Structural Asymmetry and Kinetic Limping of Single Rotary F-ATP Synthases
Abstract
:1. Introduction
2. Asymmetric Elements in the F-ATP Synthase
2.1. The F1 Domain
2.2. The FO Domain
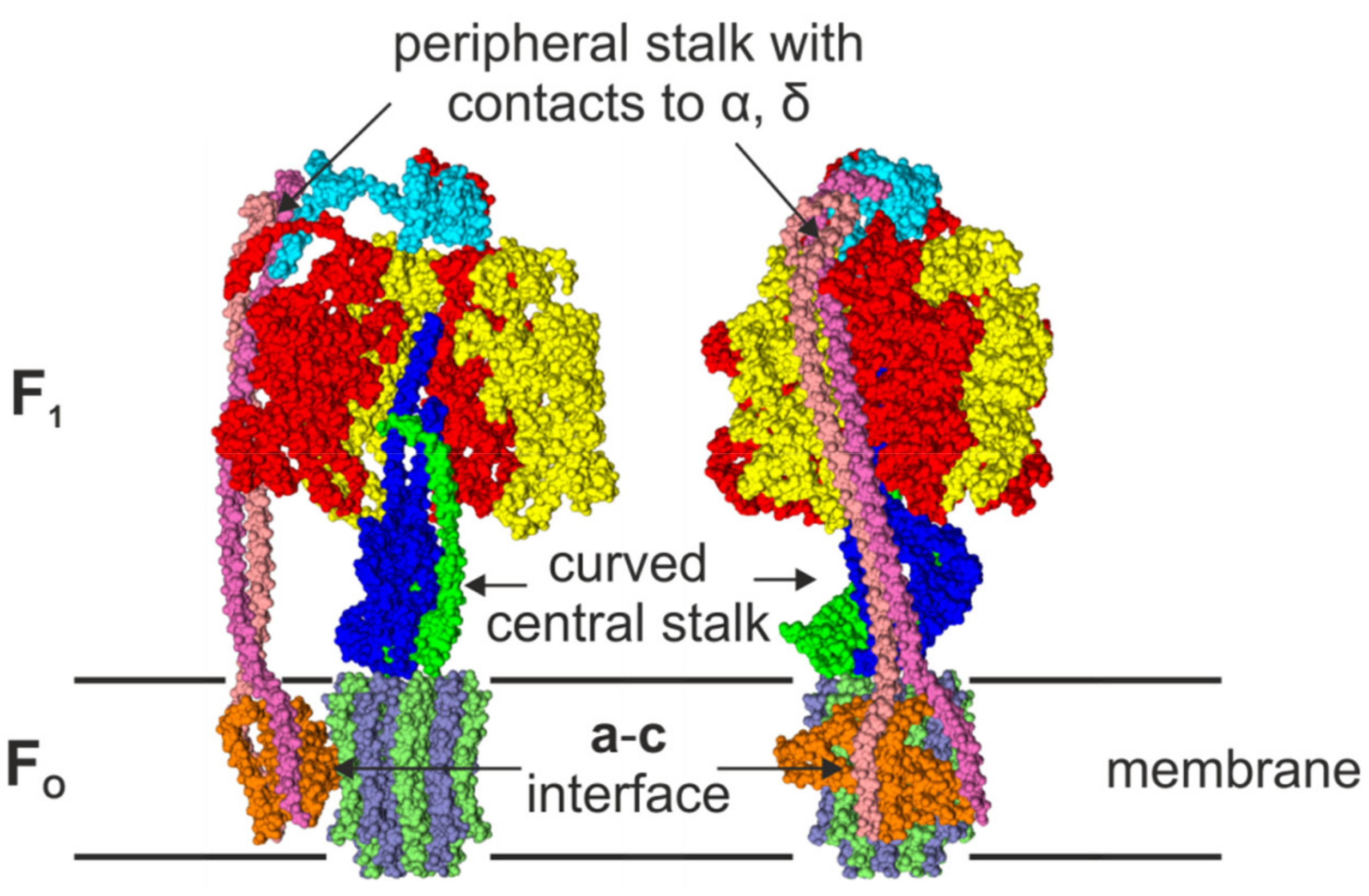
2.3. The Peripheral Stalk
2.4. The Central Stalk
2.5. Rotational Catalysis
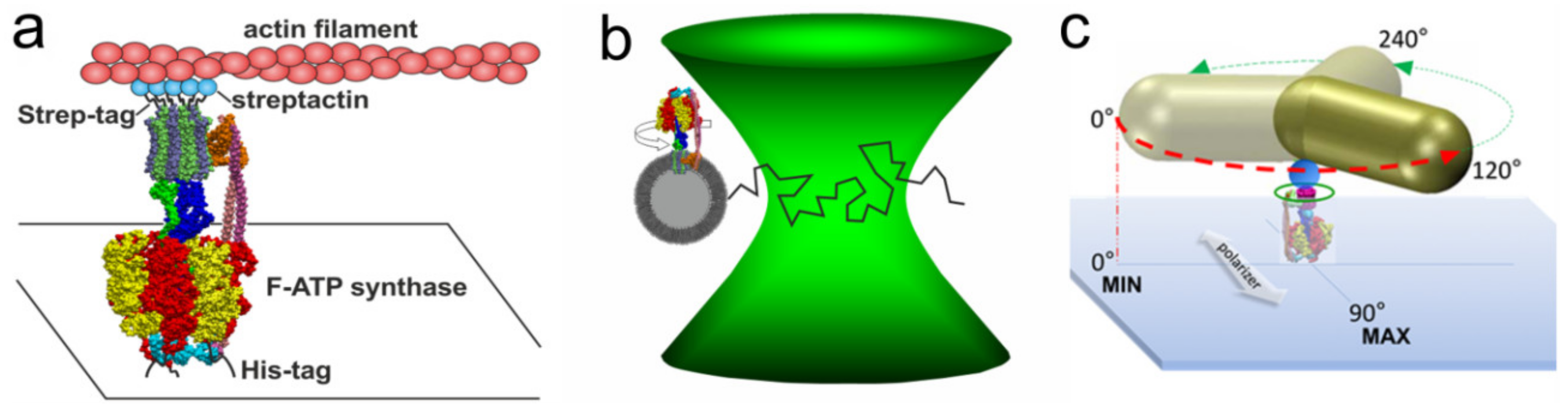
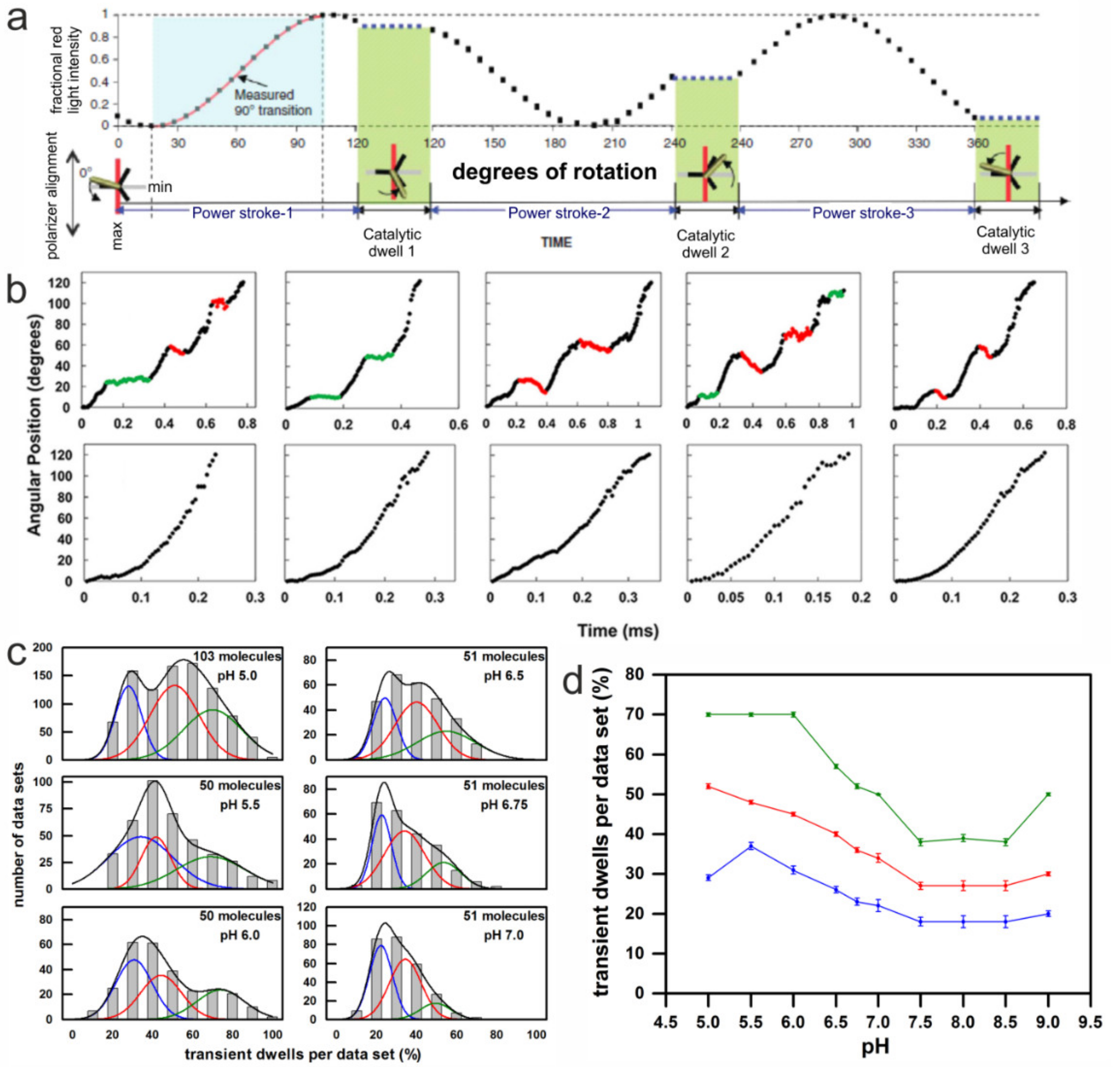
3. Single Molecule Rotation Experiments
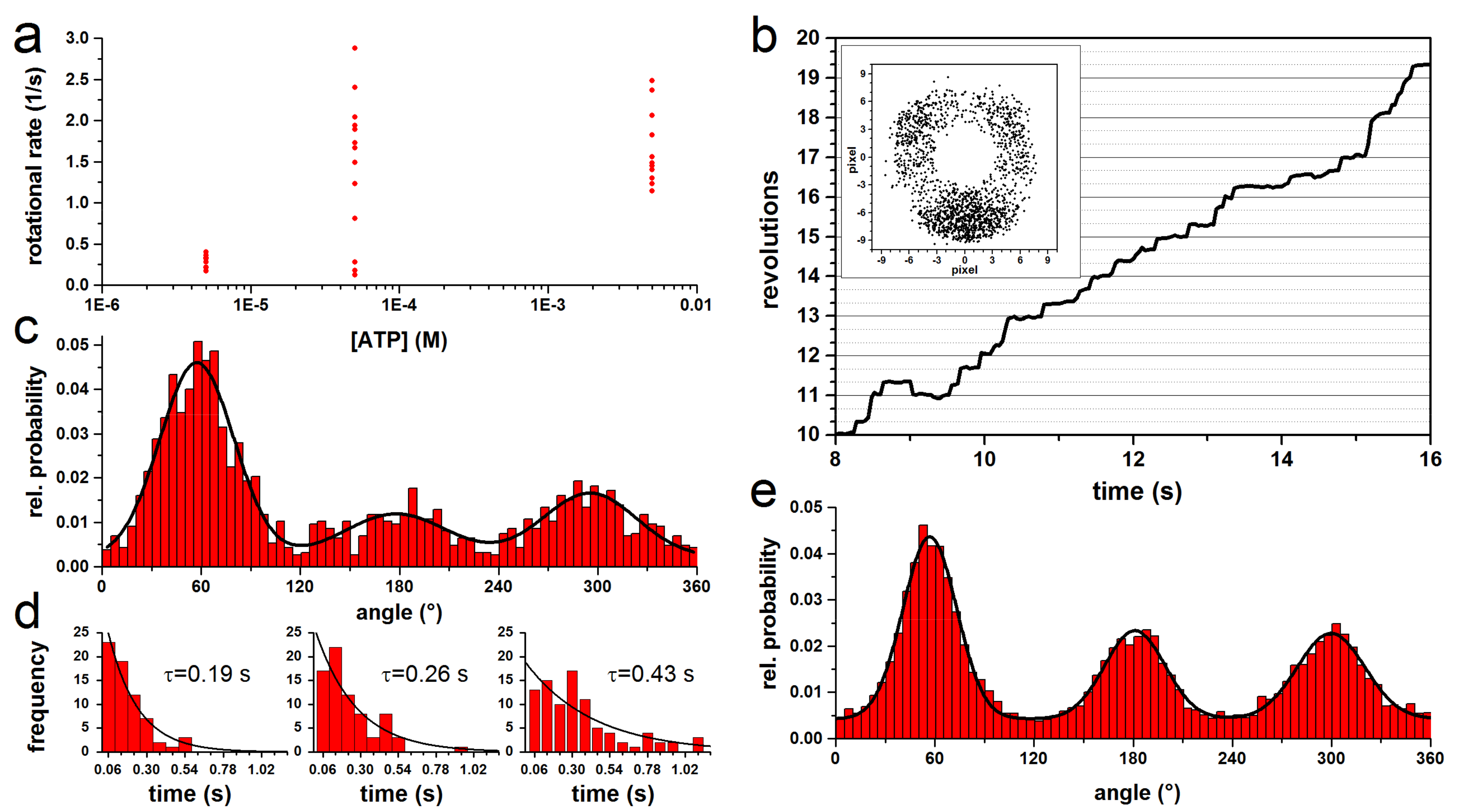
3.1. Rotation Experiments with Actin Filaments
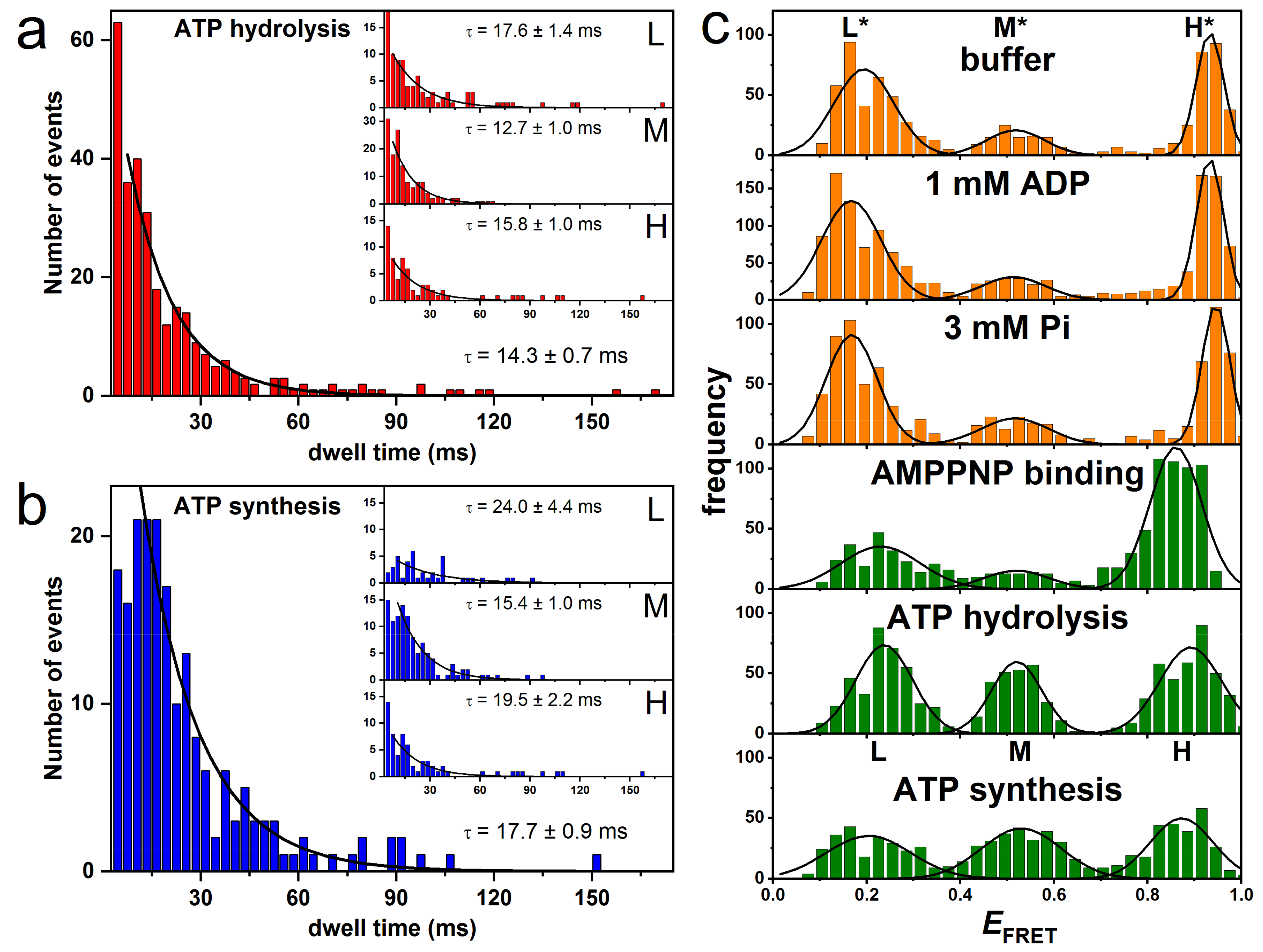
3.2. Rotation Experiments with smFRET
3.3. Rotation Experiments with Gold Nanorods
4. Discussion
4.1. Asymmetry Corroborated from Single-Molecule Rotation Experiments
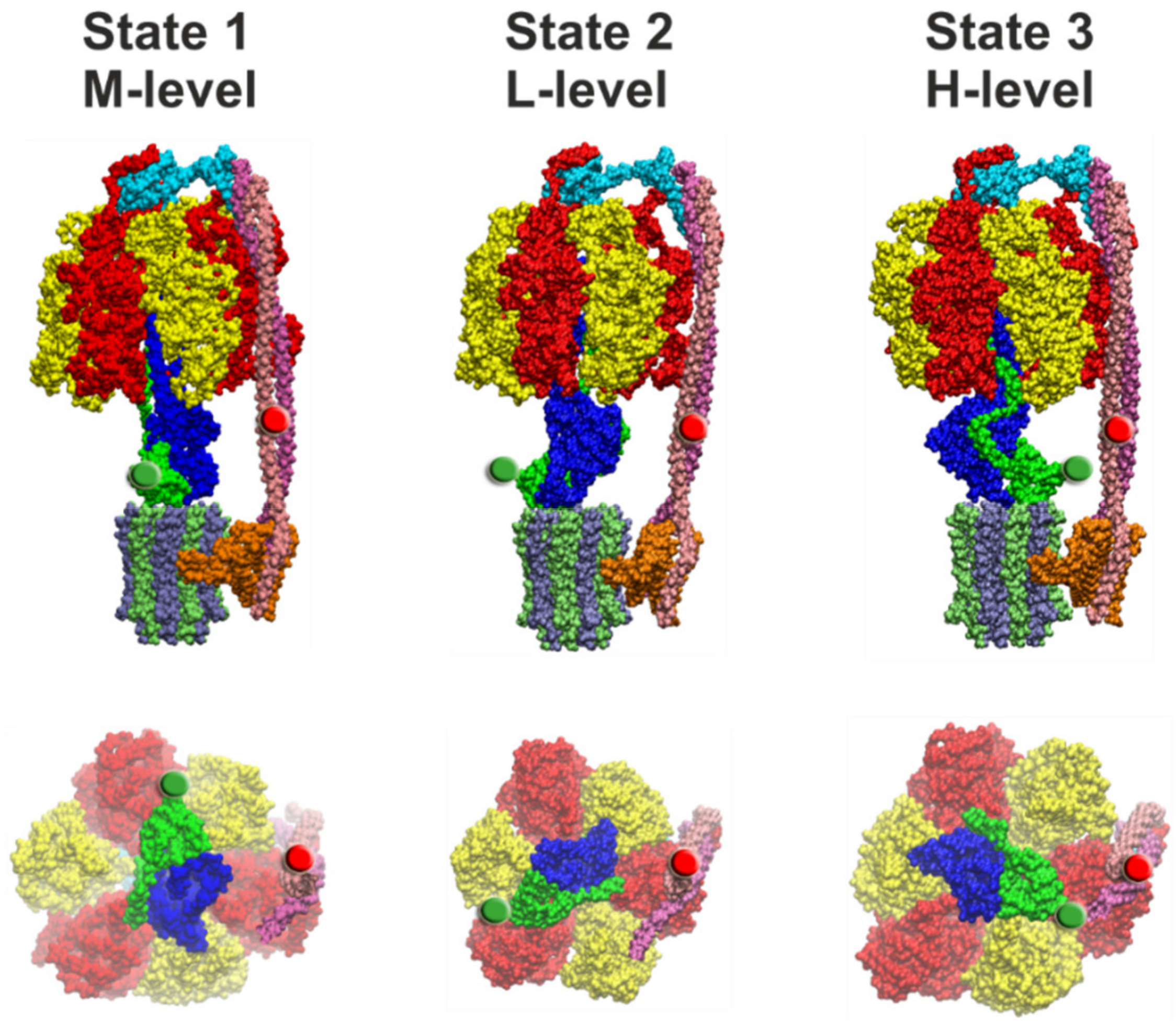
4.2. Comparison of Cryo-EM Structures
4.3. Correlation of Cryo-EM Structures with smFRET Data
4.4. Asymmetry in c-Ring Rotation
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Boyer, P.D. ATP synthase-past and future. Biochim. Biophys. Acta 1998, 1365, 3–9. [Google Scholar] [CrossRef]
- Senior, A.E.; Weber, J. Happy motoring with ATP synthase. Nat. Struct. Mol. Biol. 2004, 11, 110–112. [Google Scholar] [CrossRef] [PubMed]
- Junge, W.; Sielaff, H.; Engelbrecht, S. Torque generation and elastic power transmission in the rotary FOF1-ATPase. Nature 2009, 459, 364–370. [Google Scholar] [CrossRef] [PubMed]
- Von Ballmoos, C.; Wiedenmann, A.; Dimroth, P. Essentials for ATP synthesis by F1F0 ATP synthases. Annu. Rev. Biochem. 2009, 78, 649–672. [Google Scholar] [CrossRef] [PubMed]
- Sielaff, H.; Börsch, M. Twisting and subunit rotation in single FOF1-ATP synthase. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20120024. [Google Scholar] [CrossRef]
- Walker, J.E. The ATP synthase: The understood, the uncertain and the unknown. Biochem. Soc. Trans. 2013, 41, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Junge, W.; Nelson, N. ATP synthase. Annu. Rev. Biochem. 2015, 84, 631–657. [Google Scholar] [CrossRef]
- Noji, H.; Ueno, H.; McMillan, D.G.G. Catalytic robustness and torque generation of the F1-ATPase. Biophys. Rev. 2017, 9, 103–118. [Google Scholar] [CrossRef]
- Girvin, M.E.; Rastogi, V.K.; Abildgaard, F.; Markley, J.L.; Fillingame, R.H. Solution structure of the transmembrane H+-transporting subunit c of the F1FO ATP synthase. Biochemistry 1998, 37, 8817–8824. [Google Scholar] [CrossRef]
- Dmitriev, O.; Jones, P.C.; Jiang, W.; Fillingame, R.H. Structure of the membrane domain of subunit b of the Escherichia coli FOF1 ATP synthase. J. Biol. Chem. 1999, 274, 15598–15604. [Google Scholar] [CrossRef]
- Cingolani, G.; Duncan, T.M. Structure of the ATP synthase catalytic complex (F1) from Escherichia coli in an autoinhibited conformation. Nat. Struct. Mol. Biol. 2011, 18, 701–707. [Google Scholar] [CrossRef] [PubMed]
- Sobti, M.; Smits, C.; Wong, A.S.W.; Ishmukhametov, R.; Stock, D.; Sandin, S.; Stewart, A.G. Cryo-EM structures of the autoinhibited E. coli ATP synthase in three rotational states. eLife 2016, 5, 21598. [Google Scholar] [CrossRef] [PubMed]
- Abrahams, J.P.; Leslie, A.G.; Lutter, R.; Walker, J.E. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature 1994, 370, 621–628. [Google Scholar] [CrossRef]
- Abrahams, J.P.; Buchanan, S.K.; Van Raaij, M.J.; Fearnley, I.M.; Leslie, A.G.; Walker, J.E. The structure of bovine F1-ATPase complexed with the peptide antibiotic efrapeptin. Proc. Natl. Acad. Sci. USA 1996, 93, 9420–9424. [Google Scholar] [CrossRef] [PubMed]
- Van Raaij, M.J.; Orriss, G.L.; Montgomery, M.G.; Runswick, M.J.; Fearnley, I.M.; Skehel, J.M.; Walker, J.E. The ATPase inhibitor protein from bovine heart mitochondria: The minimal inhibitory sequence. Biochemistry 1996, 35, 15618–15625. [Google Scholar] [CrossRef] [PubMed]
- Orriss, G.L.; Leslie, A.G.; Braig, K.; Walker, J.E. Bovine F1-ATPase covalently inhibited with 4-chloro-7-nitrobenzofurazan: The structure provides further support for a rotary catalytic mechanism. Structure 1998, 6, 831–837. [Google Scholar] [CrossRef]
- Braig, K.; Menz, R.I.; Montgomery, M.G.; Leslie, A.G.; Walker, J.E. Structure of bovine mitochondrial F1-ATPase inhibited by Mg2+ ADP and aluminium fluoride. Structure 2000, 8, 567–573. [Google Scholar] [CrossRef]
- Gibbons, C.; Montgomery, M.G.; Leslie, A.G.; Walker, J.E. The structure of the central stalk in bovine F1-ATPase at 2.4 Å resolution. Nat. Struct. Biol. 2000, 7, 1055–1061. [Google Scholar]
- Menz, R.I.; Walker, J.E.; Leslie, A.G. Structure of bovine mitochondrial F1-ATPase with nucleotide bound to all three catalytic sites: Implications for the mechanism of rotary catalysis. Cell 2001, 106, 331–341. [Google Scholar] [CrossRef]
- Kagawa, R.; Montgomery, M.G.; Braig, K.; Leslie, A.G.; Walker, J.E. The structure of bovine F1-ATPase inhibited by ADP and beryllium fluoride. EMBO J. 2004, 23, 2734–2744. [Google Scholar] [CrossRef]
