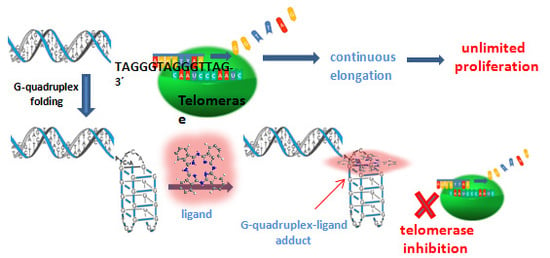
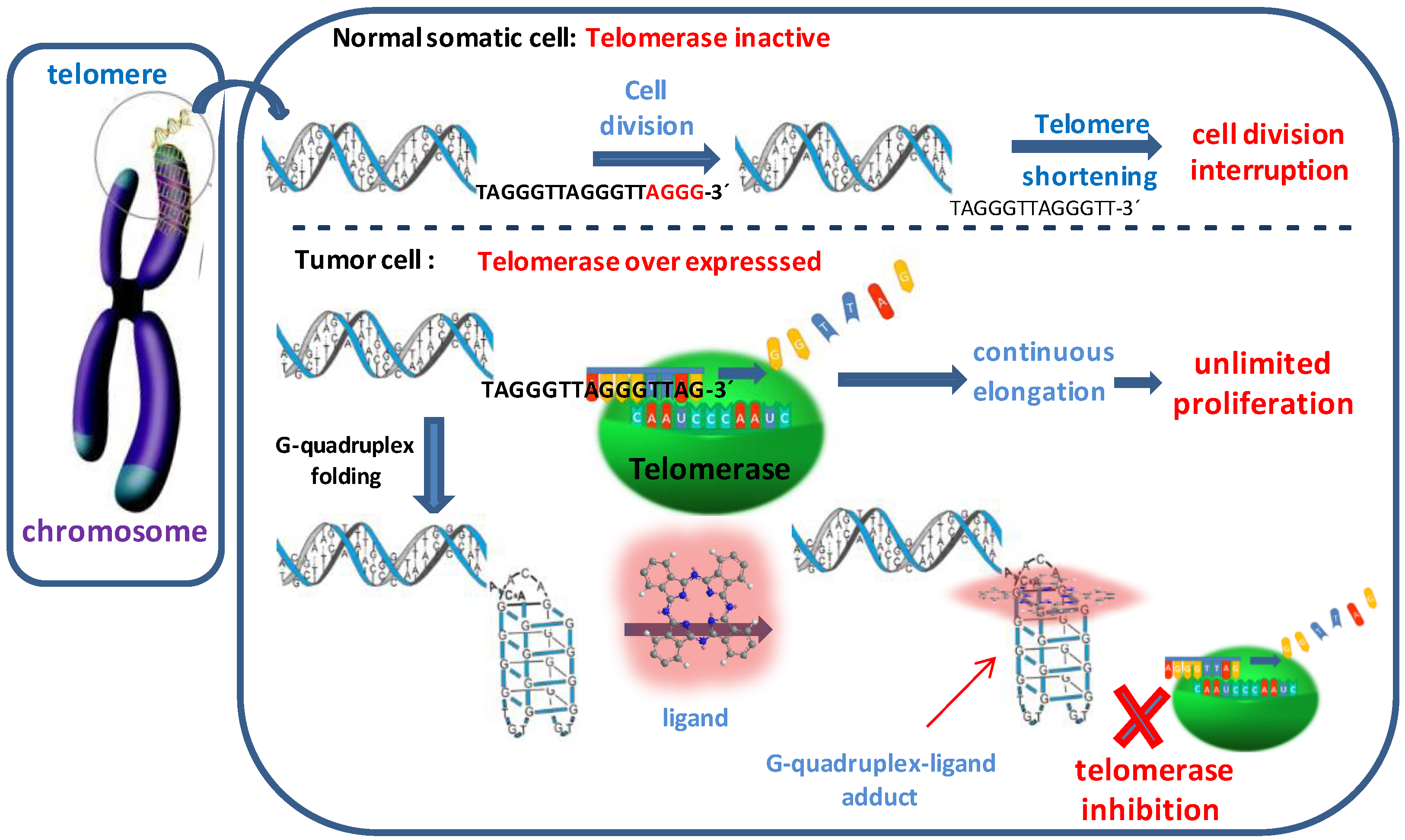
Multicharged Phthalocyanines as Selective Ligands for G-Quadruplex DNA Structures
Abstract
1. Introduction
2. Results and Discussion
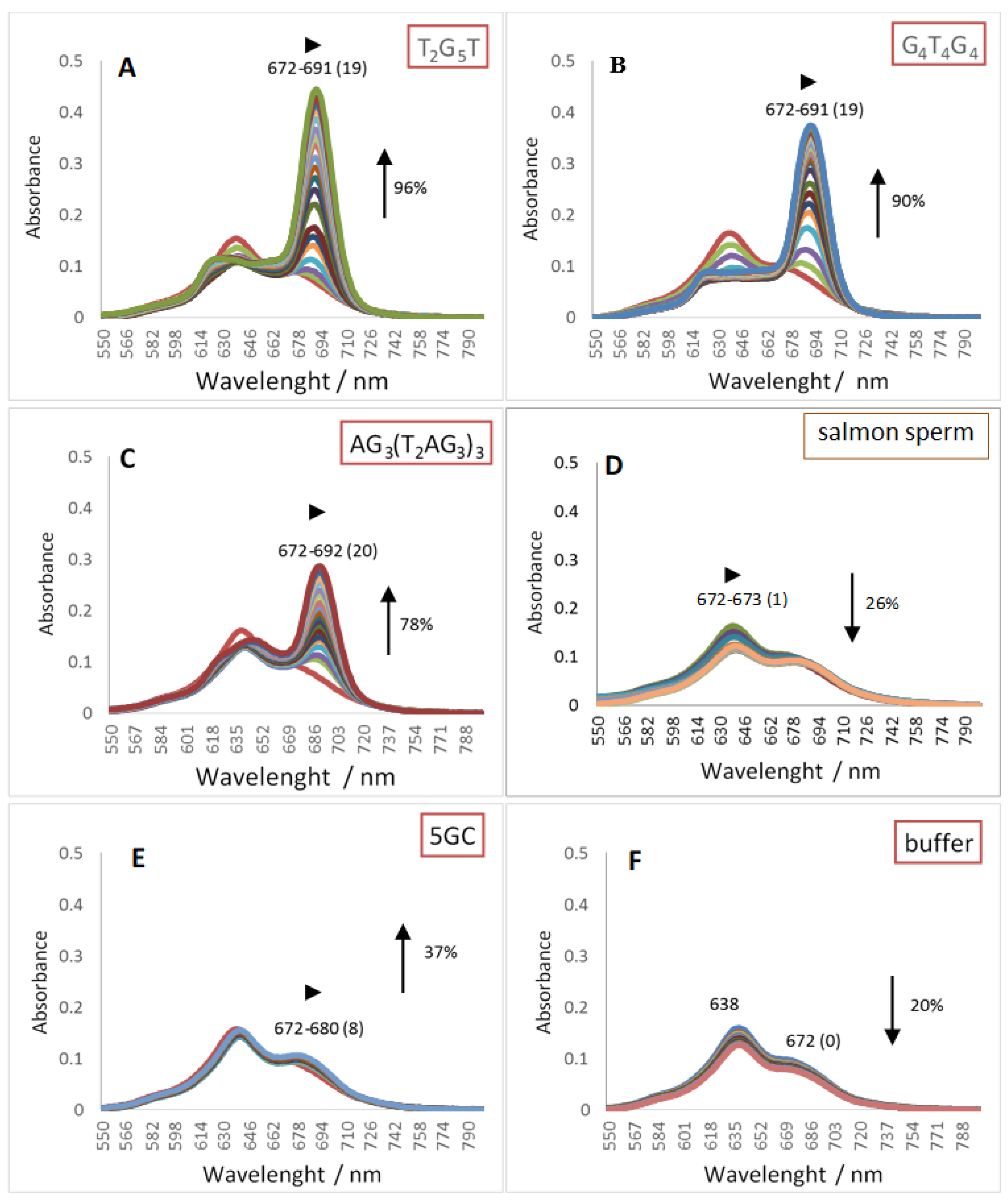
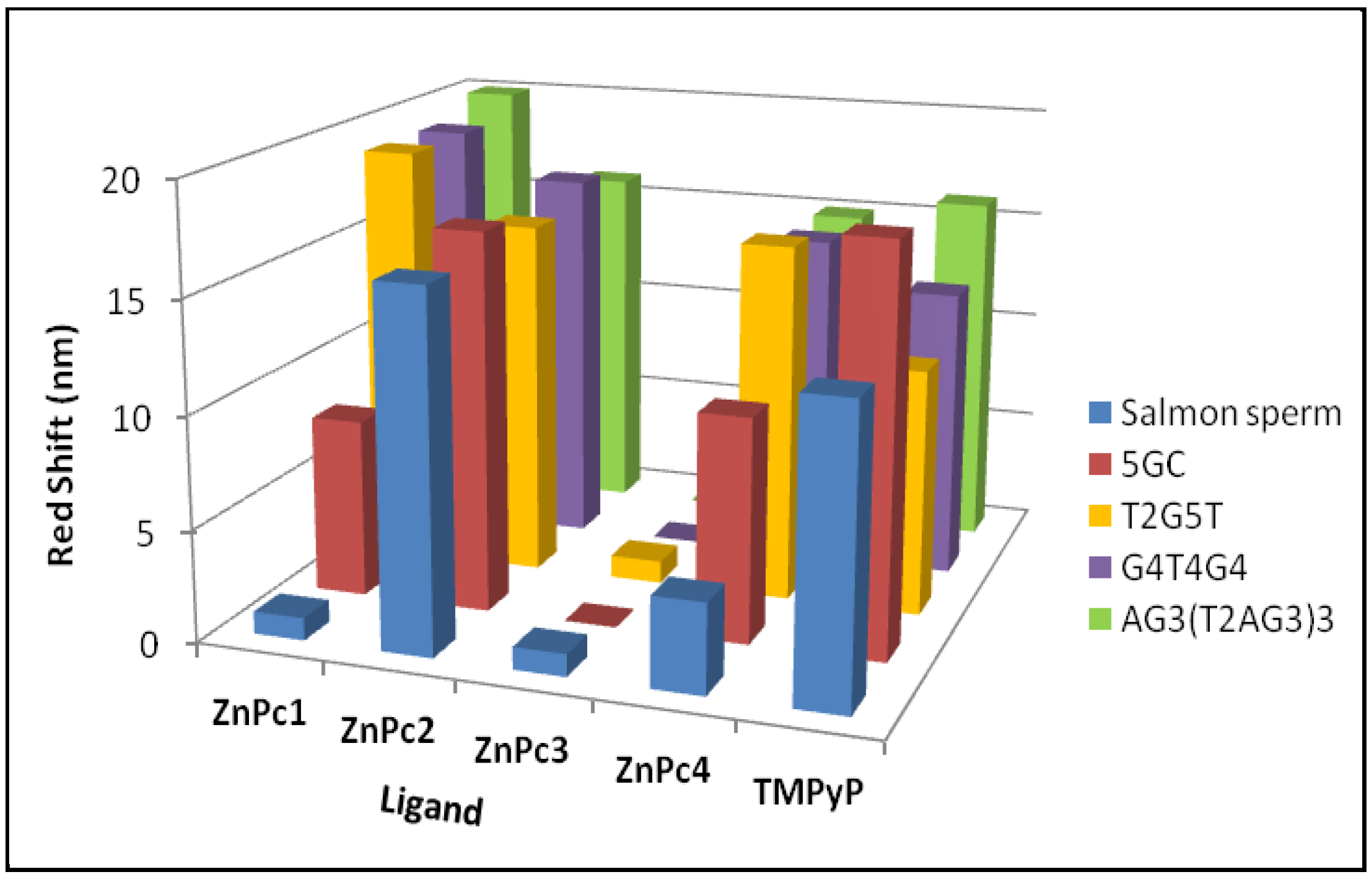
2.1. UV-Vis Spectroscopy
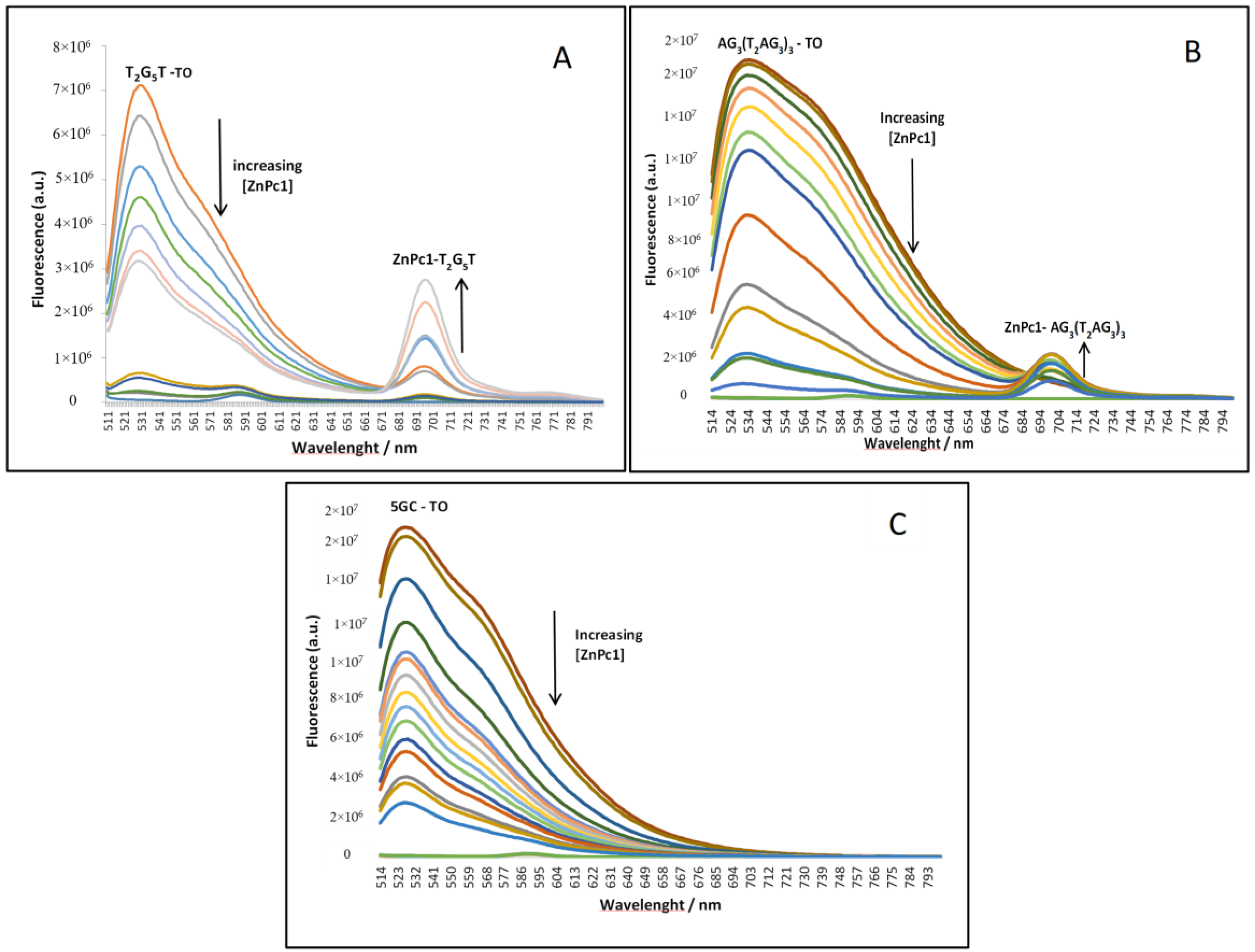
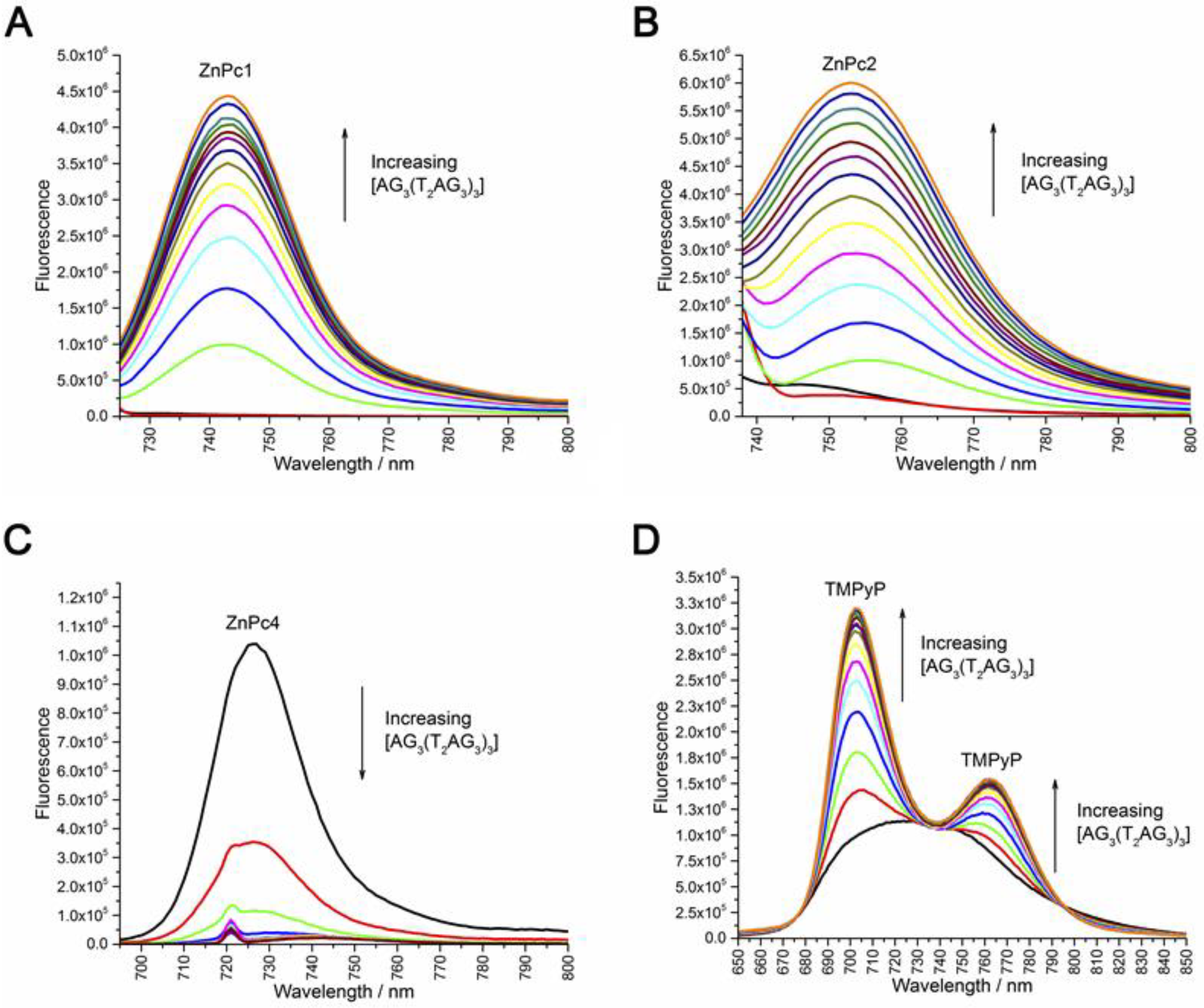
2.2. Fluorescence Experiments
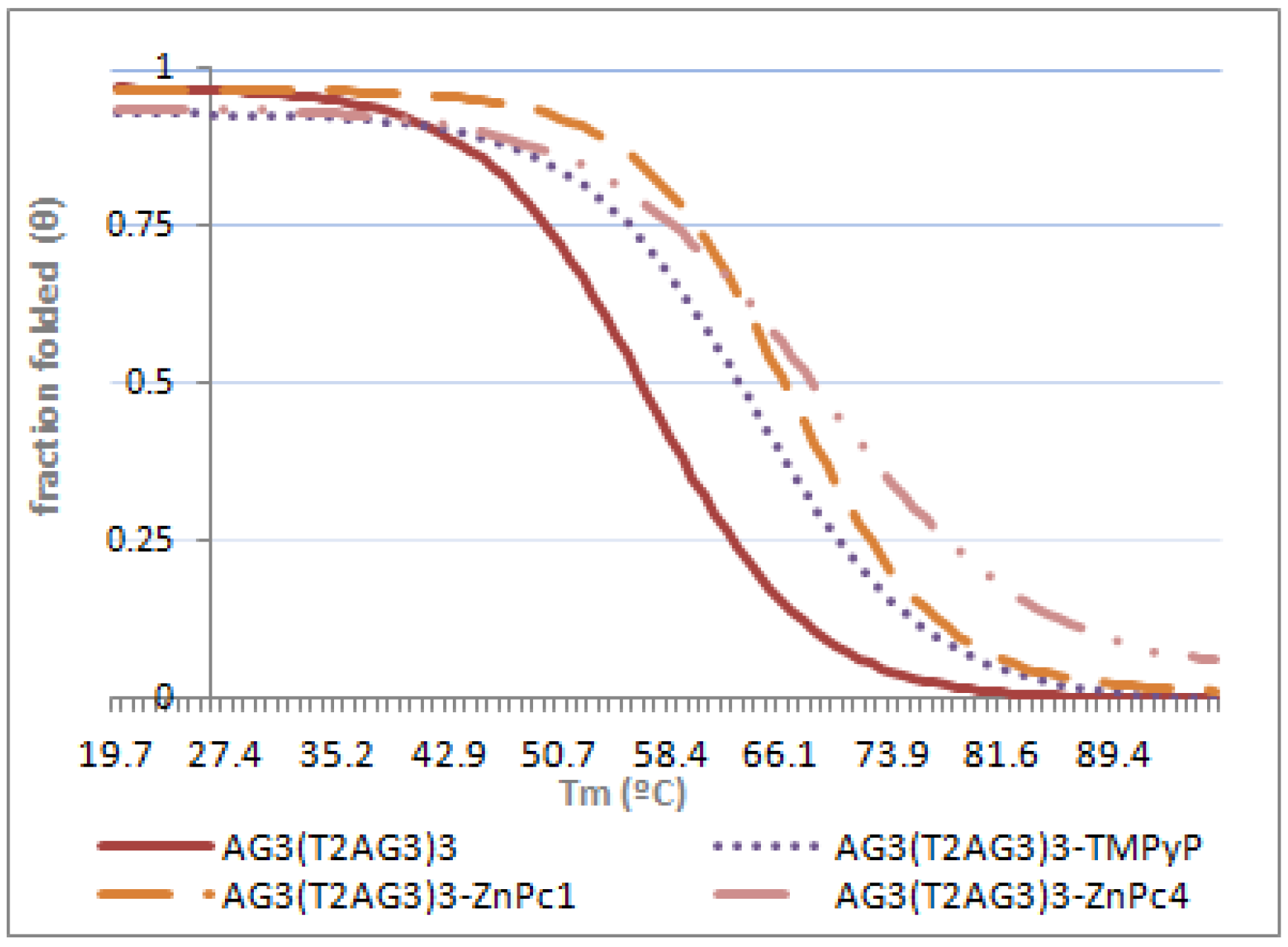
2.3. Circular Dichroism (CD)
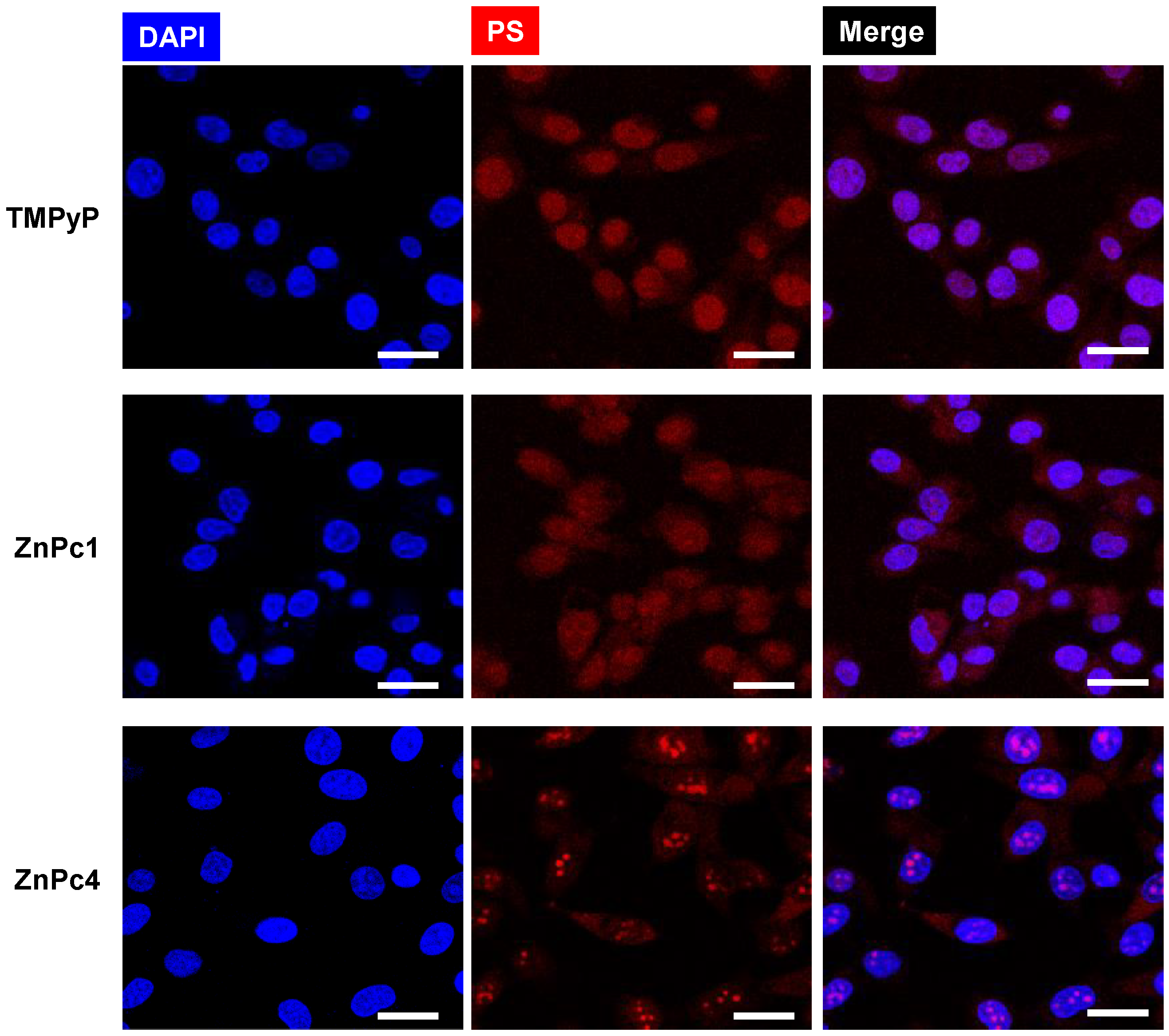
2.4. Phtalocyanine Nuclear Uptake and Toxicity in Bladder Cancer Cells
3. Materials and Methods
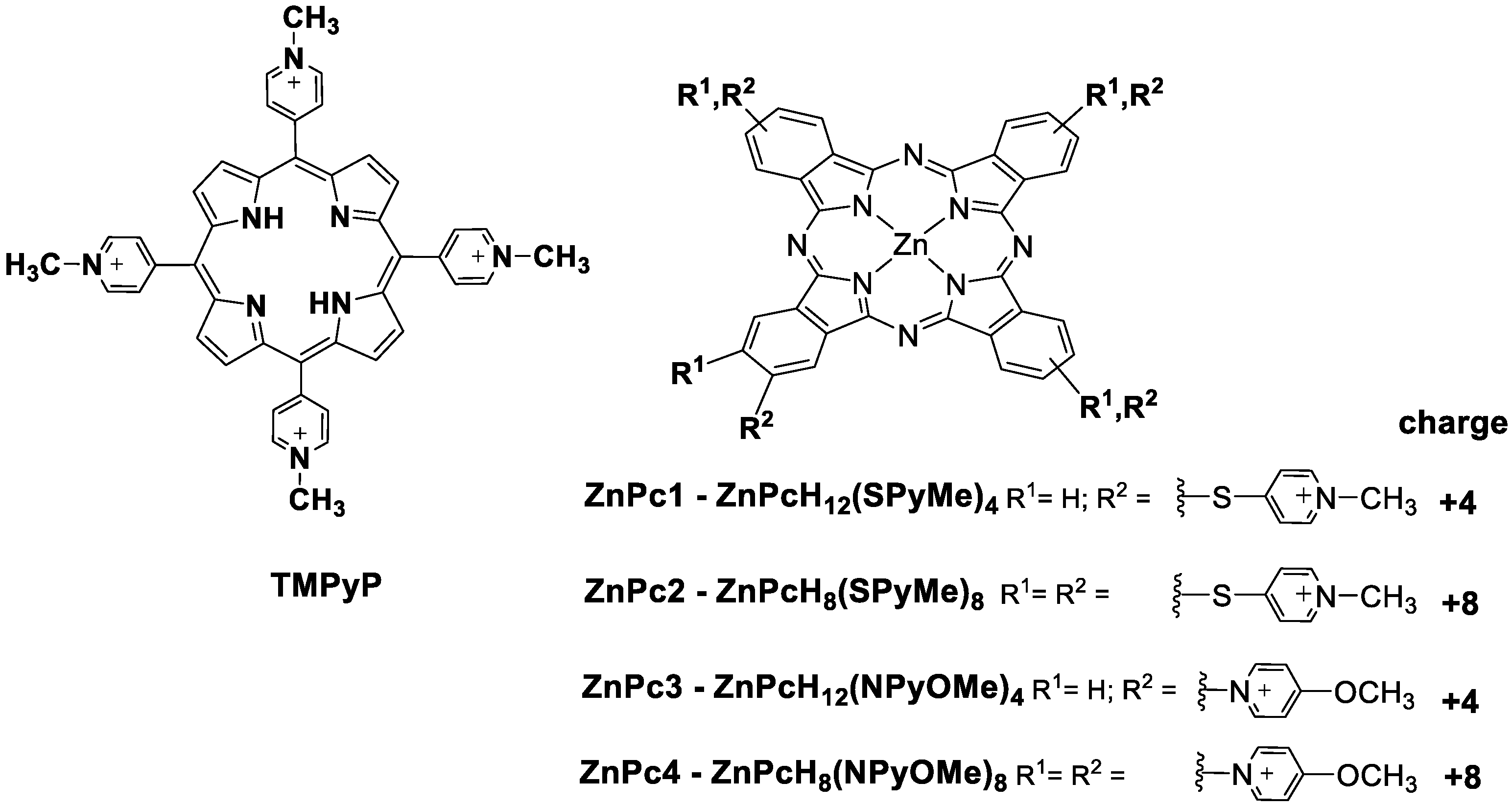
3.1. Chemicals
3.2. Preparation of DNA Structures (Double Chain and G-Quadruplexes)
3.3. Methods
3.3.1. UV-Vis Spectroscopy
3.3.2. Fluorescence Spectroscopy
3.3.3. Circular Dichroism
3.3.4. MTT Protocol
Calculations
3.3.5. Fluorescence Microscopy
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| T | thymine |
| A | adenine |
| G | guanine |
| ∆λ | wavelength deviation |
| DNA | deoxyribonucleic acid |
| GQ | G-Quadruplexes |
| Pcs | phthalocyanines |
| TO | thiazole orange |
| PBS | phosphate buffer solution |
| DC50 | concentration of ligands required to decrease the fluorescence of the probe by 50% |
| IC50 | concentration of the ligand required to reduce the cell viability by 50% |
| T2AG3 | human telomeric sequence repeat—5′-TTA GGG-3′ |
| T2G5T | tetramolecular G-quadruplex sequence—5′-TTG GGG T-3′ |
| (G4T4G4)2 | bimolecular G-quadruplex sequence—5′-GGG GTT TTG GGG-3′ |
| AG3(T2AG3)3 | unimolecular G-Quadruplex—5′-AGG GTT AGG GTTAGG GTT AGGG-3′ |
| 5GC | double strand DNA—5′-GCG CGC GCG C-3′ |
| UV-Vis | UV-Visible spectroscopy |
| G4-FID | G-Quadruplex fluorescent intercalator displacement assay |
| CD | circular dichroism |
References
- Cech, T.R. Begining to understand the end of the chromosome. Cell 2004, 116, 273–279. [Google Scholar] [CrossRef]
- Shay, J.W.; Shay, J.W.; Zou, Y.; Zou, Y.; Hiyama, E.; Hiyama, E.; Wright, W.E.; Wright, W.E. Telomerase and cancer. Hum. Mol. Genet. 2001, 10, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.W.; Piatyszek, M.A.; Prowse, K.R.; Harley, C.B.; West, M.D.; Ho, P.L.C.; Coviello, G.M.; Wright, W.E.; Weinrich, S.L.; Shay, J.W. Specific Association of Human Telomerase Activity with Immortal Cells and Cancer. Science 1994, 266, 2011–2016. [Google Scholar] [CrossRef] [PubMed]
- Schrank, Z.; Khan, N.; Osude, C.; Singh, S.; Miller, R.J.; Merrick, C.; Mabel, A.; Kuckovic, A.; Puri, N. Oligonucleotides targeting telomeres and telomerase in cancer. Molecules 2018, 9, 2267. [Google Scholar] [CrossRef] [PubMed]
