The β-N-Acetylhexosaminidase in the Synthesis of Bioactive Glycans: Protein and Reaction Engineering
Abstract
1. Introduction
2. Results
2.1. Production and Purification of TfHex Mutants
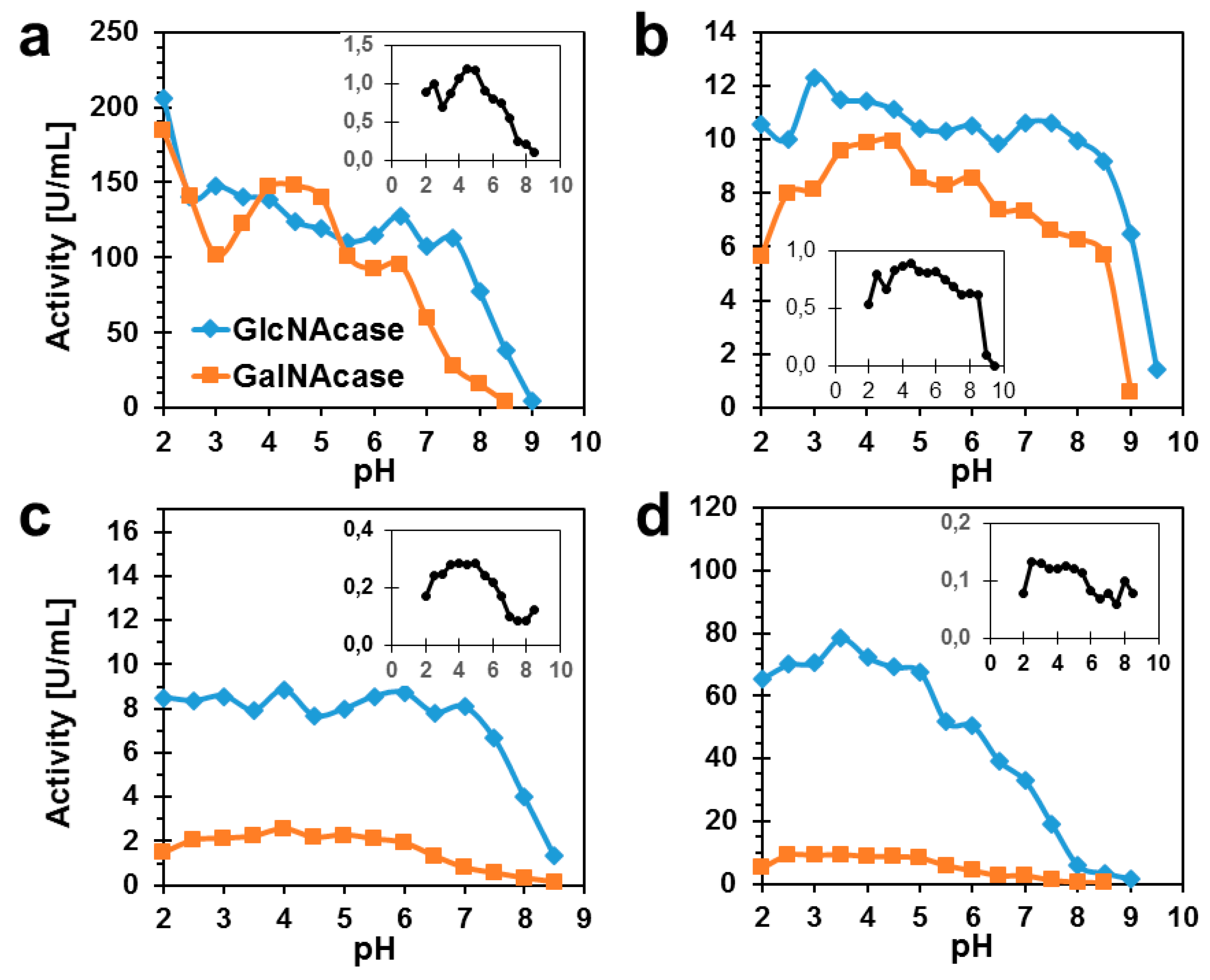
2.2. Biochemical Characterization of TfHex Mutants
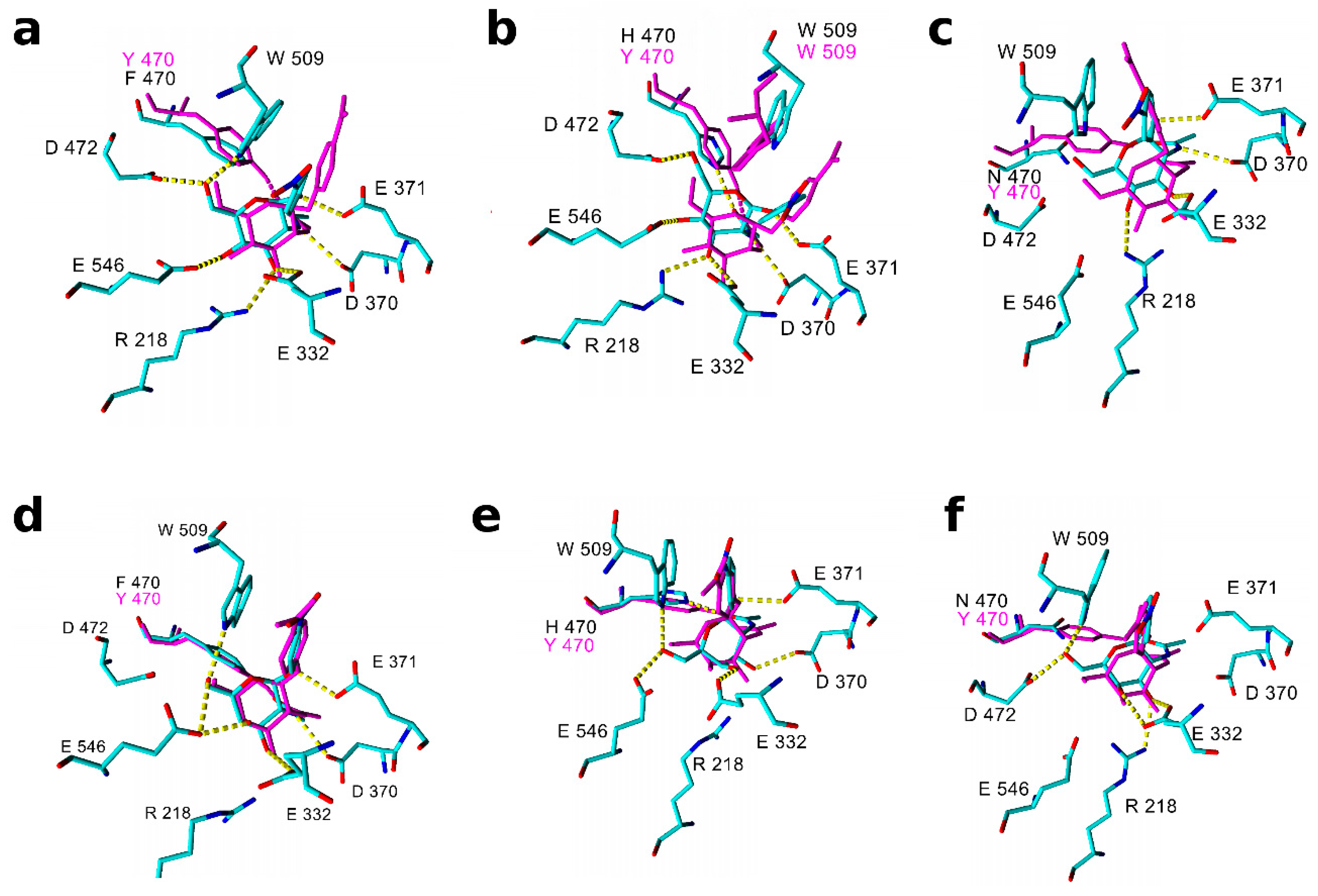
2.3. Molecular Modeling and Docking of TfHex Mutants
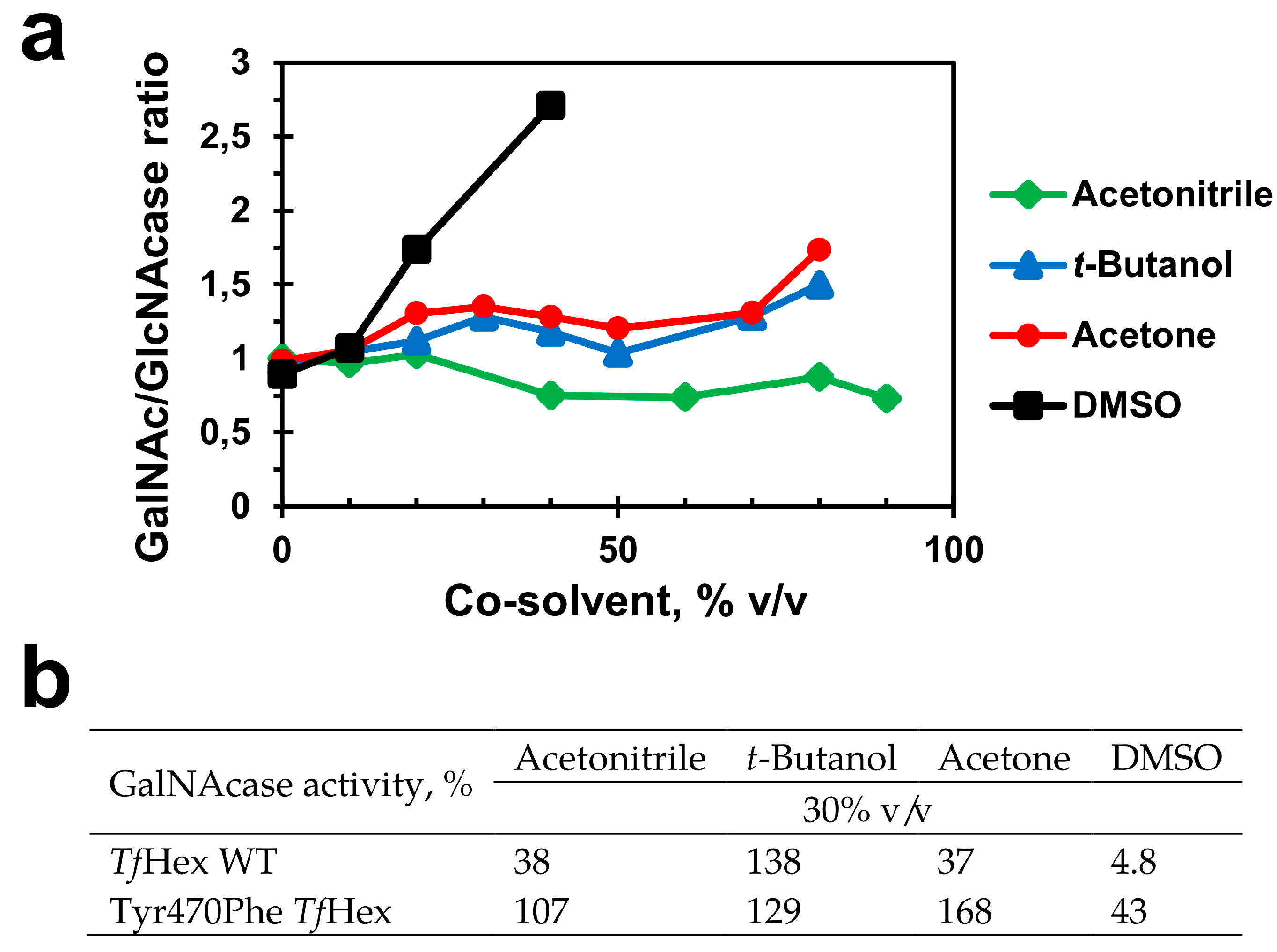
2.4. Medium Engineering
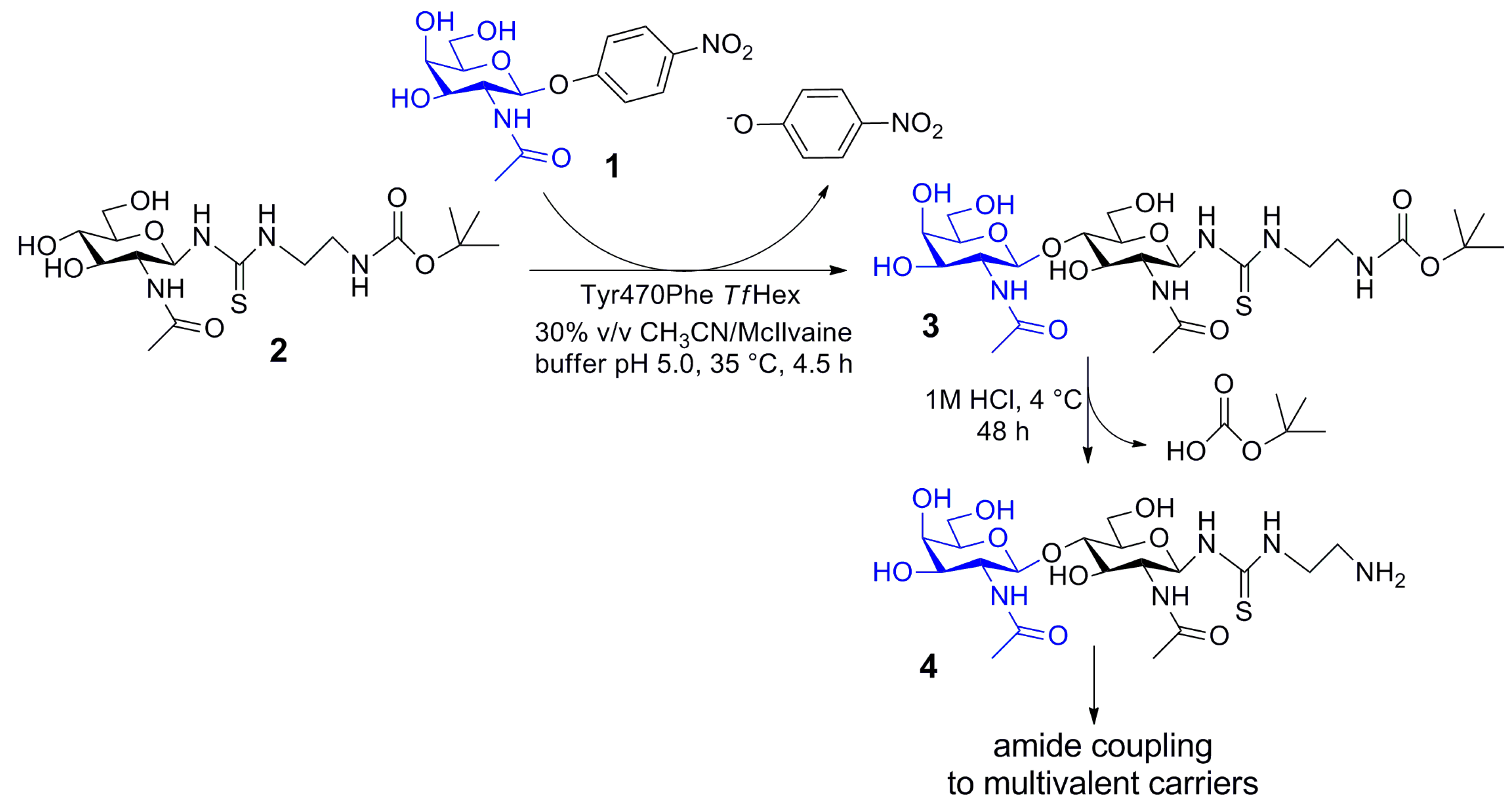
2.5. Enzymatic Synthesis of Functionalized Disaccharide 3
3. Discussion
4. Materials and Methods
4.1. Structural Characterization of Compounds
4.2. Production of TfHex Variants in Pichia pastoris
4.3. Standard Assay for Enzyme Activity
4.4. Molecular Modeling, Docking and Molecular Dynamics
4.5. Compound (t-Butoxycarbonylamino)ethylthioureidyl 2-acetamido-2-deoxy-β-d-glucopyranoside (2)
4.6. Compound (t-Butoxycarbonylamino)ethylthioureidyl 2-acetamido-2-deoxy-β-d-galactopyranosyl-(1→4)-2-acetamido-2-deoxy-β-d-glucopyranoside (3)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Laaf, D.; Bojarová, P.; Elling, L.; Křen, V. Galectin-carbohydrate interactions in biomedicine and biotechnology. Trends Biotechnol. 2018. in print. [Google Scholar] [CrossRef] [PubMed]
- Bojarová, P.; Křen, V. Sugared biomaterial binding lectins: Achievements and perspectives. Biomater. Sci. 2016, 4, 1142–1160. [Google Scholar] [CrossRef] [PubMed]