- Kabaleeswaran, V.; Puri, N.; Walker, J.E.; Leslie, A.G.; Mueller, D.M. Novel features of the rotary catalytic mechanism revealed in the structure of yeast F1 ATPase. EMBO J. 2006, 25, 5433–5442. [Google Scholar] [CrossRef] [PubMed]
- Bowler, M.W.; Montgomery, M.G.; Leslie, A.G.; Walker, J.E. Ground state structure of F1-ATPase from bovine heart mitochondria at 1.9 Å resolution. J. Biol. Chem. 2007, 282, 14238–14242. [Google Scholar] [CrossRef]
- Guo, H.; Rubinstein, J.L. Cryo-EM of ATP synthases. Curr. Opin. Struct. Biol. 2018, 52, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Zhou, A.; Rohou, A.; Schep, D.G.; Bason, J.V.; Montgomery, M.G.; Walker, J.E.; Grigorieff, N.; Rubinstein, J.L. Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM. eLife 2015, 4, 10180. [Google Scholar] [CrossRef] [PubMed]
- Vinothkumar, K.R.; Montgomery, M.G.; Liu, S.; Walker, J.E. Structure of the mitochondrial ATP synthase from Pichia angusta determined by electron cryo-microscopy. Proc. Natl. Acad. Sci. USA 2016, 113, 12709–12714. [Google Scholar] [CrossRef] [PubMed]
- Klusch, N.; Murphy, B.J.; Mills, D.J.; Yildiz, O.; Kühlbrandt, W. Structural basis of proton translocation and force generation in mitochondrial ATP synthase. eLife 2017, 6, 33274. [Google Scholar] [CrossRef] [PubMed]
- Hahn, A.; Vonck, J.; Mills, D.J.; Meier, T.; Kühlbrandt, W. Structure, mechanism, and regulation of the chloroplast ATP synthase. Science 2018, 360, 4318. [Google Scholar] [CrossRef]
- Srivastava, A.P.; Luo, M.; Zhou, W.; Symersky, J.; Bai, D.; Chambers, M.G.; Faraldo-Gomez, J.D.; Liao, M.; Mueller, D.M. High-resolution cryo-EM analysis of the yeast ATP synthase in a lipid membrane. Science 2018, 360, 9699. [Google Scholar] [CrossRef]
- Guo, H.; Suzuki, T.; Rubinstein, J.L. Structure of a bacterial ATP synthase. bioRxiv 2018. [Google Scholar] [CrossRef]
- Dunn, S.D.; McLachlin, D.T.; Revington, M. The second stalk of Escherichia coli ATP synthase. Biochim. Biophys. Acta 2000, 1458, 356–363. [Google Scholar] [CrossRef]
- Del Rizzo, P.A.; Bi, Y.; Dunn, S.D. ATP synthase b subunit dimerization domain: A right-handed coiled coil with offset helices. J. Mol. Biol. 2006, 364, 735–746. [Google Scholar] [CrossRef] [PubMed]
- Colina-Tenorio, L.; Dautant, A.; Miranda-Astudillo, H.; Giraud, M.F.; Gonzalez-Halphen, D. The Peripheral Stalk of Rotary ATPases. Front. Physiol. 2018, 9, 1243. [Google Scholar] [CrossRef] [PubMed]
- Dickson, V.K.; Silvester, J.A.; Fearnley, I.M.; Leslie, A.G.; Walker, J.E. On the structure of the stator of the mitochondrial ATP synthase. EMBO J. 2006, 25, 2911–2918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rees, D.M.; Leslie, A.G.; Walker, J.E. The structure of the membrane extrinsic region of bovine ATP synthase. Proc. Natl. Acad. Sci. USA 2009, 106, 21597–21601. [Google Scholar] [CrossRef] [PubMed]
- Wächter, A.; Bi, Y.; Dunn, S.D.; Cain, B.D.; Sielaff, H.; Wintermann, F.; Engelbrecht, S.; Junge, W. Two rotary motors in F-ATP synthase are elastically coupled by a flexible rotor and a stiff stator stalk. Proc. Natl. Acad. Sci. USA 2011, 108, 3924–3929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stock, D.; Leslie, A.G.; Walker, J.E. Molecular architecture of the rotary motor in ATP synthase. Science 1999, 286, 1700–1705. [Google Scholar] [CrossRef]
- Seelert, H.; Poetsch, A.; Dencher, N.A.; Engel, A.; Stahlberg, H.; Muller, D.J. Structural biology. Proton-powered turbine of a plant motor. Nature 2000, 405, 418–419. [Google Scholar] [CrossRef]
- Stahlberg, H.; Muller, D.J.; Suda, K.; Fotiadis, D.; Engel, A.; Meier, T.; Matthey, U.; Dimroth, P. Bacterial Na+-ATP synthase has an undecameric rotor. EMBO Rep. 2001, 2, 229–233. [Google Scholar] [CrossRef]
- Mitome, N.; Suzuki, T.; Hayashi, S.; Yoshida, M. Thermophilic ATP synthase has a decamer c-ring: Indication of noninteger 10:3 H+/ATP ratio and permissive elastic coupling. Proc. Natl. Acad. Sci. USA 2004, 101, 12159–12164. [Google Scholar] [CrossRef]
- Meier, T.; Polzer, P.; Diederichs, K.; Welte, W.; Dimroth, P. Structure of the rotor ring of F-Type Na+-ATPase from Ilyobacter tartaricus. Science 2005, 308, 659–662. [Google Scholar] [CrossRef]
- Pogoryelov, D.; Yu, J.; Meier, T.; Vonck, J.; Dimroth, P.; Muller, D.J. The c15 ring of the Spirulina platensis F-ATP synthase: F1/F0 symmetry mismatch is not obligatory. EMBO Rep. 2005, 6, 1040–1044. [Google Scholar] [CrossRef] [PubMed]
- Meier, T.; Morgner, N.; Matthies, D.; Pogoryelov, D.; Keis, S.; Cook, G.M.; Dimroth, P.; Brutschy, B. A tridecameric c ring of the adenosine triphosphate (ATP) synthase from the thermoalkaliphilic Bacillus sp. strain TA2.A1 facilitates ATP synthesis at low electrochemical proton potential. Mol. Microbiol. 2007, 65, 1181–1192. [Google Scholar] [CrossRef] [PubMed]