- Nandakumar, J.; Cech, T.R. Finding the end: Recruitment of telomerase to telomeres. Nat. Rev. Mol. Cell Biol. 2013, 14, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Zahler, A.M.; Williamson, J.R.; Cech, T.R.; Prescott, D.M. Inhibition of telomerase by G-quartet DNA structures. Nature 1991, 350, 718–720. [Google Scholar] [CrossRef]
- Mergny, J.L.; Mailliet, P.; Lavelle, F.; Riou, J.F.; Laoui, A.; Hélène, C. The development of telomerase inhibitors: The G-quartet approach. Anticancer Drug Des. 1999, 14, 327–339. [Google Scholar] [PubMed]
- Hurley, L.H.; Wheelhouse, R.T.; Sun, D.; Kerwin, S.M.; Salazar, M.; Fedoroff, O.Y.; Han, F.X.; Han, H.; Izbicka, E.; Von Hoff, D.D. G-quadruplexes as targets for drug design. Pharmacol. Ther. 2000, 85, 141–158. [Google Scholar] [CrossRef]
- Oganesian, L.; Bryan, T.M. Physiological relevance of telomeric G-quadruplex formation: A potential drug target. BioEssay 2007, 29, 155–165. [Google Scholar] [CrossRef]
- Han, H.; Hurley, L.H. G-quadruplex DNA: A potential target for anti-cancer drug design. Trends Pharmacol. Sci. 2000, 21, 136–142. [Google Scholar] [CrossRef]
- Moye, A.L.; Porter, K.C.; Cohen, S.B.; Phan, T.; Zyner, K.G.; Sasaki, N.; Lovrecz, G.O.; Beck, J.L.; Bryan, T.M. Telomeric G-quadruplexes are a substrate and site of localization for human telomerase. Nat. Commun. 2015, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, J.Q.; Chen, Z.; Zheng, K.W.; Chen, C.Y.; Hao, Y.H.; Tan, Z. G-quadruplex formation at the 3′ end of telomere DNA inhibits its extension by telomerase, polymerase and unwinding by helicase. Nucleic Acids Res. 2011, 39, 6229–6237. [Google Scholar] [CrossRef] [PubMed]
- Tanpure, A.A.; Srivatsan, S.G. Conformation-sensitive nucleoside analogues as topology-specific fluorescence turn-on probes for DNA and RNA G-quadruplexes. Nucleic Acids Res. 2015, 43, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Yaku, H.; Murashima, T.; Miyoshi, D.; Sugimoto, N. Specific binding of anionic porphyrin and phthalocyanine to the G-quadruplex with a variety of in Vitro and in Vivo applications. Molecules 2012, 17, 10586–10613. [Google Scholar] [CrossRef] [PubMed]
- Bidzinska, J.; Cimino-Reale, G.; Zaffaroni, N.; Folini, M. G-quadruplex structures in the human genome as novel therapeutic targets. Molecules 2013, 18, 12368–12395. [Google Scholar] [CrossRef]
- Artese, A.; Costa, G.; Ortuso, F.; Parrotta, L.; Alcaro, S. Identification of new natural DNA G-quadruplex binders selected by a structure-based virtual screening approach. Molecules 2013, 18, 12051–12070. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.M.; Fujii, S.; Sato, S.; Okauchi, T.; Takenaka, S. A selective G-quadruplex DNA-stabilizing ligand based on a cyclic naphthalene diimide derivative. Molecules 2015, 20, 10963. [Google Scholar] [CrossRef]
