Enhanced NADH Metabolism Involves Colistin-Induced Killing of Bacillus subtilis and Paenibacillus polymyxa
Abstract
1. Introduction
2. Results
2.1. Oxidative Stress Caused by Colistin in Gram-Positive Bacteria
2.2. Scavenging Effect of Thiourea on Colistin-Induced Oxidative Stress
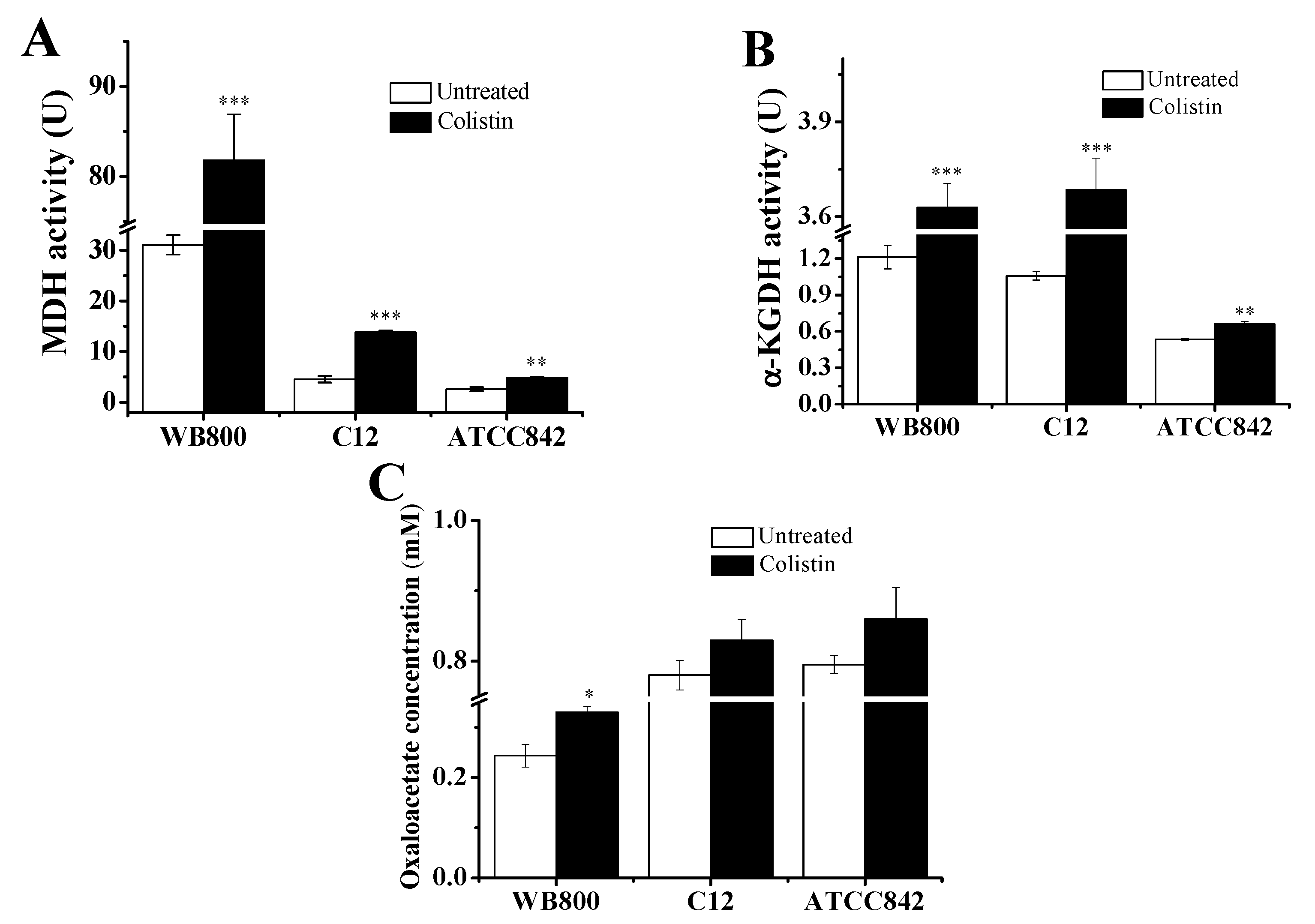
2.3. Disturbance of TCA Cycle in Colistin-Exposed Gram-Positive Bacteria
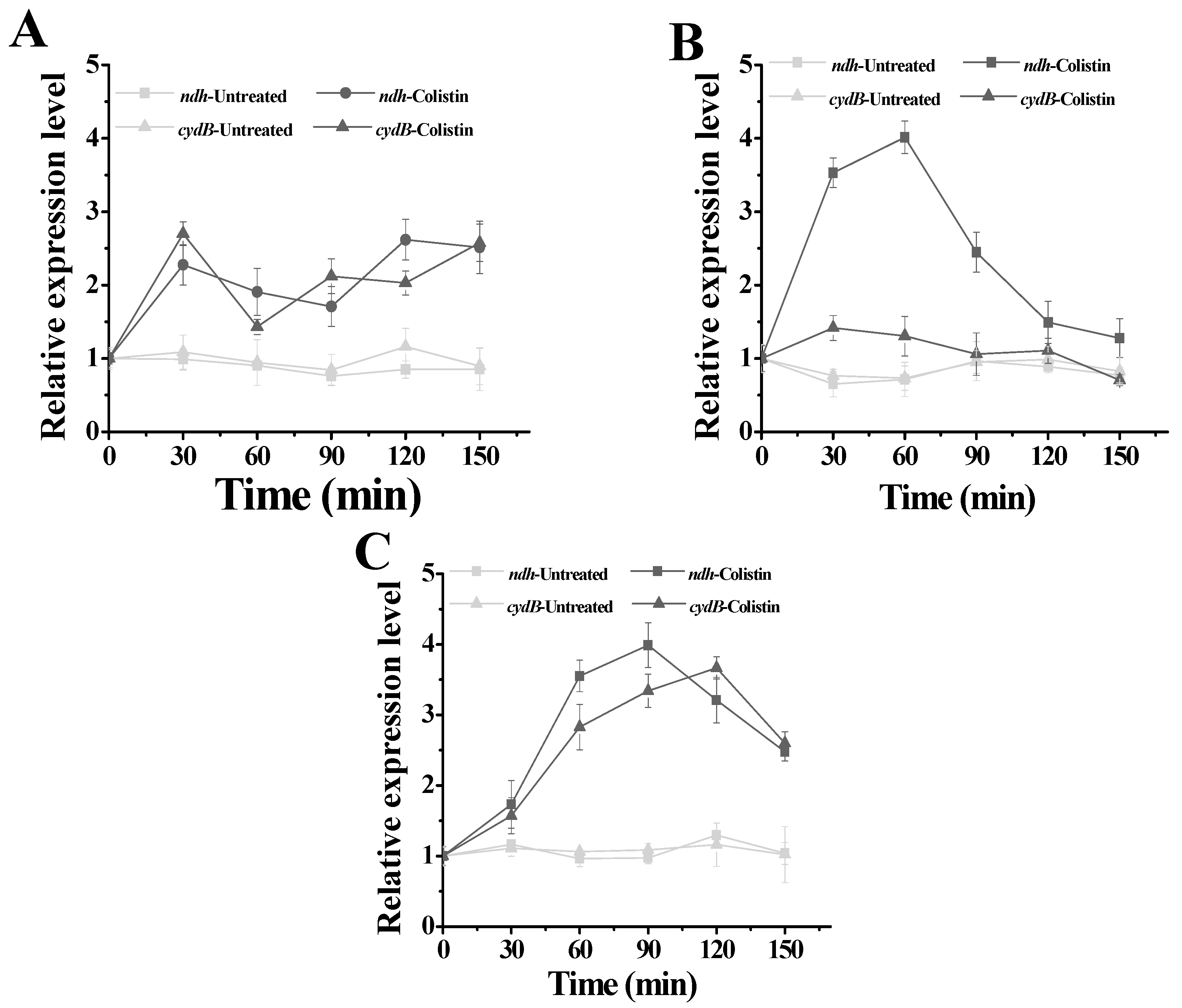
2.4. Influence of Colistin on Respiratory Chain
3. Discussion
4. Materials and Methods
4.1. Strains and Growth Conditions
4.2. Treatment of Drugs on Strains
4.3. Detection of Total Plate Count
4.4. Measurement of Hydroxyl Radicals
4.5. Determination of Protein Carbonyl Content
4.6. Determination of Lipid Peroxidation
4.7. Detection of 8-OHdG Content
4.8. Retrieval of Gene Sequences
4.9. Analysis of Gene Expression Using Quantitative Real-Time PCR (qPCR)
4.10. Detection of MDH and α-KGDH Activities, and Oxaloacetate Concentration
4.11. Detection of NAD+ and NADH Levels
4.12. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Competing interests
References
- Rice, B.L. Challenges in identifying new antimicrobial agents effective for treating infections with Acinetobacter baumannii and Pseudomonas aeruginosa. Clin. Infect. Dis. 2006, 43, 100–105. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Nation, R.; Turnidge, J.D.; Milne, R.W.; Coulthard, K.; Rayner, C.R.; Paterson, D.L. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect. Dis. 2006, 6, 589–601. [Google Scholar] [CrossRef]
- Li, J.; Nation, R. Old polymyxins are back: is resistance close? Clin. Infect. Dis. 2006, 43, 663–664. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Qin, W.; Lin, J.; Fang, S.; Qiu, J. Antibacterial mechanisms of polymyxin and bacterial resistance. Biomed Res. Int. 2015, 2015, 679109. [Google Scholar] [CrossRef] [PubMed]
- Velkov, T.; Thompson, P.E.; Nation, R.L.; Li, J. Structure-activity relationships of polymyxin antibiotics. J. Med. Chem. 2010, 53, 1898–1916. [Google Scholar] [CrossRef] [PubMed]