- Pogoryelov, D.; Reichen, C.; Klyszejko, A.L.; Brunisholz, R.; Muller, D.J.; Dimroth, P.; Meier, T. The oligomeric state of c rings from cyanobacterial F-ATP synthases varies from 13 to 15. J. Bacteriol. 2007, 189, 5895–5902. [Google Scholar] [CrossRef] [PubMed]
- Meier, T.; Krah, A.; Bond, P.J.; Pogoryelov, D.; Diederichs, K.; Faraldo-Gomez, J.D. Complete ion-coordination structure in the rotor ring of Na+-dependent F-ATP synthases. J. Mol. Biol. 2009, 391, 498–507. [Google Scholar] [CrossRef] [PubMed]
- Pogoryelov, D.; Yildiz, O.; Faraldo-Gomez, J.D.; Meier, T. High-resolution structure of the rotor ring of a proton-dependent ATP synthase. Nat. Struct. Mol. Biol. 2009, 16, 1068–1073. [Google Scholar] [CrossRef] [PubMed]
- Vollmar, M.; Schlieper, D.; Winn, M.; Büchner, C.; Groth, G. Structure of the c14 rotor ring of the proton translocating chloroplast ATP synthase. J. Biol. Chem. 2009, 284, 18228–18235. [Google Scholar] [CrossRef] [PubMed]
- Watt, I.N.; Montgomery, M.G.; Runswick, M.J.; Leslie, A.G.; Walker, J.E. Bioenergetic cost of making an adenosine triphosphate molecule in animal mitochondria. Proc. Natl. Acad. Sci. USA 2010, 107, 16823–16827. [Google Scholar] [CrossRef]
- Preiss, L.; Klyszejko, A.L.; Hicks, D.B.; Liu, J.; Fackelmayer, O.J.; Yildiz, O.; Krulwich, T.A.; Meier, T. The c-ring stoichiometry of ATP synthase is adapted to cell physiological requirements of alkaliphilic Bacillus pseudofirmus OF4. Proc. Natl. Acad. Sci. USA 2013, 110, 7874–7879. [Google Scholar] [CrossRef]
- Matthies, D.; Zhou, W.; Klyszejko, A.L.; Anselmi, C.; Yildiz, O.; Brandt, K.; Muller, V.; Faraldo-Gomez, J.D.; Meier, T. High-resolution structure and mechanism of an F/V-hybrid rotor ring in a Na(+)-coupled ATP synthase. Nat. Commun. 2014, 5, 5286. [Google Scholar] [CrossRef]
- Preiss, L.; Langer, J.D.; Hicks, D.B.; Liu, J.; Yildiz, O.; Krulwich, T.A.; Meier, T. The c-ring ion binding site of the ATP synthase from Bacillus pseudofirmus OF4 is adapted to alkaliphilic lifestyle. Mol. Microbiol. 2014, 92, 973–984. [Google Scholar] [CrossRef]
- Preiss, L.; Langer, J.D.; Yildiz, O.; Eckhardt-Strelau, L.; Guillemont, J.E.; Koul, A.; Meier, T. Structure of the mycobacterial ATP synthase Fo rotor ring in complex with the anti-TB drug bedaquiline. Sci. Adv. 2015, 1, 1500106. [Google Scholar] [CrossRef] [PubMed]
- Mazhab-Jafari, M.T.; Rohou, A.; Schmidt, C.; Bueler, S.A.; Benlekbir, S.; Robinson, C.V.; Rubinstein, J.L. Atomic model for the membrane-embedded VO motor of a eukaryotic V-ATPase. Nature 2016, 539, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Hermolin, J.; Fillingame, R.H. The preferred stoichiometry of c subunits in the rotary motor sector of Escherichia coli ATP synthase is 10. Proc. Natl. Acad. Sci. USA 2001, 98, 4966–4971. [Google Scholar] [CrossRef] [PubMed]
- Ballhausen, B.; Altendorf, K.; Deckers-Hebestreit, G. Constant c10 ring stoichiometry in the Escherichia coli ATP synthase analyzed by cross-linking. J. Bacteriol. 2009, 191, 2400–2404. [Google Scholar] [CrossRef] [PubMed]
- Düser, M.G.; Zarrabi, N.; Cipriano, D.J.; Ernst, S.; Glick, G.D.; Dunn, S.D.; Börsch, M. 36 degrees step size of proton-driven c-ring rotation in FoF1-ATP synthase. EMBO J. 2009, 28, 2689–2696. [Google Scholar] [CrossRef]
- Ishmukhametov, R.; Hornung, T.; Spetzler, D.; Frasch, W.D. Direct observation of stepped proteolipid ring rotation in E. coli FOF1-ATP synthase. EMBO J. 2010, 29, 3911–3923. [Google Scholar] [CrossRef]
- Junge, W.; Pänke, O.; Cherepanov, D.A.; Gumbiowski, K.; Müller, M.; Engelbrecht, S. Inter-subunit rotation and elastic power transmission in F0F1-ATPase. FEBS Lett. 2001, 504, 152–160. [Google Scholar] [CrossRef]
- Sielaff, H.; Rennekamp, H.; Wächter, A.; Xie, H.; Hilbers, F.; Feldbauer, K.; Dunn, S.D.; Engelbrecht, S.; Junge, W. Domain compliance and elastic power transmission in rotary FOF1-ATPase. Proc. Natl. Acad. Sci. USA 2008, 105, 17760–17765. [Google Scholar] [CrossRef]
- Ernst, S.; Düser, M.G.; Zarrabi, N.; Dunn, S.D.; Börsch, M. Elastic deformations of the rotary double motor of single FoF1-ATP synthases detected in real time by Forster resonance energy transfer. Biochim. Biophys. Acta 2012, 1817, 1722–1731. [Google Scholar] [CrossRef]
- Martin, J.L.; Ishmukhametov, R.; Spetzler, D.; Hornung, T.; Frasch, W.D. Elastic coupling power stroke mechanism of the F1-ATPase molecular motor. Proc. Natl. Acad. Sci. USA 2018, 115, 5750–5755. [Google Scholar] [CrossRef]
- Cross, R.L. The Mechanism and Regulation of Atp Synthesis by F1-Atpases. Annu. Rev. Biochem. 1981, 50, 681–714. [Google Scholar] [CrossRef] [PubMed]
- Boyer, P.D. The binding change mechanism for ATP synthase—Some probabilities and possibilities. Biochim. Biophys. Acta 1993, 1140, 215–250. [Google Scholar] [CrossRef]
- Noji, H.; Yasuda, R.; Yoshida, M.; Kinosita, K., Jr. Direct observation of the rotation of F1-ATPase. Nature 1997, 386, 299–302. [Google Scholar] [CrossRef] [PubMed]