- Murat, P.; Singh, Y.; Defrancq, E. Methods for investigating G-quadruplex DNA/ligand interactions. Chem. Soc. Rev. 2011, 40, 5293–5307. [Google Scholar] [CrossRef]
- Jaumot, J.; Gargallo, R. Experimental Methods for Studying the Interactions between G-Quadruplex Structures and Ligands. Curr. Pharm. Des. 2012, 18, 1900–1916. [Google Scholar] [CrossRef] [PubMed]
- Dupont, J.I.; Henderson, K.L.; Metz, A.; Le, V.H.; Emerson, J.P.; Lewis, E.A. Calorimetric and spectroscopic investigations of the binding of metallated porphyrins to G-quadruplex DNA. Biochim. Biophys. Acta Gen. Subj. 2016, 1860, 902–909. [Google Scholar] [CrossRef]
- Largy, E.; Hamon, F.; Teulade-Fichou, M.P. Development of a high-throughput G4-FID assay for screening and evaluation of small molecules binding quadruplex nucleic acid structures. Anal. Bioanal. Chem. 2011, 400, 3419–3427. [Google Scholar] [CrossRef] [PubMed]
- Pagano, B.; Cosconati, S.; Gabelica, V.; Petraccone, L.; De Tito, S.; Marinelli, L.; La Pietra, V.; Saverio di Leva, F.; Lauri, I.; Trotta, R.; et al. State-of-the-Art Methodologies for the Discovery and Characterization of DNA G-Quadruplex Binders. Curr. Pharm. Des. 2012, 18, 1880–1899. [Google Scholar] [CrossRef] [PubMed]
- Izbicka, E.; Wheelhouse, R.T.; Raymond, E.; Davidson, K.K.; Lawrence, R.A.; Sun, D.; Windle, B.E.; Hurley, L.H.; Von Hoff, D.D. Effects of cationic porphyrins as G-quadruplex interactive agents in human tumor cells. Cancer Res. 1999, 59, 639–644. [Google Scholar] [PubMed]
- Shi, D.F.; Wheelhouse, R.T.; Sun, D.; Hurley, L.H. Quadruplex-interactive agents as telomerase inhibitors: Synthesis of porphyrins and structure-activity relationship for the inhibition of telomerase. J. Med. Chem. 2001, 44, 4509–4523. [Google Scholar] [CrossRef] [PubMed]
- Wheelhouse, R.T.; Sun, D.; Han, H.; Han, F.X.; Hurley, L.H. Cationic Porphyrins as Telomerase Inhibitors: The Interaction of Tetra-(N-methyl-4-pyridyl)porphine with Quadruplex DNA. J. Am. Chem. Soc. 1998, 120, 3261–3262. [Google Scholar] [CrossRef]
- Boschi, E.; Davis, S.; Taylor, S.; Butterworth, A.; Chirayath, L.A.; Purohit, V.; Siegel, L.K.; Buenaventura, J.; Sheriff, A.H.; Jin, R.; et al. Interaction of a cationic porphyrin and its metal derivatives with G-quadruplex DNA. J. Phys. Chem. B 2016, 120, 12807–12819. [Google Scholar] [CrossRef] [PubMed]
- Romera, C.; Bombarde, O.; Bonnet, R.; Gomez, D.; Dumy, P.; Calsou, P.; Gwan, J.F.; Lin, J.H.; Defrancq, E.; Pratviel, G. Improvement of porphyrins for G-quadruplex DNA targeting. Biochimie 2011, 93, 1310–1317. [Google Scholar] [CrossRef]
- Haq, I.; Trent, J.O.; Chowdhry, B.Z.; Jenkins, T.C. Intercalative G-tetraplex stabilization of telomeric DNA by a cationic porphyrin. J. Am. Chem. Soc. 1999, 121, 1768–1779. [Google Scholar] [CrossRef]