- Velkov, T.; Roberts, K.D.; Nation, R.L.; Thompson, P.E.; Li, J. Pharmacology of polymyxins: new insights into an old class of antibiotics. Future Microbiol. 2013, 8, 711–724. [Google Scholar] [CrossRef] [PubMed]
- Pristovsek, P.; Kidric, J. The search for molecular determinants of LPS inhibition by proteins and peptides. Curr. Top. Med. Chem. 2004, 4, 1185–1201. [Google Scholar] [CrossRef] [PubMed]
- Abachin, E.; Poyart, C.; Pellegrini, E.; Milohanic, E.; Fiedler, F.; Berche, P.; Trieu-Cuot, P. Formation of D-alanyl-lipoteichoic acid is required for adhesion and virulence of Listeria monocytogenes. Mol. Microbiol. 2002, 43, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Abi Khattar, Z.; Rejasse, A.; Destoumieux-Garzón, D.; Escoubas, J.M.; Sanchis, V.; Lereclus, D.; Givaudan, A.; Kallassy, M.; Nielsen-Leroux, C.; Gaudriault, S. The dlt operon of Bacillus cereus is required for resistance to cationic antimicrobial peptides and for virulence in insects. J. Bacteriol. 2009, 191, 7063–7073. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Cai, Y.; Qin, W.; Lin, J.; Qiu, J. Polymyxin E induces rapid Paenibacillus polymyxa death by damaging cell membrane while Ca2+ can protect cells from damage. PLoS ONE 2015, 10, e0135198. [Google Scholar] [CrossRef]
- Kohanski, M.A.; Dwyer, D.J.; Hayete, B.; Lawrence, C.A.; Collins, J.J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 2007, 130, 797–810. [Google Scholar] [CrossRef] [PubMed]
- Lobritz, M.A.; Belenky, P.C.; Porter, B.; Gutierrez, A.; Yang, J.H.; Schwarz, E.G.; Dwyer, D.J.; Khalil, A.S.; Collins, J.J. Antibiotic efficacy is linked to bacterial cellular respiration. Proc. Natl. Acad. Sci. USA 2015, 112, 8173–8180. [Google Scholar] [CrossRef] [PubMed]
- Dwyer, D.J.; Belenky, P.A.; Yang, J.H.; Macdonald, I.C.; Martell, J.D.; Takahashi, N.; Chan, C.T.; Lobritz, M.A.; Braff, D.; Schwarz, E.G.; et al. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc. Natl. Acad. Sci. USA 2014, 111, 2100–2109. [Google Scholar] [CrossRef] [PubMed]
