Optimal Extraction Study of Gastrodin-Type Components from Gastrodia Elata Tubers by Response Surface Design with Integrated Phytochemical and Bioactivity Evaluation
Abstract
:1. Introduction
2. Results
2.1. Method Validation
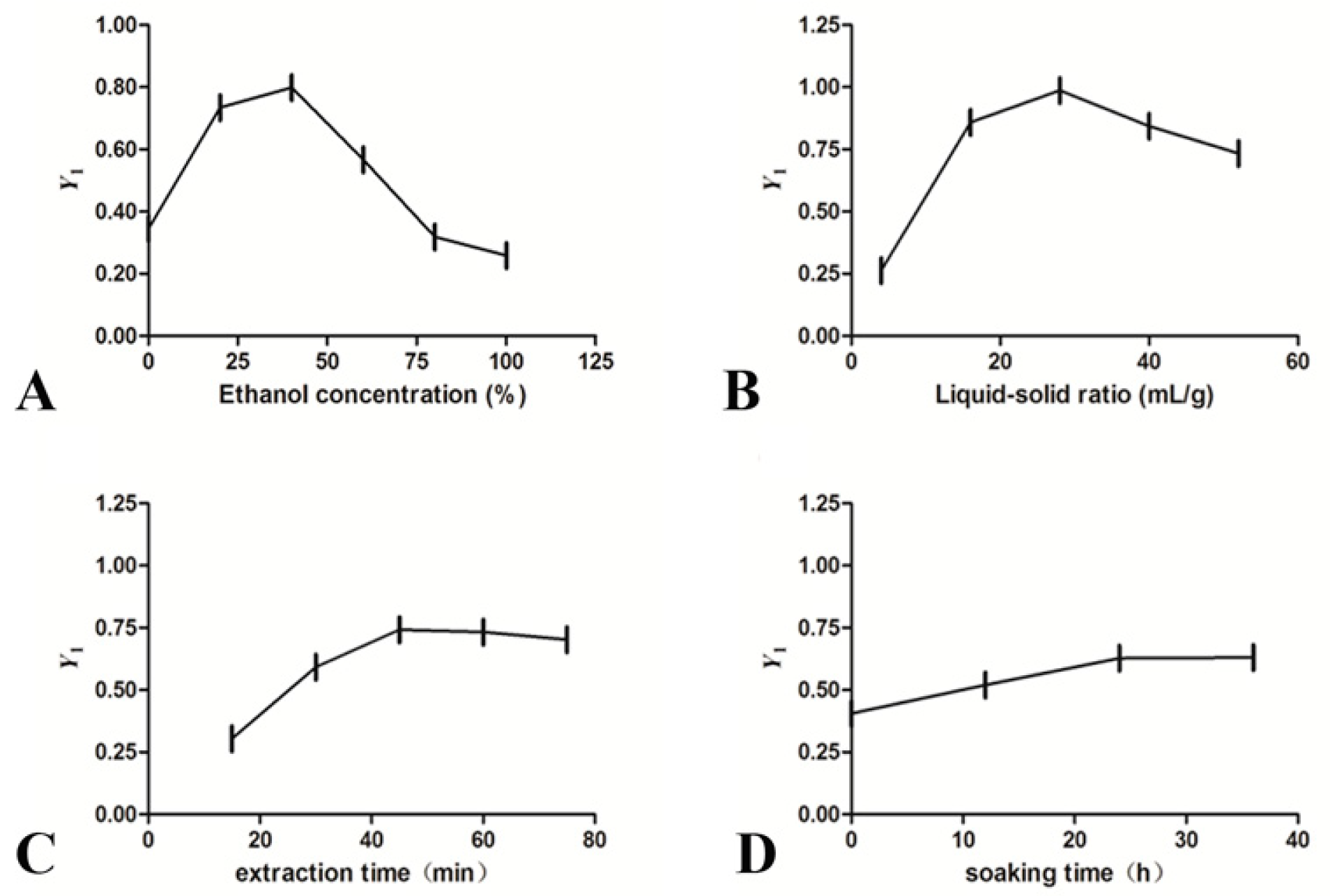
2.2. Optimization of Extraction Procedure
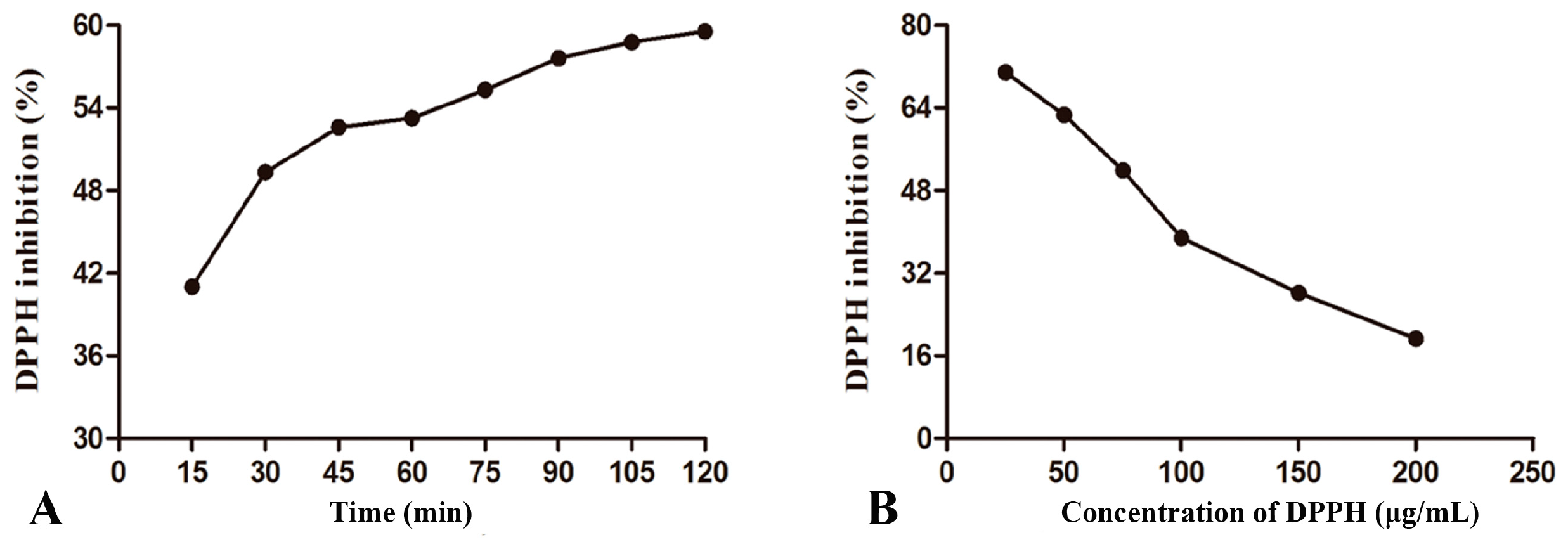
2.3. Optimization of DPPH Assay
2.4. Optimization of the Procedure
2.4.1. The Statistical Analysis and Model Fitting
- Y = 1.80 + 0.016X1 + 0.13X2 + 0.094X3 − 0.021X4 − 0.11X1X2 + 0.13X1X3 + 0.10X1X4 − 0.20X2X3 + 0.25X2X4 + 0.042X3X4 − 0.47X12 − 1.11X22 − 0.42X32 − 0.18X42
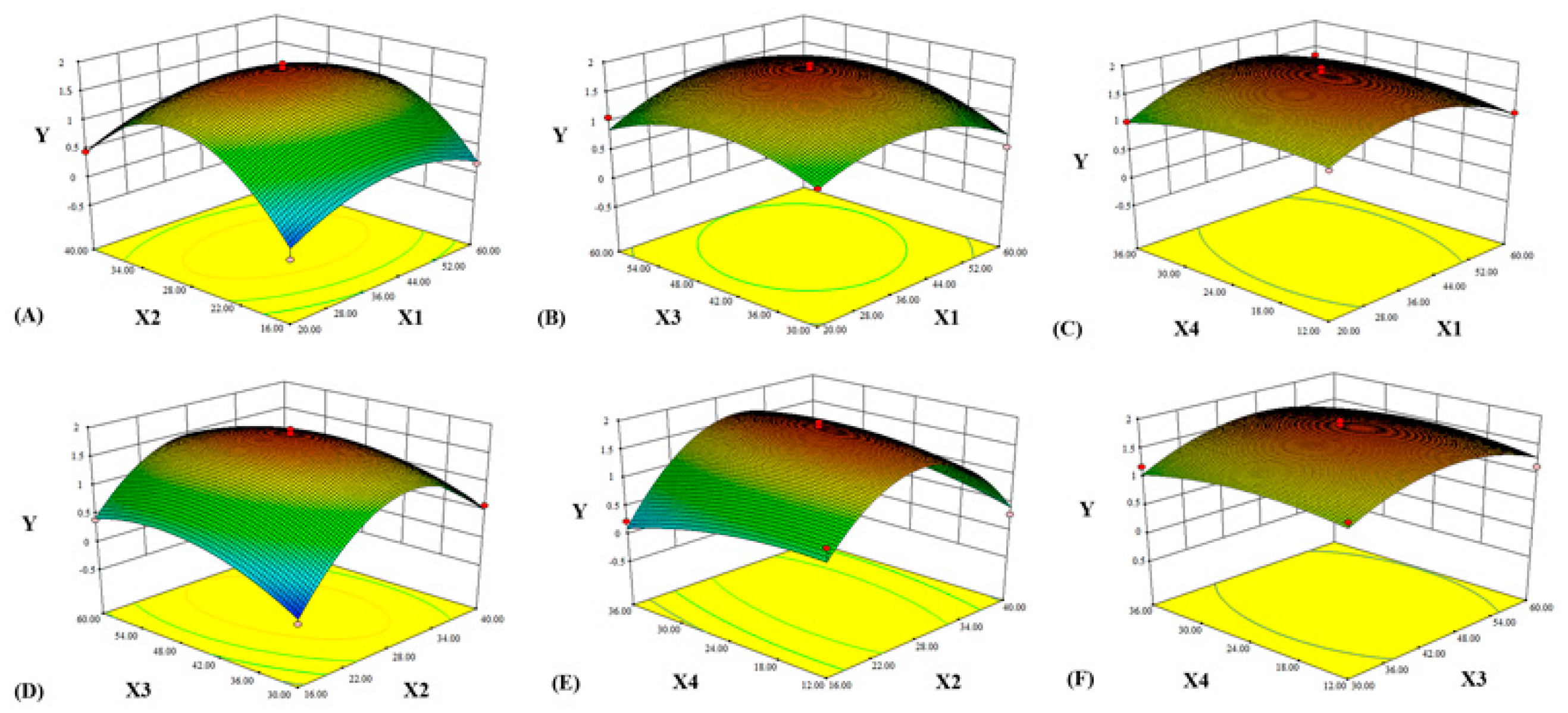
2.4.2. Analysis of the Response Surface
2.4.3. Verification of Predictive Model
3. Materials and Methods
3.1. Chemicals and Materials
3.2. Preparation of Standard Solutions
3.3. Sample Preparation
3.4. Determination of the Gastrodin-Type Components
3.4.1. Chromatographic Conditions
3.4.2. Method Validation
3.5. Detection of Antioxidant Activity
3.6. Cell Culture and Viability Assay
3.7. Experimental Design for RSM
3.8. Data Analysis
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Lee, D.K.; Lim, D.K.; Um, J.A.; Lim, C.J.; Hong, J.Y.; Yoon, Y.A.; Ryu, Y.; Kim, H.J.; Cho, H.J.; Park, J.H.; et al. Evaluation of four different analytical tools to determine the regional origin of Gastrodia elata and Rehmannia glutinosa on the basis of metabolomics study. Molecules 2014, 19, 6294–6308. [Google Scholar] [CrossRef]
- The National Pharmacopoeia Commission of P.R. China. Pharmacopoeia of the People’s Republic of China; China Medicine Science Technology Press: Beijing, China, 2015; pp. 200–201. [Google Scholar]
- Song, E.; Chung, H.; Shim, E.; Jeong, J.K.; Han, B.K.; Choi, H.J.; Hwang, J. Gastrodia elata Blume extract modulates antioxidant activity and ultraviolet A-irradiated skin aging in human dermal fibroblast cells. J. Med. Food 2016, 19, 1057–1064. [Google Scholar] [CrossRef] [PubMed]
- Ahn, E.K.; Jeon, H.J.; Lim, E.J.; Jung, H.J.; Park, E.H. Anti-inflammatory and anti-angiogenic activities of Gastrodia elata Blume. J. Ethnopharmacol. 2007, 110, 476–482. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.B.; Zhao, T.; Muna, S.S.; Jin, H.M.; Park, J.I.; Jo, K.S.; Lee, B.H.; Chae, S.W.; Kim, S.Y.; Park, S.H.; et al. Therapeutic potential of Gastrodia elata Blume for the treatment of Alzheimer’s disease. Neural Regen. Res. 2013, 8, 1061–1070. [Google Scholar] [PubMed]