- Zhu, L.N.; Wu, B.; Kong, D.M. Specific recognition and stabilization of monomeric and multimeric G-quadruplexes by cationic porphyrin TMPipEOPP under molecular crowding conditions. Nucleic Acids Res. 2013, 41, 4324–4335. [Google Scholar] [CrossRef]
- Ramos, C.I.V.; Tomé, J.P.C.; Santana-Marques, M.G. Charge and substituent effects on the stability of porphyrin/G-quadruplex adducts. J. Mass Spectrom. 2012, 47, 173–179. [Google Scholar] [CrossRef]
- Hou, J.Q.; Chen, S. Bin; Zan, L.P.; Ou, T.M.; Tan, J.H.; Luyt, L.G.; Huang, Z.S. Identification of a selective G-quadruplex DNA binder using a multistep virtual screening approach. Chem. Commun. 2015, 51, 198–201. [Google Scholar] [CrossRef] [PubMed]
- Yaku, H.; Fujimoto, T.; Murashima, T.; Miyoshi, D.; Sugimoto, N. Phthalocyanines: A new class of G-quadruplex-ligands with many potential applications. Chem. Commun. 2012, 48, 6203–6216. [Google Scholar] [CrossRef] [PubMed]
- Alzeer, J.; Luedtke, N.W. PH-mediated fluorescence and G-quadruplex binding of amido phthalocyanines. Biochemistry 2010, 49, 4339–4348. [Google Scholar] [CrossRef] [PubMed]
- Kawauchi, K.; Sugimoto, W.; Yasui, T.; Murata, K.; Itoh, K.; Takagi, K.; Tsuruoka, T.; Akamatsu, K.; Tateishi-Karimata, H.; Sugimoto, N.; et al. An anionic phthalocyanine decreases NRAS expression by breaking down its RNA G-quadruplex. Nat. Commun. 2018, 9, E2271. [Google Scholar] [CrossRef] [PubMed]
- Rowland, G.B.; Barnett, K.; Dupont, J.I.; Akurathi, G.; Le, V.H.; Lewis, E.A. The effect of pyridyl substituents on the thermodynamics of porphyrin binding to G-quadruplex DNA. Bioorganic Med. Chem. 2013, 21, 7515–7522. [Google Scholar] [CrossRef] [PubMed]
- Franceschin, M.; Lombardo, C.M.; Pascucci, E.; D’Ambrosio, D.; Micheli, E.; Bianco, A.; Ortaggi, G.; Savino, M. The number and distances of positive charges of polyamine side chains in a series of perylene diimides significantly influence their ability to induce G-quadruplex structures and inhibit human telomerase. Bioorganic Med. Chem. 2008, 16, 2292–2304. [Google Scholar] [CrossRef] [PubMed]
- Chen, S. Bin; Shi, Q.X.; Peng, D.; Huang, S.Y.; Ou, T.M.; Li, D.; Tan, J.H.; Gu, L.Q.; Huang, Z.S. The role of positive charges on G-quadruplex binding small molecules: Learning from bisaryldiketene derivatives. Biochim. Biophys. Acta Gen. Subj. 2013, 1830, 5006–5013. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, Y.C.; Kong, D.M.; Guo, D.S. Tetraphenylethene Derivatives with Different Numbers of Positively Charged Side Arms have Different Multimeric G-Quadruplex Recognition Specificity. Chem. A Eur. J. 2015, 21, 13253–13260. [Google Scholar] [CrossRef]