- Belenky, P.; Ye, J.D.; Porter, C.B.; Cohen, N.R.; Lobritz, M.A.; Ferrante, T.; Jain, S.; Korry, B.J.; Schwarz, E.G.; Walker, G.C.; et al. Bactericidal antibiotics induce toxic metabolic perturbations that lead to cellular damage. Cell Rep. 2015, 13, 968–980. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Imlay, J.A. Cell death from antibiotics without the involvement of reactive oxygen species. Science 2013, 339, 1210–1213. [Google Scholar] [CrossRef] [PubMed]
- Keren, I.; Wu, Y.; Inocencio, J.; Mulcahy, L.R.; Lewis, K. Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science 2013, 339, 1213–1216. [Google Scholar] [CrossRef] [PubMed]
- Kohanski, M.A.; Dwyer, D.J.; Collins, J.J. How antibiotics kill bacteria: from targets to networks. Nat. Rev. Microbiol. 2010, 8, 423–435. [Google Scholar] [CrossRef] [PubMed]
- Ramos, P.I.; Custódio, M.G.; Quispe Saji, G.D.; Cardoso, T.; da Silva, G.L.; Braun, G.; Martins, W.M.; Girardello, R.; de Vasconcelos, A.T.; Fernández, E.; et al. The polymyxin B-induced transcriptomic response of a clinical, multidrug-resistant Klebsiella pneumoniae involves multiple regulatory elements and intracellular targets. BMC Genomics 2016, 17, 737. [Google Scholar] [CrossRef]
- Baek, S.H.; Li, A.H.; Sassetti, C.M. Metabolic regulation of mycobacterial growth and antibiotic sensitivity. PLoS Biol. 2011, 9, e1001065. [Google Scholar] [CrossRef]
- Nandakumar, M.; Nathan, C.; Rhee, K.Y. Isocitrate lyase mediates broad antibiotic tolerance in Mycobacterium tuberculosis. Nat. Commun. 2014, 5, 4306–4316. [Google Scholar] [CrossRef]
- Van Acker, H.; Van Dijck, P.; Coenye, T. Molecular mechanisms of antimicrobial tolerance and resistance in bacterial and fungal biofilms. Trends Microbiol. 2014, 22, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Dwyer, D.J.; Kohanski, M.A.; Hayete, B.; Collins, J.J. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol. Syst. Biol. 2007, 3, 91. [Google Scholar] [CrossRef] [PubMed]