- Lin, Y.E.; Lin, S.H.; Chen, W.C.; Ho, C.T.; Lai, Y.S.; Panyod, S.; Lai, Y.S.; Ho, C.T.; Sheen, L.Y. Antidepressant-like effects of water extract of Gastrodia elata blume in rats exposed to unpredictable chronic mild stress via modulation of monoamine regulatory pathways. J. Ethnopharmacol. 2016, 187, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Kumar, H.; Kim, I.S.; More, S.V.; Kim, B.W.; Bahk, Y.Y.; Choi, D.K. Gastrodin protects apoptotic dopaminergic neurons in a toxin-induced Parkinson’s disease model. Evid.-Based Complment. Altern. 2013. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.W.; Yoon, B.H.; Oh, H.R.; Ahn, J.H.; Kim, S.Y.; Park, S.Y.; Ryu, J.F. Anxiolytic-Like effects of Gastrodia elata and its phenolic constituents in mice. Biol. Pharm. Bull. 2006, 29, 261–265. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, M.; Kang, R.X.; Shi, J.G.; Liu, G.T.; Zhang, J.J. NHBA isolated from Gastrodia elata exerts sedative and hypnotic effects in sodium pentobarbital-treated mice. Pharmacol. Biochem. Behav. 2012, 102, 450–457. [Google Scholar] [CrossRef]
- Zhao, Y.; Kang, Z.J.; Zhou, X.; Yang, S.L. An edible medicinal plant-Gastrodia elata Bl. J. Guizhou Norm. Univ. 2013, 31, 9–12. [Google Scholar]
- Kang, C.; Lai, C.J.; Zhao, D.; Zhou, T.; Liu, D.H.; Lv, C.; Wang, S.; Kang, L.; Yang, J.; Zhan, Z.L.; et al. A practical protocol for comprehensive evaluation of sulfur-fumigation of Gastrodia Rhizoma using metabolome and health risk assessment analysis. J. Hazard. Mater. 2017, 340, 221–230. [Google Scholar] [CrossRef]
- Chen, W.C.; Lai, Y.S.; Lin, S.H.; Lu, K.H.; Lin, Y.E.; Panyod, S.; Ho, C.T.; Sheen, L.Y. Anti-depressant effects of Gastrodia elata Blume and its compounds gastrodin and 4-hydroxybenzyl alcohol, via the monoaminergic system and neuronal cytoskeletal remodeling. J. Ethnopharmacol. 2016, 182, 190–199. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.L.; Hu, Y.Q.; Liu, L.L.; Cai, J.L.; Peng, C.; Li, Q.L. Gastrodin protects against MPP+-induced oxidative stress by up regulates heme oxygenase-1 expression through p38 MAPK/Nrf2 pathway in human dopaminergic cells. Neurochem. Int. 2014, 75, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Kim, S.W.; Lee, H.K.; Kim, I.D.; Lee, H.; Lee, J.K. Anti-oxidative effects of 4-hydroxybenzyl alcohol in astrocytes confer protective effects in autocrine and paracrine manners. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Haddadi, R.; Poursina, M.; Zeraati, F.; Nadi, F. Gastrodin microinjection suppresses 6-OHDA-induced motor impairments in parkinsonian rats: Insights into oxidative balance and microglial activation in SNc. Inflammopharmacology 2018, 26, 1305–1316. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ran, R.; Huang, G.L. Study on the best initial processing technology of Gastrodia elata. Pharm. Chem. J. 2018, 52, 224–230. [Google Scholar] [CrossRef]