- Ramos, C.I.V.; Barros, C.M.; Fernandes, A.M.; Santana-Marques, M.G.; Correia, A.J.F.; Tomé, J.P.C.; Carrilho, M.D.C.T.; Faustino, M.A.F.; Tomé, A.C.; Neves, M.G.P.M.S.; et al. Interactions of cationic porphyrins with double-stranded oligodeoxynucleotides: A study by electrospray ionisation mass spectrometry. J. Mass Spectrom. 2005, 40, 1439–1447. [Google Scholar] [CrossRef]
- Lourenço, L.M.O.; Iglesias, B.A.; Pereira, P.M.R.; Girão, H.; Fernandes, R.; Neves, M.G.P.M.S.; Cavaleiro, J.A.S.; Tomé, J.P.C. Synthesis, characterization and biomolecule-binding properties of novel tetra-platinum(ii)-thiopyridylporphyrins. Dalt. Trans. 2015, 44, 530–538. [Google Scholar] [CrossRef]
- Marciel, L.; Teles, L.; Moreira, B.; Pacheco, M.; Lourenço, L.M.O.; Neves, M.G.P.M.S.; Tomé, J.P.C.; Faustino, M.A.F.; Almeida, A. An effective and potentially safe blood disinfection protocol using tetrapyrrolic photosensitizers. Future Med. Chem. 2017, 9, 365–379. [Google Scholar] [CrossRef] [PubMed]
- Ramos, C.I.V.; Santana-Marques, M.G. Electrospray mass spectrometry for the study of the non-covalent interactions of porphyrins and duplex desoxyribonucleotides. J. Porphyr. Phthalocyanines 2009, 13, 518–523. [Google Scholar] [CrossRef]
- Mesquita, M.Q.; Menezes, J.C.J.M.D.S.; Neves, M.G.P.M.S.; Tomé, A.C.; Cavaleiro, J.A.S.; Cunha, Â.; Almeida, A.; Hackbarth, S.; Röder, B.; Faustino, M.A.F. Bioorganic & Medicinal Chemistry Letters Photodynamic inactivation of bioluminescent Escherichia coli by neutral and cationic pyrrolidine-fused chlorins and isobacteriochlorins. Bioorg. Med. Chem. Lett. 2014, 24, 808–812. [Google Scholar] [PubMed]
- Silva, E.M.P.; Ramos, C.I.V.; Pereira, P.M.R.; Giuntini, F.; Faustino, M.A.F.; Tomé, J.P.C.; Tomé, A.C.; Silva, A.M.S.; Santana-Marques, M.G.; Neves, M.G.P.M.S.; et al. Cationic β-vinyl substituted meso -tetraphenylporphyrins: Synthesis and non-covalent interactions with a short poly(dGdC) duplex. J. Porphyr. Phthalocyanines 2012, 16, 101–113. [Google Scholar] [CrossRef]
- Lourenço, L.M.O.; Sousa, A.; Gomes, M.C.; Faustino, M.A.F.; Almeida, A.; Silva, A.M.S.; Neves, M.G.P.M.S.; Cavaleiro, J.A.S.; Cunha, Â.; Tomé, J.P.C. Inverted methoxypyridinium phthalocyanines for PDI of pathogenic bacteria. Photochem. Photobiol. Sci. 2015, 14, 1853–1863. [Google Scholar] [CrossRef]
- Pereira, J.B.; Carvalho, E.F.A.; Faustino, M.A.F.; Fernandes, R.; Neves, M.G.P.M.S.; Cavaleiro, J.A.S.; Gomes, N.C.M.; Cunha, Â.; Almeida, A.; Tomé, J.P.C. Phthalocyanine thio-pyridinium derivatives as antibacterial photosensitizers. Photochem. Photobiol. 2012, 88, 537–547. [Google Scholar] [CrossRef]
- Monteiro, A.R.; Ramos, C.I.V.; Fateixa, S.; Moura, N.M.M.; Neves, M.G.P.M.S.; Trindade, T. Hybrids Based on Graphene Oxide and Porphyrin as Tools for Detection and Stabilization of DNA G-Quadruplexes. ACS Omega 2018, 3, 11184–11191. [Google Scholar] [CrossRef]