- Bojarová, P.; Křenek, K.; Kuzma, M.; Petrásková, L.; Bezouška, K.; Namdjou, D.-J.; Elling, L.; Křen, V. N-Acetylhexosamine triad in one molecule: Enzymatic introduction of 2-acetamido-2-deoxy-β-d-galactopyranosyluronic acid residue into a complex oligosaccharide. J. Mol. Catal. B Enzymatic 2008, 50, 69–73. [Google Scholar] [CrossRef]
- Bojarová, P.; Slámová, K.; Křenek, K.; Gažák, R.; Kulik, N.; Ettrich, R.; Pelantová, H.; Kuzma, M.; Riva, S.; Adámek, D.; Bezouška, K.; Křen, V. Charged hexosaminides as new substrates for β-N-acetylhexosaminidase-catalyzed synthesis of immunomodulatory disaccharides. Adv. Synth. Catal. 2011, 353, 2409–2420. [Google Scholar] [CrossRef]
- Slámová, K.; Gažák, R.; Bojarová, P.; Kulik, N.; Ettrich, R.; Pelantová, H.; Sedmera, P.; Křen, V. 4-Deoxy-substrates for β-N-acetylhexosaminidases: How to make use of their loose specificity. Glycobiology 2010, 20, 1002–1009. [Google Scholar] [CrossRef] [PubMed]
- Bojarová, P.; Křen, V. Glycosidases in carbohydrate synthesis: When organic chemistry falls short. Chimia 2011, 65, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Slámová, K.; Bojarová, P. Engineered N-acetylhexosamine-active enzymes in glycoscience. Biochim. Biophys. Acta 2017, 1861, 2070–2087. [Google Scholar] [CrossRef] [PubMed]
- Bojarová, P.; Chytil, P.; Mikulová, B.; Bumba, L.; Konefał, R.; Pelantová, H.; Krejzová, J.; Slámová, K.; Petrásková, L.; Kotrchová, L.; et al. Glycan-decorated HPMA copolymers as high-affinity lectin ligands. Polym. Chem. 2017, 8, 2647–2658. [Google Scholar] [CrossRef]