- Zhang, M.H.; Fan, L.; Gao, T.T. A comparison of extraction techniques on gastrodin from Gastrodia elata by response surface methodology and orthogonal design. J. Gansu Agric. Univ. 2014, 49, 60–65. [Google Scholar]
- Xu, D.Q.; Zhou, J.J.; Liu, Y.H. Research the extraction and detection method of gastrodin content in Gastrodia elata. J. Chin. Med. Mater. 2012, 35, 1799–1804. [Google Scholar]
- Yue, L.; Zhang, F.; Wang, Z.X. Study on ultrasonic extraction of gastrodin from Gastrodia elata Bl. Sep. Sci. Technol. 2010, 45, 832–838. [Google Scholar] [CrossRef]
- Shan, M.Q.; Zhang, L.; Yu, S.; Qian, Y.; Wang, J.Y.; Ding, A.W. Simultaneous determination of eight active components in Gastrodiae Rhizoma by HPLC-MS. Chin. Tradit. Herbal Drugs 2015, 46, 2087–2091. [Google Scholar]
- Ku, Y.R.; Liu, Y.C.; Hau, J.P.; Wen, K.C.; Lin, J.H.; Huang, W.F. Determination of parishin, parishins B and C in Gastrodiae Rhizoma by HPLC. J. Food Drug Anal. 1995, 3, 287–293. [Google Scholar]
- Liu, Z.H.; Wang, W.P.; Feng, N.; Wang, L.; Shi, J.G.; Wang, X.L. Parishin C’s prevention of A β(1-42)-induced inhibition of long-term potentiation is related to NMDA receptors. Acta Pharm. Sin. B 2016, 6, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.F.; Sun, Y.J.; Weng, Y.F.; Matsuura, A.; Xiang, L.; Qi, J.H. Parishin from Gastrodia elata extends the lifespan of yeast via regulation of Sir2/Uth1/TOR signaling pathway. Oxid. Med. Cell. Longev. 2016. [Google Scholar] [CrossRef] [PubMed]
- Shin, E.J.; Whang, W.K.; Kim, S.; Bach, J.H.; Kim, J.M.; Nguyen, X.K.T.; Nguyen, T.T.L.; Jung, B.D.; Yamada, K.; Nabeshima, T.; et al. Parishin C attenuates phencyclidine-induced schizophrenia-like psychosis in mice: Involvements of 5-HT1A receptor. J. Pharmacol. Sci. 2010, 113, 404–408. [Google Scholar] [CrossRef] [PubMed]
- Berek, L.; Szabo, D.; Petri, I.B.; Shoyama, Y.; Lin, Y.H.; Molnar, J. Effects of naturally occurring glucosides, solasodine glucosides, ginsenosides and parishinderivatives on multidrug resistance of lymphoma cells and leukocyte functions. In Vivo 2001, 15, 151–156. [Google Scholar] [PubMed]
- Tang, C.L.; Wang, L.; Liu, X.X.; Cheng, M.C.; Qu, Y.; Xiao, H.B. Comparative pharmacokinetics of gastrodin in rats after intragastric administration of free gastrodin, parishin and Gastrodia elata extract. J. Ethnopharmacol. 2015, 176, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.L.; Wang, L.; Li, J.J.; Liu, X.X.; Cheng, M.C.; Xiao, H.B. Analysis of the metabolic profile of parishin by ultra-performance liquid chromatography/quadrupole-time of flight mass spectrometry. Biomed. Chromatogr. 