.30 |
| DiMeOC | 92.6 | 100 | 89.4 | 96.95 | 100 | 100 |
| DIMeOC-Cu | 31.4 | 100 | 75.8 | 74.5 | 48.4 | 48.02 |
| DiBncOC | 3.0 | 10.7 | 10.9 | NC | 1.7 | 6.4 |
| DiBncOC-Cu | 6.6 | 4.1 | NC | NC | NC | 7.5 |
| PhCurcu | 47.5 | 31.2 | 54.7 | 25,0 | 48.7 | 53.0 |
| PhCurcu-Cu | 3.5 | NC | 4.2 | NC | NC | NC |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meza-Morales, W.; Machado-Rodriguez, J.C.; Alvarez-Ricardo, Y.; Obregón-Mendoza, M.A.; Nieto-Camacho, A.; Toscano, R.A.; Soriano-García, M.; Cassani, J.; Enríquez, R.G. A New Family of Homoleptic Copper Complexes of Curcuminoids: Synthesis, Characterization and Biological Properties. Molecules 2019, 24, 910. https://doi.org/10.3390/molecules24050910
Meza-Morales W, Machado-Rodriguez JC, Alvarez-Ricardo Y, Obregón-Mendoza MA, Nieto-Camacho A, Toscano RA, Soriano-García M, Cassani J, Enríquez RG. A New Family of Homoleptic Copper Complexes of Curcuminoids: Synthesis, Characterization and Biological Properties. Molecules. 2019; 24(5):910. https://doi.org/10.3390/molecules24050910
Chicago/Turabian StyleMeza-Morales, William, Juan C. Machado-Rodriguez, Yair Alvarez-Ricardo, Marco A. Obregón-Mendoza, Antonio Nieto-Camacho, Rubén. A. Toscano, Manuel Soriano-García, Julia Cassani, and Raúl G. Enríquez. 2019. "A New Family of Homoleptic Copper Complexes of Curcuminoids: Synthesis, Characterization and Biological Properties" Molecules 24, no. 5: 910. https://doi.org/10.3390/molecules24050910
APA StyleMeza-Morales, W., Machado-Rodriguez, J. C., Alvarez-Ricardo, Y., Obregón-Mendoza, M. A., Nieto-Camacho, A., Toscano, R. A., Soriano-García, M., Cassani, J., & Enríquez, R. G. (2019). A New Family of Homoleptic Copper Complexes of Curcuminoids: Synthesis, Characterization and Biological Properties. Molecules, 24(5), 910. https://doi.org/10.3390/molecules24050910