- Sambongi, Y.; Iko, Y.; Tanabe, M.; Omote, H.; Iwamoto-Kihara, A.; Ueda, I.; Yanagida, T.; Wada, Y.; Futai, M. Mechanical rotation of the c subunit oligomer in ATP synthase (F0F1): Direct observation. Science 1999, 286, 1722–1724. [Google Scholar] [CrossRef] [PubMed]
- Pänke, O.; Gumbiowski, K.; Junge, W.; Engelbrecht, S. F-ATPase: Specific observation of the rotating c subunit oligomer of EFoEF1. FEBS Lett. 2000, 472, 34–38. [Google Scholar] [CrossRef]
- Tsunoda, S.P.; Aggeler, R.; Yoshida, M.; Capaldi, R.A. Rotation of the c subunit oligomer in fully functional F1Fo ATP synthase. Proc. Natl. Acad. Sci. USA 2001, 98, 898–902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Börsch, M.; Diez, M.; Zimmermann, B.; Reuter, R.; Gräber, P. Stepwise rotation of the gamma-subunit of EFoF1-ATP synthase observed by intramolecular single-molecule fluorescence resonance energy transfer. FEBS Lett. 2002, 527, 147–152. [Google Scholar] [CrossRef]
- Börsch, M.; Diez, M.; Zimmermann, B.; Trost, M.; Steigmiller, S.; Gräber, P. Stepwise rotation of the γ-subunit of EFoF1-ATP synthase during ATP synthesis: A single-molecule FRET approach. Proc. SPIE 2003, 4962. [Google Scholar] [CrossRef]
- Diez, M.; Zimmermann, B.; Börsch, M.; Konig, M.; Schweinberger, E.; Steigmiller, S.; Reuter, R.; Felekyan, S.; Kudryavtsev, V.; Seidel, C.A.; et al. Proton-powered subunit rotation in single membrane-bound FoF1-ATP synthase. Nat. Struct. Mol. Biol. 2004, 11, 135–141. [Google Scholar] [CrossRef]
- Nishizaka, T.; Oiwa, K.; Noji, H.; Kimura, S.; Muneyuki, E.; Yoshida, M.; Kinosita, K., Jr. Chemomechanical coupling in F1-ATPase revealed by simultaneous observation of nucleotide kinetics and rotation. Nat. Struct. Mol. Biol. 2004, 11, 142–148. [Google Scholar] [CrossRef]
- Watanabe, R.; Iino, R.; Noji, H. Phosphate release in F1-ATPase catalytic cycle follows ADP release. Nat. Chem. Biol. 2010, 6, 814–820. [Google Scholar] [CrossRef] [PubMed]
- Hirata, T.; Iwamoto-Kihara, A.; Sun-Wada, G.H.; Okajima, T.; Wada, Y.; Futai, M. Subunit rotation of vacuolar-type proton pumping ATPase: Relative rotation of the G and C subunits. J. Biol. Chem. 2003, 278, 23714–23719. [Google Scholar] [CrossRef] [PubMed]
- Imamura, H.; Takeda, M.; Funamoto, S.; Shimabukuro, K.; Yoshida, M.; Yokoyama, K. Rotation scheme of V1-motor is different from that of F1-motor. Proc. Natl. Acad. Sci. USA 2005, 102, 17929–17933. [Google Scholar] [CrossRef] [PubMed]
- Furuike, S.; Nakano, M.; Adachi, K.; Noji, H.; Kinosita, K., Jr.; Yokoyama, K. Resolving stepping rotation in Thermus thermophilus H+-ATPase/synthase with an essentially drag-free probe. Nat. Commun. 2011, 2, 233. [Google Scholar] [CrossRef] [PubMed]
- Minagawa, Y.; Ueno, H.; Hara, M.; Ishizuka-Katsura, Y.; Ohsawa, N.; Terada, T.; Shirouzu, M.; Yokoyama, S.; Yamato, I.; Muneyuki, E.; et al. Basic properties of rotary dynamics of the molecular motor Enterococcus hirae V1-ATPase. J. Biol. Chem. 2013, 288, 32700–32707. [Google Scholar] [CrossRef] [PubMed]
- Sielaff, H.; Martin, J.; Singh, D.; Biukovic, G.; Grüber, G.; Frasch, W.D. Power Stroke Angular Velocity Profiles of Archaeal A-ATP Synthase Versus Thermophilic and Mesophilic F-ATP Synthase Molecular Motors. J. Biol. Chem. 2016, 291, 25351–25363. [Google Scholar] [CrossRef]
- Singh, D.; Sielaff, H.; Börsch, M.; Grüber, G. Conformational dynamics of the rotary subunit F in the A3B3DF complex of Methanosarcina mazei Gö1 A-ATP synthase monitored by single-molecule FRET. FEBS Lett. 2017, 591, 854–862. [Google Scholar] [CrossRef]
- Yasuda, R.; Noji, H.; Yoshida, M.; Kinosita, K., Jr.; Itoh, H. Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature 2001, 410, 898–904. [Google Scholar] [CrossRef]
- Shimabukuro, K.; Yasuda, R.; Muneyuki, E.; Hara, K.Y.; Kinosita, K., Jr.; Yoshida, M. Catalysis and rotation of F1 motor: Cleavage of ATP at the catalytic site occurs in 1 ms before 40° substep rotation. Proc. Natl. Acad. Sci. USA 2003, 100, 14731–14736. [Google Scholar] [CrossRef]
- Adachi, K.; Oiwa, K.; Nishizaka, T.; Furuike, S.; Noji, H.; Itoh, H.; Yoshida, M.; Kinosita, K., Jr. Coupling of rotation and catalysis in F1-ATPase revealed by single-molecule imaging and manipulation. Cell 2007, 130, 309–321. [Google Scholar] [CrossRef]
- Arai, S.; Saijo, S.; Suzuki, K.; Mizutani, K.; Kakinuma, Y.; Ishizuka-Katsura, Y.; Ohsawa, N.; Terada, T.; Shirouzu, M.; Yokoyama, S.; et al. Rotation mechanism of Enterococcus hirae V1-ATPase based on asymmetric crystal structures. Nature 2013, 493, 703–707. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Tanaka, K.; Wakabayashi, C.; Saita, E.; Yoshida, M. Chemomechanical coupling of human mitochondrial F1-ATPase motor. Nat. Chem. Biol. 2014, 10, 930–936. [Google Scholar] [CrossRef] [PubMed]
- Börsch, M.; Duncan, T.M. Spotlighting motors and controls of single FoF1-ATP synthase. Biochem. Soc. Trans. 2013, 41, 1219–1226. [Google Scholar] [CrossRef] [PubMed]