2015, 29, 1913–1920. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Hwang, S.M.; Kang, D.G.; Kim, J.S.; Lee, H.S. Effect of Gastrodia elata on tumor necrosis factor-alpha-induced matrix metalloproteinase activity in endothelial cells. J. Nat. Med. 2009, 63, 463–467. [Google Scholar] [CrossRef]
- Hwang, S.M.; Lee, Y.J.; Kang, D.G.; Lee, H.S. Anti-Inflammatory effect of Gastrodia elata rhizome in human umbilical vein endothelial cells. Am. J. Chin. Med. 2009, 37, 395–406. [Google Scholar] [CrossRef]
- Seok, P.R.; Kim, J.H.; Kwon, H.R.; Heo, J.S.; Choi, J.R.; Shin, J.H. Protective effects of Gastrodia elata Blume on acetaminophen-induced liver and kidney toxicity in rats. Food Sci. Biotechnol. 2018, 27, 1445–1454. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, Y.P.; Jin, L.X. Preparation, characterization and anti-ageing activity of Gastrodia elata blume polysaccharide. Acta Aliment. 2018, 47, 210–219. [Google Scholar] [CrossRef]
- Shi, A.H.; Xiang, J.M.; He, F.Y.; Zhu, Y.P.; Zhu, G.B.; Lin, Y.H.; Zhou, N.N. The phenolic components of Gastrodia elata improve prognosis in rats after cerebral ischemia/reperfusion by enhancing the endogenous antioxidant mechanisms. Oxid. Med. Cell. Longev. 2018. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Zhang, W.J.; Zhang, J.C.; Yang, M.; Zhang, F.X.; Zhang, J.P. Anti-hypertensive effect of Gastrodia elata Bl leaf extract in rats. Trop. J. Pharm. Res. 2017, 16, 1887–1891. [Google Scholar] [CrossRef]
- Huang, J.Y.; Yuan, Y.H.; Yan, J.Q.; Wang, Y.N.; Chu, S.F.; Zhu, C.G.; Guo, Q.L.; Shi, J.G.; Chen, N.H. 20C, a bibenzyl compound isolated from Gastrodia elata, protects PC12 cells against rotenone-induced apoptosis via activation of the Nrf2/ARE/HO-1 signaling pathway. Acta Pharmacol. Sin. 2016, 37, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.S.; Choi, D.K.; Jung, H.J. Neuroprotective Effects of vanillyl alcohol in Gastrodia elata Blume through suppression of oxidative stress and anti-apoptotic activity in toxin-induced dopaminergic MN9D cells. Molecules 2011, 16, 5349–5361. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.J.; Kim, J.R.; Lee, C.K.; Han, J.E.; Lee, J.H.; Kim, H.S.; Hong, J.H.; Kang, S.G. Gastrodia elata blume and an active component, p-hydroxybenzyl alcohol reduce focal ischemic brain injury through antioxidant related gene expressions. Biol. Pharm. Bull. 2005, 28, 1016–1020. [Google Scholar] [CrossRef] [PubMed]