- Mesquita, M.Q.; Dias, C.J.; Neves, M.G.P.M.S.; Almeida, A.; Faustino, M.A.F. Revisiting current photoactive materials for antimicrobial photodynamic therapy. Molecules 2018, 23, 2424. [Google Scholar] [CrossRef]
- Lourenço, L.M.O.; Pereira, P.M.R.; MacIel, E.; Válega, M.; Domingues, F.M.J.; Domingues, M.R.M.; Neves, M.G.P.M.S.; Cavaleiro, J.A.S.; Fernandes, R.; Tomé, J.P.C. Amphiphilic phthalocyanine-cyclodextrin conjugates for cancer photodynamic therapy. Chem. Commun. 2014, 50, 8363–8366. [Google Scholar] [CrossRef]
- Wang, Y.; Patel, D.J. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure 1993, 1, 263–282. [Google Scholar] [CrossRef]
- Schultze, P.; Smith, F.W.; Feigon, J. Refined solution structure of the dimeric quadruplex formed from the Oxytricha telomeric oligonucleotide d(GGGGTTTTGGGG). Structure 1994, 2, 221–233. [Google Scholar] [CrossRef]
- Sirajuddin, M.; Ali, S.; Badshah, A. Drug-DNA interactions and their study by UV-Visible, fluorescence spectroscopies and cyclic voltametry. J. Photochem. Photobiol. B Biol. 2013, 124, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.Y.; Jia, G.Q.; Yuan, J.L.; Feng, Z.C.; Li, C. A spectroscopic study on the interactions of porphyrin with G- quadruplex DNAs. Biochem. USA 2006, 45, 6681–6691. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Ji, F.; Liu, R.; Lin, J.; Xu, Q.; Gao, C. Interaction mechanism of 2-aminobenzothiazole with herring sperm DNA. J. Lumin. 2012, 132, 507–512. [Google Scholar] [CrossRef]
- Bhattacharjee, S.; Sengupta, P.K.; Bhowmik, S. Exploring the preferential interaction of quercetin with VEGF promoter G-quadruplex DNA and construction of a pH-dependent DNA-based logic gate. RSC Adv. 2017, 7, 37230–37240. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, X.L.; Shi, J.; Wang, S.F.; Yin, Y.; Yang, Y.S.; Zhang, W.M.; Zhu, H.L. Synthesis, molecular modeling and biological evaluation of N-benzylidene-2-((5-(pyridin-4-yl)-1,3,4-oxadiazol-2-yl)thio)acetohydrazide derivatives as potential anticancer agents. Bioorganic Med. Chem. 2014, 22, 468–477. [Google Scholar] [CrossRef] [PubMed]
- Bağda, E.; Bağda, E.; Durmuş, M. G-quadruplex and calf thymus DNA interaction of quaternized tetra and octa pyridyloxy substituted indium (III) phthalocyanines. J. Photochem. Photobiol. B Biol. 2017, 175, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Correia, R.F.; Andrade, S.M.; Viseu, M.I. Aggregation and disaggregation of anionic aluminum phthalocyanines in cationic pre-micelle and micelle media: A fluorescence study. J. Photochem. Photobiol. A Chem. 2012, 235, 21–28. [Google Scholar] [CrossRef]