- Bumba, L.; Laaf, D.; Spiwok, V.; Elling, L.; Křen, V.; Bojarová, P. Poly-N-acetyllactosamine neo-glycoproteins as nanomolar ligands of human galectin-3: Binding kinetics and modeling. Int. J. Mol. Sci. 2018, 19, 372. [Google Scholar] [CrossRef]
- Slámová, K.; Krejzová, J.; Marhol, P.; Kalachova, L.; Kulik, N.; Pelantová, H.; Cvačka, J.; Křen, V. Synthesis of derivatized chitooligomers using transglycosidases engineered from the fungal GH20 β-N-acetylhexosaminidase. Adv. Synth. Catal. 2015, 357, 1941–1950. [Google Scholar] [CrossRef]
- Horsch, M.; Mayer, C.; Sennhauser, U.; Rast, D.M. β-N-Acetylhexosaminidase: A target for the design of antifungal agents. Pharmacol Ther. 1997, 76, 187–218. [Google Scholar] [CrossRef]
- Bojarová, P.; Kulik, N.; Slámová, K.; Hubálek, M.; Kotik, M.; Cvačka, J.; Pelantová, H.; Křen, V. Selective β-N-acetylhexosaminidase from Aspergillus versicolor—A tool for producing bioactive carbohydrates. Appl. Microbiol. Biotechnol 2019. in print. [Google Scholar] [CrossRef]
- Slámová, K.; Bojarová, P.; Gerstorferová, D.; Fliedrová, B.; Hofmeisterová, J.; Fiala, M.; Pompach, P.; Křen, V. Sequencing, cloning and high-yield expression of a fungal β-N-acetylhexosaminidase in Pichia pastoris. Prot. Express. Purif. 2012, 82, 212–217. [Google Scholar] [CrossRef] [PubMed]
- Fialová, P.; Carmona, A.T.; Robina, I.; Ettrich, R.; Sedmera, P.; Přikrylová, V.; Petrásková-Hušáková, L.; Křen, V. Glycosyl azide—A novel substrate for enzymatic transglycosylations. Tetrahedron Lett. 2005, 46, 8715–8718. [Google Scholar] [CrossRef]