- Granato, D.; Grevink, R.; Zielinski, A.A.F.; Nunes, D.S.; van Ruth, S.M. Analytical strategy coupled with Response Surface Methodology to maximize the extraction of antioxidants from ternary mixtures of Green, Yellow and Red Teas (Camellia sinensis var. sinensis). J. Agric. Food Chem. 2014, 62, 10283–10296. [Google Scholar] [CrossRef] [PubMed]
- Pinho, C.; Melo, A.; Mansilha, C.; Ferreira, I.M.P.L.V.O. Optimization of conditions for anthocyanin hydrolysis from red wine using Response Surface Methodology (RSM). J. Agric. Food Chem. 2011, 59, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Sharif, I.; Adewale, P.; Dalli, S.S.; Rakshit, S. Microwave pretreatment and optimization of osmotic dehydration of wild blueberries using response surface methodology. Food Chem. 2018, 269, 300–310. [Google Scholar] [CrossRef] [PubMed]
- Masghati, S.; Ghoreishi, S.M. Supercritical CO2 extraction of cinnamaldehyde and eugenol from cinnamon bark: Optimization of operating conditions via response surface methodology. J. Supercrit. Fluid 2018, 140, 62–71. [Google Scholar] [CrossRef]
- Yoon, J.H.; Kim, J.H.; Ham, S.S.; Gang, B.Y.; Lee, S.H.; Choi, G.; Kim, Y.S.; Lee, G.; Ju, Y.S. Optimal processing conditions of Boswellia carteri Birdw. using Response Surface Methodology. Phamacogn. Mag. 2018, 14, 235–241. [Google Scholar]
- Ha, W.R.; Park, J.H.; Kim, J.H. Optimization for decocting later of Menthae Herba in Eungyo-San, a herbal formula, using Response Surface Methodology with gas chromatography/mass spectrometry. Phamacogn. Mag. 2018, 14, 17–21. [Google Scholar]
- Kumarasamy, Y.; Byres, M.; Cox, P.J.; Jaspars, M.; Nahar, L.; Sarker, S.D. Screening seeds of some Scottish plants for free radical scavenging activity. Phytother. Res. 2007, 21, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.L.; Chen, Y.L.; Liao, L.Z.; Wu, W.K. Resveratrol protects HUVECs from oxidized-LDL induced oxidative damage by autophagy upregulation via the AMPK/SIRT1 pathway. Cardiovasc. Drugs Ther. 2013, 27, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.C.; Tao, L.; Li, W.K.; Zhang, Y.Y.; Luo, H.; Xia, Y.Y. Evidence-based antioxidant activity of the essential oil from Fructus A. zerumbet on cultured human umbilical vein endothelial cells’ injury induced by ox-LDL. BMC Complemt. Altern. Med. 2012, 12. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |
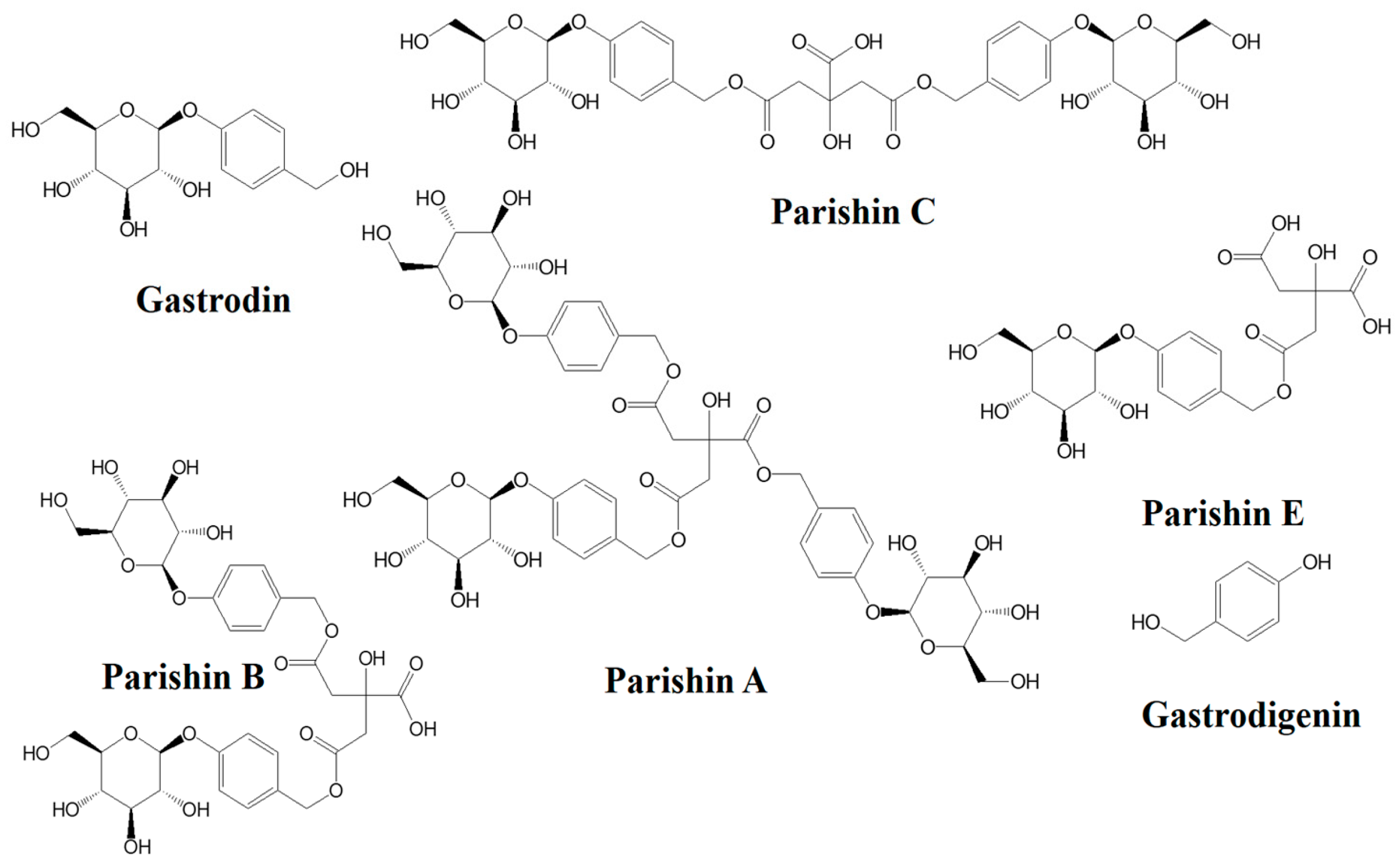
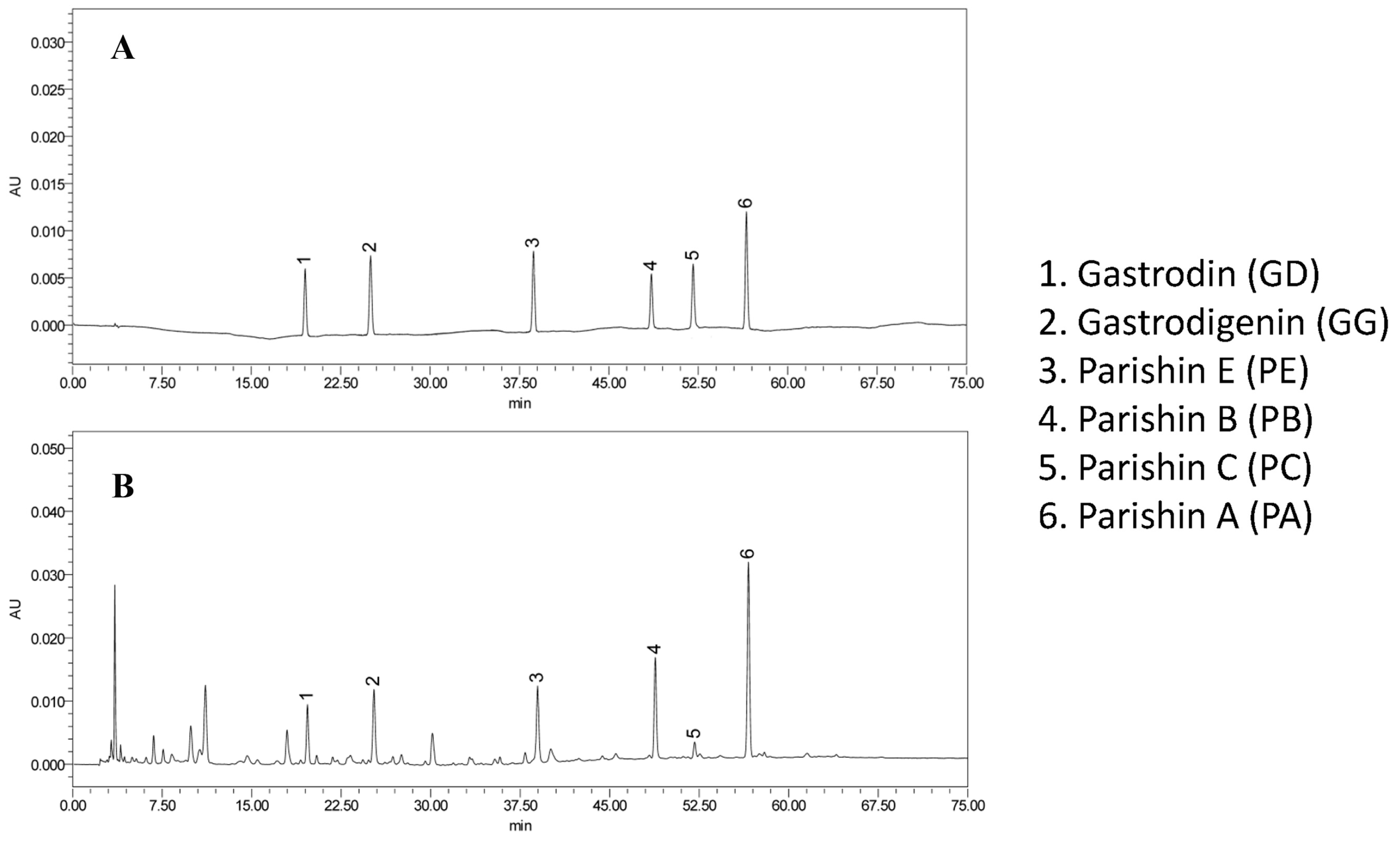
| Analyte | Regression Equation | Linear Range (μg/mL) | R2 | LOD (μg/mL) | LOQ (μg/mL) |
|---|---|---|---|---|---|
| Gastrodin (GD) | Y = 1781.62X + 1448.30 | 5.21–166.64 | 0.9997 | 2.19 | 6.95 |
| Gastrodigenin (GG) | Y = 7335.04X − 1097.67 | 2.15–68.68 | 0.9999 | 1.38 | 4.91 |