- Alzeer, J.; Vummidi, B.R.; Roth, P.J.C.; Luedtke, N.W. Guanidinium-modified phthalocyanines as high-affinity G-quadruplex fluorescent probes and transcriptional regulators. Angew. Chemie Int. Ed. 2009, 48, 9362–9365. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Jia, G.; Zhou, J.; Han, G.; Li, C. Evidence for the binding mode of porphyrins to G-quadruplex DNA. Phys. Chem. Chem. Phys. 2009, 11, 4025–4032. [Google Scholar] [CrossRef] [PubMed]
- Randazzo, A.; Spada, G.P.; Webba Da Silva, M. Circular dichroism of quadruplex structures. Top. Curr. Chem. 2013, 330, 67–86. [Google Scholar]
- Kypr, J.; Kejnovská, I.; Renčiuk, D.; Vorlíčková, M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009, 37, 1713–1725. [Google Scholar] [CrossRef] [PubMed]
- Ambrus, A.; Chen, D.; Dai, J.; Bialis, T.; Jones, R.A.; Yang, D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006, 34, 2723–2735. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Shafer, R.H. Covalent ligation studies on the human telomere quadruplex. Nucleic Acids Res. 2005, 33, 3185–3192. [Google Scholar] [CrossRef] [PubMed]
- Paramasivan, S.; Rujan, I.; Bolton, P.H. Circular dichroism of quadruplex DNAs: Applications to structure, cation effects and ligand binding. Methods 2007, 43, 324–331. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, J.; Queiroz, J.A.; Cruz, C. Circular dichroism of G-Quadruplex: A laboratory experiment for the study of topology and ligand binding. J. Chem. Educ. 2017, 94, 1547–1551. [Google Scholar] [CrossRef]
- Ruan, T.L.; Davis, S.J.; Powell, B.M.; Harbeck, C.P.; Habdas, J.; Habdas, P.; Yatsunyk, L.A. Lowering the overall charge on TMPyP4 improves its selectivity for G-quadruplex DNA. Biochimie 2017, 132, 121–130. [Google Scholar] [CrossRef]
- Nagesh, N.; Sharma, V.K.; Ganesh Kumar, A.; Lewis, E.A. Effect of ionic strength on porphyrin drugs interaction with quadruplex DNA formed by the promoter region of C-myc and Bcl2 oncogenes. J. Nucleic Acids 2010, 2010. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds ZnPc1, ZnPc2, ZnPc3 and ZnPc4 are available from the authors. |

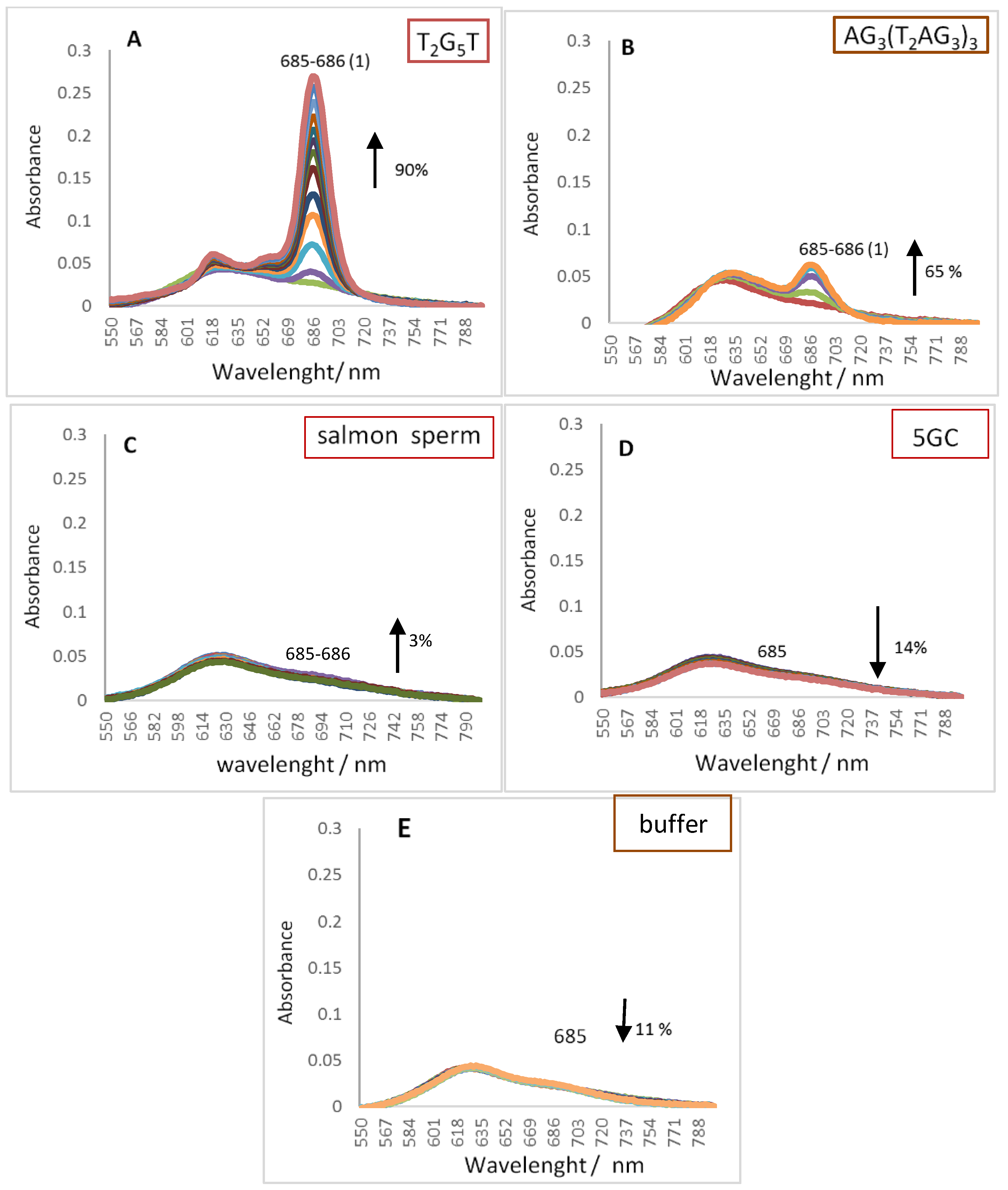
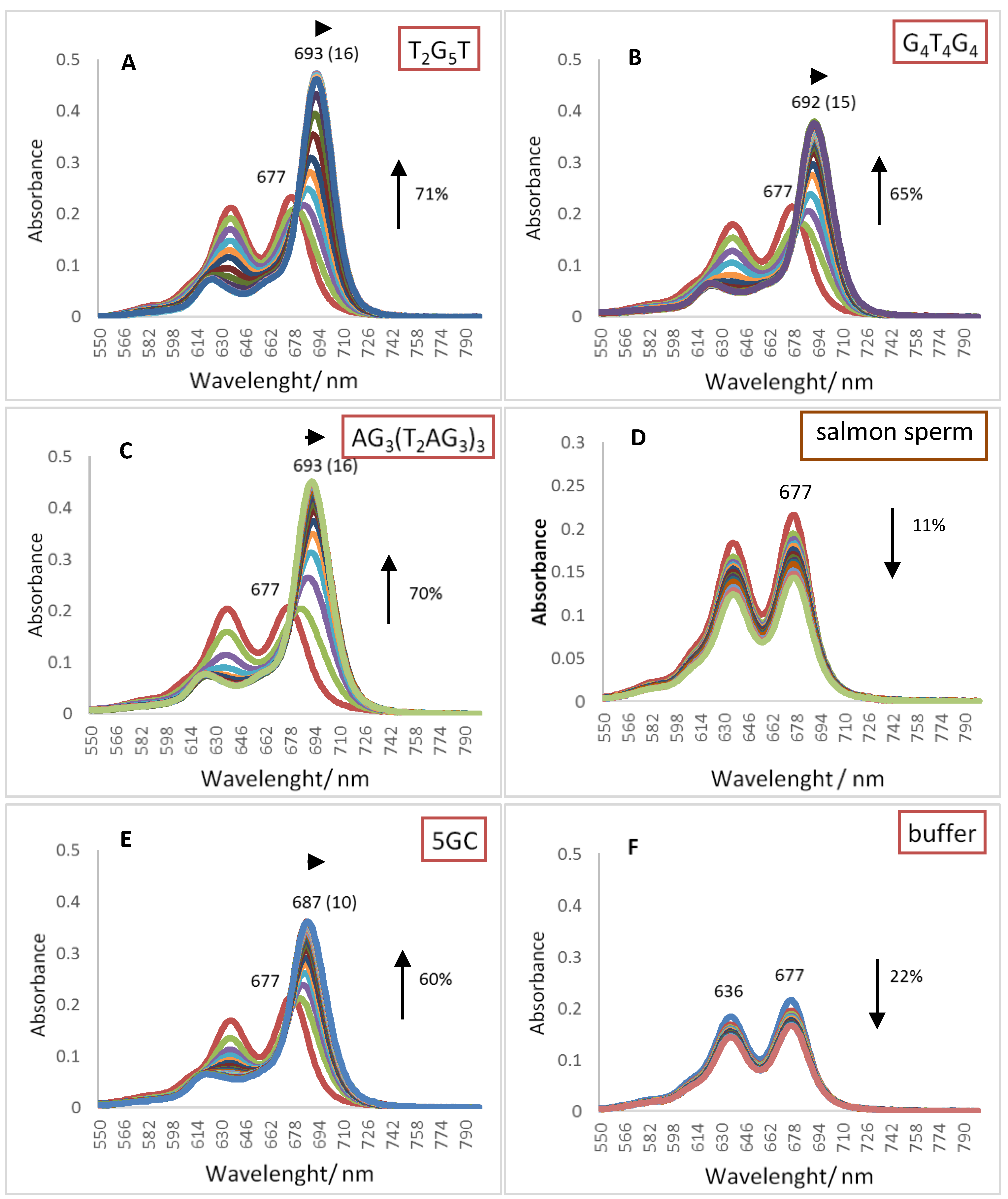
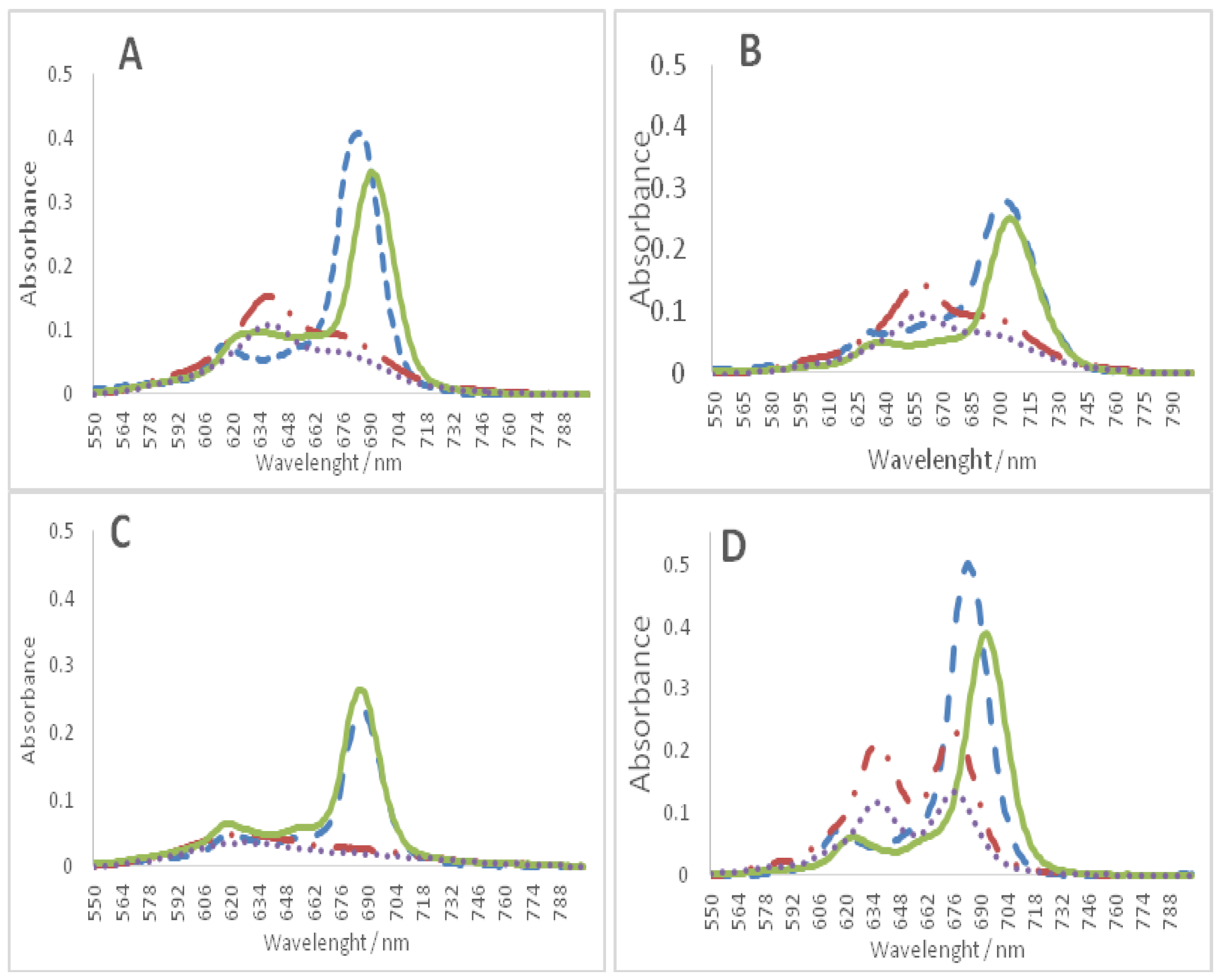
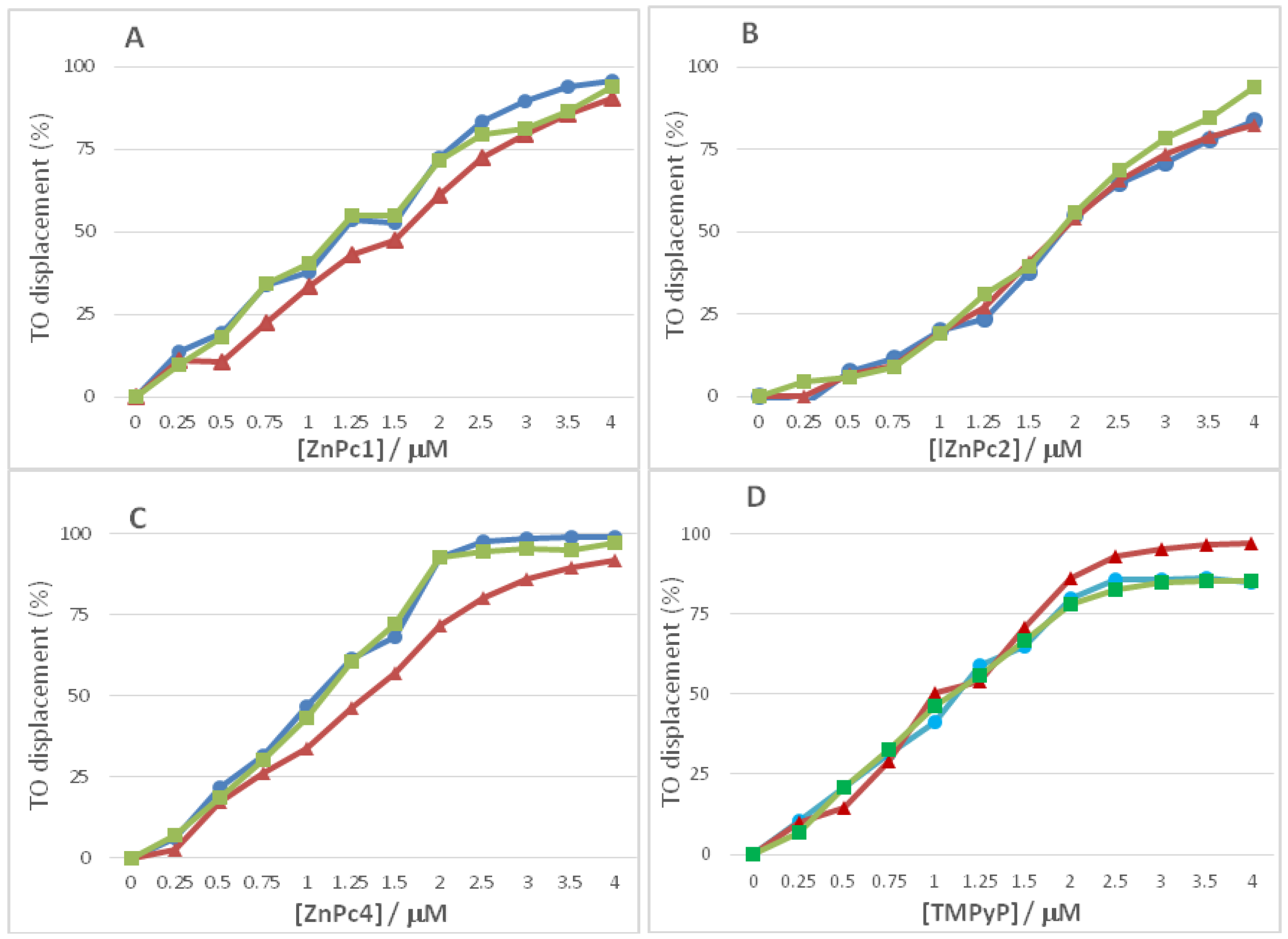
| Oligonucleotide Sequence | Topology | Abbreviation |
|---|---|---|
| 5′-TTGGG GGT-3′ | Tetramolecular G-Quadruplex | (T2G5T)4 |
| 5′-GGG GTT TT GGG G-3′ (Oxytricha repeat oligonucleotide) | Bimolecular G-Quadruplex | (G4T4G4)2 |
| 5′-AGG GTT AGG GTTAGG GTT AGGG-3′ (human telomeric repeat) | Unimolecular G-Quadruplex | AG3(T2AG3)3 |
| 5′-GCGCG CGC GC-3′ | Double strand DNA | 5GC |
| Long single strand | Double strand DNA | Salmon sperm DNA |
| Entry | G-Quadruplexes | Duplexes | Ligand | ||||
|---|---|---|---|---|---|---|---|
| (T2G5T)4 | (G4T4G4)2 | AG3 (T2AG3)3 | 5 GC | Salmon Sperm | |||