- Breloy, I.; Söte, S.; Ottis, P.; Bonar, D.; Grahn, A.; Hanisch, F.-G. O-Linked LacdiNAc-modified glycans in extracellular matrix glycoproteins are specifically phosphorylated at the subterminal GlcNAc. J. Biol. Chem. 2012, 287, 18275–18286. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Kenny, D.T.; Skoog, E.C.; Padra, M.; Adamczyk, B.; Vitizeva, V.; Thorell, A.; Venkatakrishnan, V.; Lindén, S.K.; Karlsson, N.G. Structural diversity of human gastric mucin glycans. Mol. Cell. Proteom. 2017, 16, 743–758. [Google Scholar] [CrossRef] [PubMed]
- Wuhrer, M.; Koeleman, C.A.M.; Deelder, A.M.; Hokke, C.H. Repeats of LacdiNAc and fucosylated LacdiNAc on N-glycans of the human parasite Schistosoma mansoni. FEBS J. 2006, 273, 347–361. [Google Scholar] [CrossRef]
- Hirano, K.; Matsuda, A.; Shirai, T.; Furukawa, K. Expression of LacdiNAc groups on N-glycans among human tumors is complex. Biomed. Res. Int. 2014, 2014, 981627. [Google Scholar] [CrossRef]
- Laaf, D.; Bojarová, P.; Pelantová, H.; Křen, V.; Elling, L. Tailored multivalent neo-glycoproteins: Synthesis, evaluation, and application of a library of galectin-3-binding glycan ligands. Bioconjug. Chem. 2017, 28, 2832–2840. [Google Scholar] [CrossRef]
- Laaf, D.; Bojarová, P.; Mikulová, B.; Pelantová, H.; Křen, V.; Elling, L. Two-step enzymatic synthesis of β-d-N-acetylgalactosamine-(1→4)-d-N-acetylglucosamine (LacdiNAc) chitooligomers for deciphering galectin binding behavior. Adv. Synth. Catal. 2017, 359, 2101–2108. [Google Scholar] [CrossRef]