| Parishin E (PE) | Y = 1265.20X − 1840.13 | 9.75–312.00 | 0.9999 | 0.47 | 1.72 |
| Parishin B (PB) | Y = 1842.78X − 6412.23 | 8.42–269.28 | 0.9993 | 3.76 | 13.80 |
| Parishin C (PC) | Y = 764.82X − 70.51 | 5.04–161.36 | 0.9999 | 2.81 | 6.43 |
| Parishin A (PA) | Y = 956.37X − 6952.21 | 23.83–762.56 | 0.9997 | 3.92 | 10.60 |
| Analyte | Precision | Stability RSD (%) | Repeatability RSD (%) | Recovery | ||
|---|---|---|---|---|---|---|
| Intra-Day RSD (%) | Inter-Day RSD (%) | Mean (%) | RSD (%) | |||
| GD | 2.42 | 2.89 | 2.01 | 2.28 | 98.15 | 1.60 |
| GG | 1.06 | 2.06 | 1.22 | 1.61 | 98.09 | 2.32 |
| PE | 1.22 | 3.27 | 1.03 | 1.21 | 99.72 | 2.14 |
| PB | 2.65 | 2.80 | 1.89 | 2.75 | 101.72 | 1.68 |
| PC | 2.15 | 1.95 | 2.45 | 2.03 | 96.99 | 0.92 |
| PA | 2.26 | 3.04 | 2.30 | 2.23 | 99.57 | 2.25 |
| No. | X1 | X2 | X3 | X4 | GD (mg/g) | GG (mg/g) | PE (mg/g) | PB (mg/g) | PC (mg/g) | PA (mg/g) | IC50 (mg/mL) | SMTT (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 1 | 0 | 1 | 1.88 | 0.57 | 2.79 | 3.85 | 1.43 | 13.75 | 37.80 | 29.34 |
| 2 | −1 | 0 | 1 | 0 | 1.09 | 0.43 | 2.90 | 2.20 | 0.81 | 7.00 | 26.05 | 38.65 |
| 3 | 0 | 0 | 1 | 1 | 1.74 | 0.63 | 4.66 | 3.65 | 1.39 | 13.59 | 28.11 | 30.87 |
| 4 | 0 | 0 | −1 | −1 | 1.91 | 0.61 | 5.05 | 3.98 | 1.47 | 14.32 | 32.68 | 34.22 |
| 5 | −1 | 0 | 0 | −1 | 1.64 | 0.65 | 4.30 | 3.27 | 1.20 | 10.79 | 29.92 | 35.75 |
| 6 | −1 | 0 | 0 | 1 | 1.57 | 0.23 | 4.26 | 3.30 | 1.26 | 9.88 | 28.27 | 33.28 |
| 7 | 0 | 0 | 1 | −1 | 1.83 | 0.65 | 4.87 | 3.83 | 1.45 | 14.02 | 28.93 | 28.52 |
| 8 | 0 | 0 | −1 | 1 | 1.73 | 0.62 | 4.64 | 3.68 | 1.34 | 13.4 | 30.31 | 32.74 |
| 9 | 1 | 0 | 0 | −1 | 1.73 | 0.63 | 3.17 | 2.84 | 0.80 | 10.71 | 29.61 | 36.81 |
| 10 | 0 | 0 | 0 | 0 | 1.84 | 0.65 | 4.93 | 3.86 | 1.43 | 14.27 | 22.07 | 37.63 |
| 11 | 1 | −1 | 0 | 0 | 1.64 | 0.61 | 2.73 | 2.89 | 0.83 | 11.12 | 34.26 | 22.15 |
| 12 | 0 | 1 | −1 | 0 | 1.85 | 0.65 | 4.95 | 3.96 | 1.45 | 14.29 | 36.24 | 23.58 |
| 13 | 1 | 0 | −1 | 0 | 1.53 | 0.12 | 2.81 | 2.67 | 0.74 | 10.14 | 28.97 | 28.30 |
| 14 | 1 | 0 | 0 | 1 | 1.74 | 0.62 | 3.66 | 3.44 | 1.06 | 12.47 | 26.3 | 35.37 |
| 15 | −1 | 1 | 0 | 0 | 1.60 | 0.58 | 4.18 | 3.16 | 1.17 | 10.57 | 31.76 | 22.81 |
| 16 | 0 | 0 | 0 | 0 | 1.94 | 0.61 | 5.01 | 3.88 | 1.48 | 14.39 | 21.87 | 38.62 |
| 17 | 0 | −1 | 0 | −1 | 1.76 | 0.62 | 4.69 | 3.76 | 1.35 | 13.61 | 34.55 | 29.55 |
| 18 | 0 | −1 | 1 | 0 | 1.55 | 0.55 | 4.13 | 3.26 | 1.21 | 12.18 | 39.91 | 28.23 |
| 19 | −1 | 0 | −1 | 0 | 1.44 | 0.55 | 3.90 | 3.08 | 1.13 | 10.37 | 27.04 | 29.91 |
| 20 | 0 | 0 | 0 | 0 | 1.86 | 0.67 | 4.96 | 3.83 | 1.42 | 14.26 | 21.23 | 38.28 |