- Börsch, M. Microscopy of single FoF1-ATP synthases—The unraveling of motors, gears, and controls. IUBMB Life 2013, 65, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Junge, W.; Lill, H.; Engelbrecht, S. ATP synthase: An electrochemical transducer with rotatory mechanics. Trends Biochem. Sci. 1997, 22, 420–423. [Google Scholar] [CrossRef]
- Wang, H.; Oster, G. Ratchets, power strokes, and molecular motors. Appl. Phys. A 2002, 75, 315–323. [Google Scholar] [CrossRef]
- Leone, V.; Pogoryelov, D.; Meier, T.; Faraldo-Gomez, J.D. On the principle of ion selectivity in Na+/H+-coupled membrane proteins: Experimental and theoretical studies of an ATP synthase rotor. Proc. Natl. Acad. Sci. USA 2015, 112, E1057–E1066. [Google Scholar] [CrossRef]
- Grüber, G.; Wieczorek, H.; Harvey, W.R.; Müller, V. Structure-function relationships of A-, F- and V-ATPases. J. Exp. Biol. 2001, 204, 2597–2605. [Google Scholar]
- Yamato, I.; Kakinuma, Y.; Murata, T. Operating principles of rotary molecular motors: Differences between F1 and V1 motors. Biophys. Physicobiol. 2016, 13, 37–44. [Google Scholar] [CrossRef]
- Cross, R.L.; Müller, V. The evolution of A-, F-, and V-type ATP synthases and ATPases: Reversals in function and changes in the H+/ATP coupling ratio. FEBS Lett. 2004, 576, 1–4. [Google Scholar] [CrossRef]
- Muench, S.P.; Trinick, J.; Harrison, M.A. Structural divergence of the rotary ATPases. Q. Rev. Biophys. 2011, 44, 311–356. [Google Scholar] [CrossRef] [PubMed]
- Stewart, A.G.; Sobti, M.; Harvey, R.P.; Stock, D. Rotary ATPases: Models, machine elements and technical specifications. Bioarchitecture 2013, 3, 2–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grüber, G.; Manimekalai, M.S.; Mayer, F.; Müller, V. ATP synthases from archaea: The beauty of a molecular motor. Biochim. Biophys. Acta 2014, 1837, 940–952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.; Benlekbir, S.; Rubinstein, J.L. Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase. Nature 2015, 521, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Davies, R.B.; Smits, C.; Wong, A.S.W.; Stock, D.; Christie, M.; Sandin, S.; Stewart, A.G. Cryo-EM analysis of a domain antibody bound rotary ATPase complex. J. Struct. Biol. 2017, 197, 350–353. [Google Scholar] [CrossRef]
- Nakanishi, A.; Kishikawa, J.I.; Tamakoshi, M.; Mitsuoka, K.; Yokoyama, K. Cryo EM structure of intact rotary H+-ATPase/synthase from Thermus thermophilus. Nat. Commun. 2018, 9, 89. [Google Scholar] [CrossRef] [PubMed]
- Iwata, M.; Imamura, H.; Stambouli, E.; Ikeda, C.; Tamakoshi, M.; Nagata, K.; Makyio, H.; Hankamer, B.; Barber, J.; Yoshida, M.; et al. Crystal structure of a central stalk subunit C and reversible association/dissociation of vacuole-type ATPase. Proc. Natl. Acad. Sci. USA 2004, 101, 59–64. [Google Scholar] [CrossRef]
- Numoto, N.; Hasegawa, Y.; Takeda, K.; Miki, K. Inter-subunit interaction and quaternary rearrangement defined by the central stalk of prokaryotic V1-ATPase. EMBO Rep. 2009, 10, 1228–1234. [Google Scholar] [CrossRef]
- Saijo, S.; Arai, S.; Hossain, K.M.; Yamato, I.; Suzuki, K.; Kakinuma, Y.; Ishizuka-Katsura, Y.; Ohsawa, N.; Terada, T.; Shirouzu, M.; et al. Crystal structure of the central axis DF complex of the prokaryotic V-ATPase. Proc. Natl. Acad. Sci. USA 2011, 108, 19955–19960. [Google Scholar] [CrossRef] [Green Version]
- Kishikawa, J.; Ibuki, T.; Nakamura, S.; Nakanishi, A.; Minamino, T.; Miyata, T.; Namba, K.; Konno, H.; Ueno, H.; Imada, K.; et al. Common evolutionary origin for the rotor domain of rotary ATPases and flagellar protein export apparatus. PLoS ONE 2013, 8, 64695. [Google Scholar] [CrossRef]
- Singh, D.; Sielaff, H.; Sundararaman, L.; Bhushan, S.; Grüber, G. The stimulating role of subunit F in ATPase activity inside the A-complex of the Methanosarcina mazei Gö1 A1AO ATP synthase. Biochim. Biophys. Acta 2016, 1857, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Uchihashi, T.; Iino, R.; Ando, T.; Noji, H. High-speed atomic force microscopy reveals rotary catalysis of rotorless F1-ATPase. Science 2011, 333, 755–758. [Google Scholar] [CrossRef] [PubMed]
- Senior, A.E.; Nadanaciva, S.; Weber, J. The molecular mechanism of ATP synthesis by F1F0-ATP synthase. Biochim. Biophys. Acta 2002, 1553, 188–211. [Google Scholar] [CrossRef]
- Mao, H.Z.; Weber, J. Identification of the betaTP site in the x-ray structure of F1-ATPase as the high-affinity catalytic site. Proc. Natl. Acad. Sci. USA 2007, 104, 18478–18483. [Google Scholar] [CrossRef] [PubMed]
- Allegretti, M.; Klusch, N.; Mills, D.J.; Vonck, J.; Kühlbrandt, W.; Davies, K.M. Horizontal membrane-intrinsic alpha-helices in the stator a-subunit of an F-type ATP synthase. Nature 2015, 521, 237–240. [Google Scholar] [CrossRef]