| Red shift (nm) | 19 | 19 | 20 | 8 | 1 | ZnPc1 | |
| (1) | Hypo/Hyper chromism (%) | +96 | +90 | +78 | +37 | −26 | |
| Red shift (nm) | 16 | 17 | 16 | 17 | 17 | ZnPc2 | |
| (2) | Hypo/Hyper chromism (%) | +85 | +79 | +82 | +51 | +54 | |
| Red shift (nm) | 1 | n.a. | 0 | 0 | 1 | ZnPc3 | |
| (3) | Hypo/Hyper chromism (%) | +90 | n.a. | +65 | +3 | −14 | |
| Red shift (nm) | 16 | 15 | 15 | 10 | 4 | ZnPc4 | |
| (4) | Hypo/Hyper chromism (%) | +71 | +65 | +70 | +60 | −11 | |
| Red shift (nm) | 11 | 13 | 16 | 18 | 13 | TMPyP | |
| (5) | Hypo/Hyper chromism (%) | −25 | −27 | −33 | −29 | −36 | |
| KD (M) | ||
|---|---|---|
| AG3(T2AG3)3 | 5GC | |
| ZnPc1 | (1.62 ± 0.12) × 10−7 | (1.38 ± 0.15) × 10−4 |
| ZnPc2 | (9.58 ± 0.09) × 10−6 | (1.28 ± 0.17) × 10−5 |
| ZnPc4 | (7.61 ± 0.11) × 10−8 | (4.74 ± 0.13) × 10−4 |
| DC50 (μM) | |||
|---|---|---|---|
| 5GC | (T2G5T)4 | AG3(T2AG3)3 | |
| ZnPc1 | 1.59 ± 0.06 | 1.14 ± 0.12 | 1.17 ± 0.07 |
| ZnPc2 | 1.93 ± 0.48 | 1.84 ± 0.03 | 1.95 ± 0.26 |
| ZnPc4 | 1.30 ± 0.21 | 1.08 ± 0.08 | 1.04 ± 0.28 |
| TMPyP | 0.94 ± 0.20 | 1.17 ± 0.20 | 1.07 ± 0.09 |
| Tm (°C) | ΔTm (°C) | |
|---|---|---|
| AG3(T2AG3)3 | 56.1 ± 1.4 | --- |
| AG3(T2AG3)3 + ZnPc1 | 65.9 ± 2.1 | 9.8 |
| AG3(T2AG3)3 + ZnPc4 | 67.9 ± 2.4 | 11.8 |
| AG3(T2AG3)3 + TMPyP | 63.1 ± 1.9 | 7.0 |
| Ligand | IC50 (μM) |
|---|---|
| ZnPc1 | 29.44 ± 1.47 |
| ZnPc4 | 58.09 ± 1.76 |
| TMPyP | 43.48 ± 1.64 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramos, C.I.V.; Almeida, S.P.; Lourenço, L.M.O.; Pereira, P.M.R.; Fernandes, R.; Faustino, M.A.F.; Tomé, J.P.C.; Carvalho, J.; Cruz, C.; Neves, M.G.P.M.S. Multicharged Phthalocyanines as Selective Ligands for G-Quadruplex DNA Structures. Molecules 2019, 24, 733. https://doi.org/10.3390/molecules24040733
Ramos CIV, Almeida SP, Lourenço LMO, Pereira PMR, Fernandes R, Faustino MAF, Tomé JPC, Carvalho J, Cruz C, Neves MGPMS. Multicharged Phthalocyanines as Selective Ligands for G-Quadruplex DNA Structures. Molecules. 2019; 24(4):733. https://doi.org/10.3390/molecules24040733
Chicago/Turabian StyleRamos, Catarina I. V., Susana P. Almeida, Leandro M. O. Lourenço, Patrícia M. R. Pereira, Rosa Fernandes, M. Amparo F. Faustino, João P. C. Tomé, Josué Carvalho, Carla Cruz, and M. Graça P. M. S. Neves. 2019. "Multicharged Phthalocyanines as Selective Ligands for G-Quadruplex DNA Structures" Molecules 24, no. 4: 733. https://doi.org/10.3390/molecules24040733
APA StyleRamos, C. I. V., Almeida, S. P., Lourenço, L. M. O., Pereira, P. M. R., Fernandes, R., Faustino, M. A. F., Tomé, J. P. C., Carvalho, J., Cruz, C., & Neves, M. G. P. M. S. (2019). Multicharged Phthalocyanines as Selective Ligands for G-Quadruplex DNA Structures. Molecules, 24(4), 733. https://doi.org/10.3390/molecules24040733