| 21 | 0 | 0 | 0 | 0 | 1.69 | 0.62 | 4.57 | 3.59 | 1.30 | 13.14 | 22.08 | 32.14 |
| 22 | 0 | 1 | 0 | −1 | 1.62 | 0.49 | 4.24 | 3.45 | 1.26 | 11.85 | 38.77 | 24.55 |
| 23 | 0 | 0 | 0 | 0 | 1.86 | 0.59 | 4.84 | 3.76 | 1.40 | 13.92 | 21.58 | 36.16 |
| 24 | 0 | −1 | −1 | 0 | 1.62 | 0.59 | 4.27 | 3.38 | 1.22 | 12.66 | 44.65 | 18.13 |
| 25 | 0 | −1 | 0 | 1 | 1.71 | 0.63 | 4.55 | 3.62 | 1.31 | 13.21 | 42.43 | 24.20 |
| 26 | −1 | −1 | 0 | 0 | 0.81 | 0.32 | 2.16 | 1.67 | 0.60 | 5.53 | 35.93 | 25.01 |
| 27 | 0 | 1 | 1 | 0 | 1.80 | 0.65 | 4.86 | 3.90 | 1.35 | 14.04 | 41.54 | 25.66 |
| 28 | 1 | 0 | 1 | 0 | 1.73 | 0.55 | 3.18 | 3.12 | 0.86 | 11.53 | 29.56 | 36.61 |
| 29 | 1 | 1 | 0 | 0 | 1.64 | 0.64 | 3.37 | 3.10 | 0.93 | 11.24 | 33.75 | 24.90 |
| No. | Y1 | Y2 | Y3 | Y | No. | Y1 | Y2 | Y3 | Y |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.7996 | 0.7075 | 0.5466 | 0.6387 | 16 | 0.9723 | 0.0273 | 0.9986 | 1.9436 |
| 2 | 0.2836 | 0.2058 | 1.0000 | 1.0778 | 17 | 0.8824 | 0.5687 | 0.5568 | 0.8704 |
| 3 | 0.8800 | 0.2938 | 0.6208 | 1.2070 | 18 | 0.7084 | 0.7976 | 0.4924 | 0.4032 |
| 4 | 0.9742 | 0.4889 | 0.7844 | 1.2696 | 19 | 0.6167 | 0.2481 | 0.5741 | 0.9427 |
| 5 | 0.7345 | 0.3711 | 0.8586 | 1.2220 | 20 | 0.9584 | 0.0000 | 0.9821 | 1.9405 |
| 6 | 0.5910 | 0.3006 | 0.7386 | 1.0290 | 21 | 0.8346 | 0.0363 | 0.6831 | 1.4813 |
| 7 | 0.9439 | 0.3288 | 0.5065 | 1.1216 | 22 | 0.7239 | 0.7489 | 0.3131 | 0.2880 |
| 8 | 0.8634 | 0.3877 | 0.7119 | 1.1876 | 23 | 0.9120 | 0.0149 | 0.8786 | 1.7756 |
| 9 | 0.5682 | 0.3578 | 0.9103 | 1.1207 | 24 | 0.7585 | 1.0000 | 0.0000 | −0.2415 |
| 10 | 0.9519 | 0.0359 | 0.9505 | 1.8666 | 25 | 0.8448 | 0.9052 | 0.2960 | 0.2356 |
| 11 | 0.5405 | 0.5564 | 0.1959 | 0.1801 | 26 | 0.0606 | 0.6277 | 0.3354 | −0.2317 |
| 12 | 0.9659 | 0.6409 | 0.2658 | 0.5907 | 27 | 0.9254 | 0.8672 | 0.3669 | 0.4251 |
| 13 | 0.3291 | 0.3305 | 0.4955 | 0.4941 | 28 | 0.5915 | 0.3557 | 0.9009 | 1.1368 |
| 14 | 0.7206 | 0.2165 | 0.8403 | 1.3444 | 29 | 0.6229 | 0.5346 | 0.3300 | 0.4183 |
| 15 | 0.6827 | 0.4496 | 0.2281 | 0.4612 |
| Source | Sum of Squares | Degree of Freedom | Mean Square | F-Value | p-Value |
|---|---|---|---|---|---|
| Model | 9.6300 | 14 | 0.6900 | 17.61 | <0.0001 |
| X1 | 0.0031 | 1 | 0.0031 | 0.08 | 0.7817 |
| X2 | 0.2100 | 1 | 0.2100 | 5.5 | 0.0342 |
| X3 | 0.1100 | 1 | 0.1100 | 2.72 | 0.1215 |
| X4 | 0.0052 | 1 | 0.0052 | 0.13 | 0.7204 |
| X1X2 | 0.0520 | 1 | 0.0520 | 1.32 | 0.2692 |
| X1X3 | 0.0640 | 1 | 0.0640 | 1.65 | 0.2199 |
| X1X4 | 0.0430 | 1 | 0.0430 | 1.11 | 0.3096 |
| X2X3 | 0.1600 | 1 | 0.1600 | 4.2 | 0.0595 |