- Šimonová, A.; Kupper, C.E.; Böcker, S.; Müller, A.; Hofbauerová, K.; Pelantová, H.; Elling, L.; Křen, V.; Bojarová, P. Chemo-enzymatic synthesis of LacdiNAc dimers of varying length as novel galectin ligands. J. Mol. Catal. B Enzym. 2014, 101, 47–55. [Google Scholar] [CrossRef]
- Kulik, N.; Slámová, K.; Ettrich, R.; Křen, V. Computational study of β-N-acetylhexosaminidase from Talaromyces flavus, a glycosidase with high substrate flexibility. BMC Bioinformatics 2015, 16, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Bas, D.C.; Rogers, D.M.; Jensen, J.H. Very fast prediction and rationalization of pKa values for protein-ligand complexes. Proteins 2008, 73, 765–783. [Google Scholar] [CrossRef] [PubMed]
- Whitworth, G.E.; Macauley, M.S.; Stubbs, K.A.; Dennis, R.J.; Taylor, E.J.; Davies, G.J.; Greig, I.R.; Vocadlo, D.J. Analysis of PUGNAc and NAG-thiazoline as transition state analogues for human O-GlcNAcase: Mechanistic and structural insights into inhibitor selectivity and transition state poise. J. Am. Chem. Soc. 2007, 129, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Jeffrey, G.A. An introduction to hydrogen bonding; Oxford University Press: Oxford, UK, 1997; ISBN 0-19-509549-9. [Google Scholar]
- Simerská, P.; Kuzma, M.; Monti, D.; Riva, S.; Křen, V.; Macková, M. Unique transglycosylation potential of extracellular α-d-galactosidase from Talaromyces flavus. J. Mol. Catal. B Enzymatic 2006, 39, 128–134. [Google Scholar] [CrossRef]
- Vokhmyanina, O.A.; Rapoport, E.M.; André, S.; Severov, V.V.; Ryzhov, I.; Pazynina, G.V.; Korchagina, E.; Gabius, H.J.; Bovin, N.V. Comparative study of the glycan specificities of cell-bound human tandem-repeat-type galectin-4, -8 and -9. Glycobiology 2012, 22, 1207–1217. [Google Scholar] [CrossRef] [PubMed]