- Nesci, S.; Trombetti, F.; Ventrella, V.; Pagliarani, A. The a subunit asymmetry dictates the two opposite rotation directions in the synthesis and hydrolysis of ATP by the mitochondrial ATP synthase. Med. Hypotheses 2015, 84, 53–57. [Google Scholar] [CrossRef]
- Nesci, S.; Trombetti, F.; Ventrella, V.; Pagliarani, A. Opposite Rotation Directions in the Synthesis and Hydrolysis of ATP by the ATP Synthase: Hints from a Subunit Asymmetry. J. Membr. Biol. 2015, 248, 163–169. [Google Scholar] [CrossRef]
- Yanagisawa, S.; Frasch, W.D. Protonation-dependent stepped rotation of the F-type ATP synthase c-ring observed by single-molecule measurements. J. Biol. Chem. 2017, 292, 17093–17100. [Google Scholar] [CrossRef]
- McLachlin, D.T.; Coveny, A.M.; Clark, S.M.; Dunn, S.D. Site-directed cross-linking of b to the alpha, beta, and a subunits of the Escherichia coli ATP synthase. J. Biol. Chem. 2000, 275, 17571–17577. [Google Scholar] [CrossRef]
- Brandt, K.; Maiwald, S.; Herkenhoff-Hesselmann, B.; Gnirss, K.; Greie, J.C.; Dunn, S.D.; Deckers-Hebestreit, G. Individual interactions of the b subunits within the stator of the Escherichia coli ATP synthase. J. Biol. Chem. 2013, 288, 24465–24479. [Google Scholar] [CrossRef]
- Benlekbir, S.; Bueler, S.A.; Rubinstein, J.L. Structure of the vacuolar-type ATPase from Saccharomyces cerevisiae at 11-Å resolution. Nat. Struct. Mol. Biol. 2012, 19, 1356–1362. [Google Scholar] [CrossRef] [PubMed]
- Greene, M.D.; Frasch, W.D. Interactions among gamma R268, gamma Q269, and the beta subunit catch loop of Escherichia coli F1-ATPase are important for catalytic activity. J. Biol. Chem. 2003, 278, 51594–51598. [Google Scholar] [CrossRef] [PubMed]
- Hilbers, F.; Junge, W.; Sielaff, H. The torque of rotary F-ATPase can unfold subunit gamma if rotor and stator are cross-linked. PLoS ONE 2013, 8, 53754. [Google Scholar] [CrossRef] [PubMed]
- Iino, R.; Noji, H. Rotary catalysis of the stator ring of F1-ATPase. Biochim. Biophys. Acta 2012, 1817, 1732–1739. [Google Scholar] [CrossRef] [PubMed]
- Feniouk, B.A.; Suzuki, T.; Yoshida, M. The role of subunit epsilon in the catalysis and regulation of FOF1-ATP synthase. Biochim. Biophys. Acta 2006, 1757, 326–338. [Google Scholar] [CrossRef]
- Krah, A.; Zarco-Zavala, M.; McMillan, D.G.G. Insights into the regulatory function of the varepsilon subunit from bacterial F-type ATP synthases: A comparison of structural, biochemical and biophysical data. Open Biol. 2018, 8, 170275. [Google Scholar] [CrossRef]
- Sielaff, H.; Duncan, T.M.; Börsch, M. The regulatory subunit epsilon in Escherichia coli FOF1-ATP synthase. Biochim. Biophys. Acta-Bioenerg. 2018, 1859, 775–788. [Google Scholar] [CrossRef]
- Adachi, K.; Yasuda, R.; Noji, H.; Itoh, H.; Harada, Y.; Yoshida, M.; Kinosita, K., Jr. Stepping rotation of F1-ATPase visualized through angle-resolved single-fluorophore imaging. Proc. Natl. Acad. Sci. USA 2000, 97, 7243–7247. [Google Scholar] [CrossRef]
- Sielaff, H.; Rennekamp, H.; Engelbrecht, S.; Junge, W. Functional halt positions of rotary FOF1-ATPase correlated with crystal structures. Biophys. J. 2008, 95, 4979–4987. [Google Scholar] [CrossRef]
- Spetzler, D.; York, J.; Daniel, D.; Fromme, R.; Lowry, D.; Frasch, W. Microsecond time scale rotation measurements of single F1-ATPase molecules. Biochemistry 2006, 45, 3117–3124. [Google Scholar] [CrossRef]
- Martin, J.L.; Ishmukhametov, R.; Hornung, T.; Ahmad, Z.; Frasch, W.D. Anatomy of F1-ATPase powered rotation. Proc. Natl. Acad. Sci. USA 2014, 111, 3715–3720. [Google Scholar] [CrossRef] [PubMed]
- Iino, R.; Ueno, H.; Minagawa, Y.; Suzuki, K.; Murata, T. Rotational mechanism of Enterococcus hirae V-ATPase by crystal-structure and single-molecule analyses. Curr. Opin. Struct. Biol. 2015, 31, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Senior, A.E.; Weber, J.; al-Shawi, M.K. Catalytic mechanism of Escherichia coli F1-ATPase. Biochem. Soc. Trans. 1995, 23, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.; Senior, A.E. Catalytic mechanism of F1-ATPase. Biochim. Biophys. Acta 1997, 1319, 19–58. [Google Scholar] [CrossRef] [Green Version]
- Weber, J.; Nadanaciva, S.; Senior, A.E. ATP-driven rotation of the gamma subunit in F1-ATPase. FEBS Lett. 2000, 483, 1–5. [Google Scholar] [CrossRef]
- Weber, J.; Muharemagic, A.; Wilke-Mounts, S.; Senior, A.E. Analysis of sequence determinants of F1Fo-ATP synthase in the N-terminal region of alpha subunit for binding of delta subunit. J. Biol. Chem. 2004, 279, 25673–25679. [Google Scholar] [CrossRef] [PubMed]
- Hirono-Hara, Y.; Noji, H.; Nishiura, M.; Muneyuki, E.; Hara, K.Y.; Yasuda, R.; Kinosita, K., Jr.; Yoshida, M. Pause and rotation of F1-ATPase during catalysis. Proc. Natl. Acad. Sci. USA 2001, 98, 13649–13654. [Google Scholar] [CrossRef]
- Hornung, T.; Ishmukhametov, R.; Spetzler, D.; Martin, J.; Frasch, W.D. Determination of torque generation from the power stroke of Escherichia coli F1-ATPase. Biochim. Biophys. Acta 2008, 1777, 579–582. [Google Scholar] [CrossRef]