| X2X4 | 0.2400 | 1 | 0.2400 | 6.22 | 0.0258 |
| X3X4 | 0.0070 | 1 | 0.0070 | 0.18 | 0.6783 |
| X12 | 1.4200 | 1 | 1.4200 | 36.33 | <0.0001 |
| X22 | 8.0300 | 1 | 8.0300 | 205.52 | <0.0001 |
| X32 | 1.1200 | 1 | 1.1200 | 28.64 | 0.0001 |
| X42 | 0.2000 | 1 | 0.2000 | 5.1 | 0.0404 |
| Residual | 0.5500 | 14 | 0.0390 | ||
| Lack of Fit | 0.4000 | 10 | 0.0400 | 1.09 | 0.5101 |
| Pure Error | 0.1500 | 4 | 0.0370 | ||
| Cor Total | 10.1700 | 28 | |||
| R2 | 0.9463 | ||||
| Adjusted R2 | 0.8925 | ||||
| Predicted R2 | 0.7511 | ||||
| Adequate precision | 13.777 |
| No. | GD (mg/g) | GG (mg/g) | PE (mg/g) | PB (mg/g) | PC (mg/g) | PA (mg/g) | IC50 (mg/mL) | SMTT (%) |
|---|---|---|---|---|---|---|---|---|
| 1 | 1.946 | 0.676 | 4.335 | 4.844 | 1.428 | 14.050 | 24.50 | 37.44 |
| 2 | 1.946 | 0.707 | 4.310 | 5.070 | 1.370 | 13.958 | 24.04 | 37.15 |
| 3 | 1.945 | 0.688 | 4.277 | 5.082 | 1.416 | 14.054 | 24.68 | 37.21 |
| Independent Variables | Coded Symbols | Levels | ||
|---|---|---|---|---|
| −1 | 0 | 1 | ||
| Ethanol concentration (%) | X1 | 20 | 40 | 60 |
| Liquid-solid ratio (mL/g) | X2 | 16:1 | 28:1 | 40:1 |
| Extraction time (min) | X3 | 30 | 45 | 60 |
| Soaking time (h) | X4 | 12 | 24 | 36 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, M.; Yan, H.; Fu, Y.; Jiang, Y.; Yao, W.; Yu, S.; Zhang, L.; Wu, Q.; Ding, A.; Shan, M. Optimal Extraction Study of Gastrodin-Type Components from Gastrodia Elata Tubers by Response Surface Design with Integrated Phytochemical and Bioactivity Evaluation. Molecules 2019, 24, 547. https://doi.org/10.3390/molecules24030547
Hu M, Yan H, Fu Y, Jiang Y, Yao W, Yu S, Zhang L, Wu Q, Ding A, Shan M. Optimal Extraction Study of Gastrodin-Type Components from Gastrodia Elata Tubers by Response Surface Design with Integrated Phytochemical and Bioactivity Evaluation. Molecules. 2019; 24(3):547. https://doi.org/10.3390/molecules24030547
Chicago/Turabian StyleHu, Minhui, Hui Yan, Yuanyuan Fu, Yulan Jiang, Weifeng Yao, Sheng Yu, Li Zhang, Qinan Wu, Anwei Ding, and Mingqiu Shan. 2019. "Optimal Extraction Study of Gastrodin-Type Components from Gastrodia Elata Tubers by Response Surface Design with Integrated Phytochemical and Bioactivity Evaluation" Molecules 24, no. 3: 547. https://doi.org/10.3390/molecules24030547
APA StyleHu, M., Yan, H., Fu, Y., Jiang, Y., Yao, W., Yu, S., Zhang, L., Wu, Q., Ding, A., & Shan, M. (2019). Optimal Extraction Study of Gastrodin-Type Components from Gastrodia Elata Tubers by Response Surface Design with Integrated Phytochemical and Bioactivity Evaluation. Molecules, 24(3), 547. https://doi.org/10.3390/molecules24030547