- Laaf, D.; Steffens, H.; Pelantová, H.; Bojarová, P.; Křen, V.; Elling, L. Chemo-enzymatic synthesis of branched N-acetyllactosamine glycan oligomers for galectin-3 inhibition. Adv. Synth. Catal. 2017, 359, 4015–4024. [Google Scholar] [CrossRef]
- Drozdová, A.; Bojarová, P.; Křenek, K.; Weignerová, L.; Henssen, B.; Elling, L.; Christensen, H.; Jensen, H.H.; Pelantová, H.; Kuzma, M.; et al. Enzymatic synthesis of dimeric glycomimetic ligands of NK cell activation receptors. Carbohydr. Res. 2011, 346, 1599–1609. [Google Scholar] [CrossRef] [PubMed]
- Fialová, P.; Weignerová, L.; Rauvolfová, J.; Přikrylová, V.; Pišvejcová, A.; Ettrich, R.; Kuzma, M.; Sedmera, P.; Křen, V. Hydrolytic and transglycosylation reactions of N-acyl modified substrates catalysed by β-N-acetylhexosaminidases. Tetrahedron 2004, 60, 693–701. [Google Scholar] [CrossRef]
- Bradford, M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Krieger, E.; Koraimann, G.; Vriend, G. Increasing the precision of comparative models with YASARA NOVA a self-parameterizing force field. Proteins 2002, 47, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra precision Glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef] [PubMed]