- Börsch, M.; Gräber, P. Subunit movement in individual H+-ATP synthases during ATP synthesis and hydrolysis revealed by fluorescence resonance energy transfer. Biochem. Soc. Trans. 2005, 33, 878–882. [Google Scholar] [CrossRef]
- Zimmermann, B.; Diez, M.; Zarrabi, N.; Gräber, P.; Börsch, M. Movements of the epsilon-subunit during catalysis and activation in single membrane-bound H+-ATP synthase. EMBO J. 2005, 24, 2053–2063. [Google Scholar] [CrossRef]
- Zarrabi, N.; Zimmermann, B.; Diez, M.; Gräber, P.; Wrachtrup, J.; Börsch, M. Asymmetry of rotational catalysis of single membrane-bound FoF1-ATP synthase. Proc. SPIE 2005, 5699. [Google Scholar] [CrossRef]
- Düser, M.G.; Zarrabi, N.; Bi, Y.; Zimmermann, B.; Dunn, S.D.; Börsch, M. 3D-localization of the alpha-subunit in FoF1-ATP synthase by time resolved single-molecule FRET. Proc. SPIE 2006, 6092. [Google Scholar] [CrossRef]
- Düser, M.G.; Bi, Y.; Zarrabi, N.; Dunn, S.D.; Börsch, M. The proton-translocating a subunit of FOF1-ATP synthase is allocated asymmetrically to the peripheral stalk. J. Biol. Chem. 2008, 283, 33602–33610. [Google Scholar] [CrossRef]
- Ernst, S.; Düser, M.G.; Zarrabi, N.; Börsch, M. Three-color Förster resonance energy transfer within single F0F1-ATP synthases: Monitoring elastic deformations of the rotary double motor in real time. J. Biomed. Opt. 2012, 17, 011004. [Google Scholar] [CrossRef] [PubMed]
- Gumbiowski, K.; Pänke, O.; Junge, W.; Engelbrecht, S. Rotation of the c subunit oligomer in EF0EF1 mutant cD61N. J. Biol. Chem. 2002, 277, 31287–31290. [Google Scholar] [CrossRef]
- Weber, J.; Wilke-Mounts, S.; Lee, R.S.; Grell, E.; Senior, A.E. Specific placement of tryptophan in the catalytic sites of Escherichia coli F1-ATPase provides a direct probe of nucleotide binding: Maximal ATP hydrolysis occurs with three sites occupied. J. Biol. Chem. 1993, 268, 20126–20133. [Google Scholar] [PubMed]
- Martin, J.; Hudson, J.; Hornung, T.; Frasch, W.D. Fo-driven Rotation in the ATP Synthase Direction against the Force of F1 ATPase in the FoF1 ATP Synthase. J. Biol. Chem. 2015, 290, 10717–10728. [Google Scholar] [CrossRef] [PubMed]
- Ueno, H.; Suzuki, T.; Kinosita, K., Jr.; Yoshida, M. ATP-driven stepwise rotation of FoF1-ATP synthase. Proc. Natl. Acad. Sci. USA 2005, 102, 1333–1338. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, R.; Tabata, K.V.; Iino, R.; Ueno, H.; Iwamoto, M.; Oiki, S.; Noji, H. Biased Brownian stepping rotation of FoF1-ATP synthase driven by proton motive force. Nat. Commun. 2013, 4, 1631. [Google Scholar] [CrossRef] [PubMed]
- Toei, M.; Noji, H. Single-molecule analysis of F0F1-ATP synthase inhibited by N,N-dicyclohexylcarbodiimide. J. Biol. Chem. 2013, 288, 25717–25726. [Google Scholar] [CrossRef]
- Ueno, H.; Minagawa, Y.; Hara, M.; Rahman, S.; Yamato, I.; Muneyuki, E.; Noji, H.; Murata, T.; Iino, R. Torque generation of Enterococcus hirae V-ATPase. J. Biol. Chem. 2014, 289, 31212–31223. [Google Scholar] [CrossRef] [PubMed]
- Hellenkamp, B.; Schmid, S.; Doroshenko, O.; Opanasyuk, O.; Kuhnemuth, R.; Rezaei Adariani, S.; Ambrose, B.; Aznauryan, M.; Barth, A.; Birkedal, V.; et al. Precision and accuracy of single-molecule FRET measurements-a multi-laboratory benchmark study. Nat. Methods 2018, 15, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Noble, A.J.; Wei, H.; Dandey, V.P.; Zhang, Z.; Tan, Y.Z.; Potter, C.S.; Carragher, B. Reducing effects of particle adsorption to the air-water interface in cryo-EM. Nat. Methods 2018, 15, 793–795. [Google Scholar] [CrossRef] [PubMed]
- Noble, A.J.; Dandey, V.P.; Wei, H.; Brasch, J.; Chase, J.; Acharya, P.; Tan, Y.Z.; Zhang, Z.; Kim, L.Y.; Scapin, G.; et al. Routine single particle CryoEM sample and grid characterization by tomography. eLife 2018, 7, 34257. [Google Scholar] [CrossRef]
- Cohen, A.E.; Moerner, W.E. Method for trapping and manipulating nanoscale objects in solution. Appl. Phys. Lett. 2005, 86, 093109. [Google Scholar] [CrossRef]
- Cohen, A.E.; Moerner, W.E. The anti-Brownian electrophoretic trap (ABEL Trap): Fabrication and software. Proc. SPIE 2005, 5699. [Google Scholar] [CrossRef]
- Cohen, A.E.; Moerner, W.E. Suppressing Brownian motion of individual biomolecules in solution. Proc. Natl. Acad. Sci. USA 2006, 103, 4362–4365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, A.E.; Moerner, W.E. Internal mechanical response of a polymer in solution. Phys. Rev. Lett. 2007, 98, 116001. [Google Scholar] [CrossRef]
- Cohen, A.E.; Moerner, W.E. Controlling Brownian motion of single protein molecules and single fluorophores in aqueous buffer. Opt. Exp. 2008, 16, 6941–6956. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, Q.; Cohen, A.E.; Douglas, N.; Frydman, J.; Moerner, W.E. Hardware-based anti-Brownian electrokinetic trap (ABEL trap) for single molecules: Control loop simulations and application to ATP binding stoichiometry in multi-subunit enzymes. Proc. SPIE 2008, 7038. [Google Scholar] [CrossRef]