- Jakalian, A.; Jack, D.B.; Bayly, C.I. Fast, efficient generation of high quality atomic charges. AM1BCC model: II. Parameterization and validation. J. Comput. Chem. 2002, 23, 1623–1641. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.; Li, X.; Li, Y.; Liu, Z.; Wang, R. Comparative assessment of scoring functions on a diverse test set. J. Chem. Inf. Model. 2009, 49, 1079–1093. [Google Scholar] [CrossRef] [PubMed]
- Sauerzapfe, B.; Křenek, K.; Schmiedel, J.; Wakarchuk, W.W.; Pelantová, H.; Křen, V.; Elling, L. Chemo-enzymatic synthesis of poly-N-acetyllactosamine (poly-LacNAc) structures and their characterization for CGL2-galectin-mediated binding of ECM glycoproteins to biomaterial surfaces. Glycoconj. J. 2009, 26, 141–159. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds 2 and 3 are available from the authors. |
| Enzyme | Protein Yield [mg] 1 | GlcNAcase [U/mg] | GalNAcase [U/mg] | GalNAcase/GlcNAcase Ratio |
|---|---|---|---|---|
| WT TfHex | 188 | 37.4 | 45.1 | 1.2 |
| Tyr470Phe TfHex | 308 | 0.3 | 0.3 | 1.0 |
| Tyr470His TfHex | 84 | 0.4 | 0.1 | 0.3 |
| Tyr470Asn TfHex | 225 | 1.6 | 0.2 | 0.1 |
| Enzyme | Substrate | Km [mM] | kcat [s−1] | kcat/Km [s−1 mM−1] |
|---|---|---|---|---|
| WT TfHex | pNP-GlcNAc | 0.11 ± 0.02 | 47 ± 1 | 434 |
| pNP-GalNAc | 0.69 ± 0.09 | 104 ± 5 | 150 | |
| Tyr470Phe TfHex | pNP-GlcNAc | 0.023 ± 0.008 | 0.27 ± 0.01 | 12 |
| pNP-GalNAc | 0.050 ± 0.005 | 0.166 ± 0.003 | 3.3 | |
| Tyr470His TfHex | pNP-GlcNAc | 0.053 ± 0.008 | 0.67 ± 0.02 | 13 |
| pNP-GalNAc | 0.35 ± 0.03 | 0.188 ± 0.004 | 0.53 | |
| Tyr470Asn TfHex | pNP-GlcNAc | 0.89 ± 0.13 | 11.6 ± 0.6 | 13 |
| pNP-GalNAc | 0.89 ± 0.09 | 0.35 ± 0.01 | 0.40 |