- Goldsmith, R.H.; Moerner, W.E. Watching conformational- and photo-dynamics of single fluorescent proteins in solution. Nat. Chem. 2010, 2, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Bockenhauer, S.; Fürstenberg, A.; Yao, X.J.; Kobilka, B.K.; Moerner, W.E. Conformational dynamics of single G protein-coupled receptors in solution. J. Phys. Chem. B 2011, 115, 13328–13338. [Google Scholar] [CrossRef]
- Wang, Q.; Moerner, W.E. An Adaptive Anti-Brownian Electrokinetic Trap with Real-Time Information on Single-Molecule Diffusivity and Mobility. ACS Nano 2011, 5, 5792–5799. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Moerner, W.E. Lifetime and Spectrally Resolved Characterization of the Photodynamics of Single Fluorophores in Solution Using the Anti-Brownian Electrokinetic Trap. J. Phys. Chem. B 2013, 117, 4641–4648. [Google Scholar] [CrossRef] [PubMed]
- Bockenhauer, S.D.; Duncan, T.M.; Moerner, W.E.; Börsch, M. The regulatory switch of F1-ATPase studied by single-molecule FRET in the ABEL Trap. Proc. SPIE 2014, 8950. [Google Scholar] [CrossRef]
- Su, B.; Düser, M.G.; Zarrabi, N.; Heitkamp, T.; Starke, I.; Börsch, M. Observing conformations of single FoF1-ATP synthases in a fast anti-Brownian electrokinetic trap. Proc. SPIE 2015, 9329. [Google Scholar] [CrossRef]
- Dienerowitz, M.; Ilchenko, M.; Su, B.; Deckers-Hebestreit, G.; Mayer, G.; Henkel, T.; Heitkamp, T.; Börsch, M. Optimized green fluorescent protein fused to FoF1-ATP synthase for single-molecule FRET using a fast anti-Brownian electrokinetic trap. Proc. SPIE 2016, 9714. [Google Scholar] [CrossRef]
- Dienerowitz, M.; Heitkamp, T.; Gottschall, T.; Limpert, J.; Börsch, M. Confining Brownian motion of single nanoparticles in an ABELtrap. Proc. SPIE 2017, 10120. [Google Scholar] [CrossRef]
- Dienerowitz, M.; Dienerowitz, F.; Börsch, M. Measuring nanoparticle diffusion in an ABELtrap. J. Opt. 2018, 20, 034006. [Google Scholar] [CrossRef] [Green Version]
- Lill, H.; Engelbrecht, S.; Schönknecht, G.; Junge, W. The proton channel, CF0, in thylakoid membranes—Only a low proportion of CF1-lacking CF0 is active with a high unit conductance (169 fS). Eur. J. Biochem. 1986, 160, 627–634. [Google Scholar] [CrossRef]
- Schönknecht, G.; Junge, W.; Lill, H.; Engelbrecht, S. Complete tracking of proton flow in thylakoids-the unit conductance of CF0 is greater than 10 fS. FEBS Lett. 1986, 203, 289–294. [Google Scholar] [CrossRef]
- Pänke, O.; Cherepanov, D.A.; Gumbiowski, K.; Engelbrecht, S.; Junge, W. Viscoelastic dynamics of actin filaments coupled to rotary F-ATPase: Angular torque profile of the enzyme. Biophys. J. 2001, 81, 1220–1233. [Google Scholar] [CrossRef]
- Cherepanov, D.A.; Mulkidjanian, A.Y.; Junge, W. Transient accumulation of elastic energy in proton translocating ATP synthase. FEBS Lett. 1999, 449, 1–6. [Google Scholar] [CrossRef] [Green Version]

| Structure | State 1 | State 2 | State 3 | Nucleotide Occupancy | Reference |
|---|---|---|---|---|---|
| EcFOF1 | 46.3% | 30.0% | 23.7% | ADP in αβclosed | [12] |
| BPFOF1 | 45.3% | 35.1% | 19.6% | Mg-ADP in αβTP, Pi in αβE | [29] |
| CFOF1 | 8.1% | 7.5% | 84.4% | Mg-ADP in αβDP and αβTP | [27] |
| MFOF1 | 31.3% | 35.0% | 33.7% | Unknown | [24] |
| TtAOA1 | 72.5% | 18.7% | 8.8% | ADP in ABclosed and ABsemi-closed | [96] |
| YVOV1 | 47% | 36% | 17% | unknown | [94] |
| FRET-Level Method | M, Dwell Time 1 (ms) | L, Dwell Time 2 (ms) | H, Dwell Time 3 (ms) | Direction of Rotation |
|---|---|---|---|---|
| rotation assay* | 300.0 | 200.0 | 200.0 | ATP hydrolysis |
| smFRET ε56/b64 | 12.7 | 17.6 | 15.8 | ATP hydrolysis |
| smFRET ε56/b64 | 15.4 | 24.0 | 19.5 | ATP synthesis |
| Conformation | State 1 | State 2 | State 3 | |
| cryo-EM* | 47% | 30% | 24% | N.A. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sielaff, H.; Yanagisawa, S.; Frasch, W.D.; Junge, W.; Börsch, M. Structural Asymmetry and Kinetic Limping of Single Rotary F-ATP Synthases. Molecules 2019, 24, 504. https://doi.org/10.3390/molecules24030504
Sielaff H, Yanagisawa S, Frasch WD, Junge W, Börsch M. Structural Asymmetry and Kinetic Limping of Single Rotary F-ATP Synthases. Molecules. 2019; 24(3):504. https://doi.org/10.3390/molecules24030504
Chicago/Turabian StyleSielaff, Hendrik, Seiga Yanagisawa, Wayne D. Frasch, Wolfgang Junge, and Michael Börsch. 2019. "Structural Asymmetry and Kinetic Limping of Single Rotary F-ATP Synthases" Molecules 24, no. 3: 504. https://doi.org/10.3390/molecules24030504
APA StyleSielaff, H., Yanagisawa, S., Frasch, W. D., Junge, W., & Börsch, M. (2019). Structural Asymmetry and Kinetic Limping of Single Rotary F-ATP Synthases. Molecules, 24(3), 504. https://doi.org/10.3390/molecules24030504