| TfHex variant | Binding XP scores [kJ/mol] (averaged number of hydrogen bonds) | |||
|---|---|---|---|---|
| pNP-GlcNAc | pNP-GalNAc | GlcNAc | GalNAc | |
| WT | −38.5 (6.0) | −34.9 (5.5) | −23.5 | −29.5 |
| Tyr470Phe | −33.2 (4.7) | −36.5 (5.0) | −32.0 | −23.4 |
| Tyr70His (positive) | −32.2 (5.6) | −25.7 (3.3) | −27.1 | −29.4 |
| Tyr470His (neutral) | −37.0 (4.6) | −32.0 (4.6) | −22.6 | −31.5 |
| Tyr470Asn | −31.1 (3.8) | −30.8 (4.7) | −27.6 | −23.5 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bojarová, P.; Kulik, N.; Hovorková, M.; Slámová, K.; Pelantová, H.; Křen, V. The β-N-Acetylhexosaminidase in the Synthesis of Bioactive Glycans: Protein and Reaction Engineering. Molecules 2019, 24, 599. https://doi.org/10.3390/molecules24030599
Bojarová P, Kulik N, Hovorková M, Slámová K, Pelantová H, Křen V. The β-N-Acetylhexosaminidase in the Synthesis of Bioactive Glycans: Protein and Reaction Engineering. Molecules. 2019; 24(3):599. https://doi.org/10.3390/molecules24030599
Chicago/Turabian StyleBojarová, Pavla, Natalia Kulik, Michaela Hovorková, Kristýna Slámová, Helena Pelantová, and Vladimír Křen. 2019. "The β-N-Acetylhexosaminidase in the Synthesis of Bioactive Glycans: Protein and Reaction Engineering" Molecules 24, no. 3: 599. https://doi.org/10.3390/molecules24030599
APA StyleBojarová, P., Kulik, N., Hovorková, M., Slámová, K., Pelantová, H., & Křen, V. (2019). The β-N-Acetylhexosaminidase in the Synthesis of Bioactive Glycans: Protein and Reaction Engineering. Molecules, 24(3), 599. https://doi.org/10.3390/molecules24030599