- Dwyer, D.J.; Collins, J.J.; Walker, G.C. Unraveling the physiological complexities of antibiotic lethality. Annu. Rev. Pharmacol. Toxicol. 2014, 55, 313–332. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Zhu, Y.; Qin, W.; Yin, J.; Qiu, J. Oxidative stress induced by polymyxin E involves in rapid killing of Paenibacillus polymyxa. Biomed Res. Int. 2017, 2017, 5437139. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Quyen, T.D.; Le, H.T. Cloning and enhancing production of a detergent- and organic-solvent-resistant nattokinase from Bacillus subtilis VTCC-DVN-12-01 by using an eight-protease-gene-deficient Bacillus subtilis WB800. Microb. Cell Fact. 2013, 12, 79. [Google Scholar] [CrossRef]
- Jeong, H.; Park, S.Y.; Chung, W.H.; Kim, S.H.; Kim, N.; Park, S.H.; Kim, J.F. Draft genome sequence of the Paenibacillus polymyxa type strain (ATCC 842T), a plant growth-promoting bacterium. J. Bacteriol. 2011, 193, 5026–5027. [Google Scholar] [CrossRef]
- Sampson, T.R.; Liu, X.; Schroeder, M.R.; Kraft, C.S.; Burd, E.M.; Weiss, D.S. Rapid killing of Acinetobacter baumannii by polymyxins is mediated by a hydroxyl radical death pathway. Antimicrob. Agents Chemother. 2012, 56, 5642–5649. [Google Scholar] [CrossRef]
- Fedorova, M.; Bollineni, R.C.; Hoffmann, R. Protein carbonylation as a major hallmark of oxidative damage: update of analytical strategies. Mass Spectrom. Rev. 2014, 33, 79–97. [Google Scholar] [CrossRef]
- Porter, N.A.; Caldwell, S.E.; Mills, K.A. Mechanisms of free radical oxidation of unsaturated lipids. Lipids 1995, 30, 277–290. [Google Scholar] [CrossRef]
- Belenky, P.; Camacho, D.; Collins, J.J. Fungicidal drugs induce a common oxidative-damage cellular death pathway. Cell Rep. 2013, 3, 350–358. [Google Scholar] [CrossRef]
- Niebisch, A.; Bott, M. Purification of a cytochrome bc1-aa3 supercomplex with quinol oxidase activity from Corynebacterium glutamicum. J. Biol. Chem. 2003, 278, 4339–4346. [Google Scholar] [CrossRef] [PubMed]
- Deris, Z.Z.; Akter, J.; Sivanesan, S.; Roberts, K.D.; Thompson, P.E.; Nation, R.L.; Li, J.; Velkov, T. A secondary mode of action of polymyxins against Gram-negative bacteria involves the inhibition of NADH-quinone oxidoreductase activity. J. Antibiot. (Tokyo) 2014, 67, 147–151. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Dong, Z.; Liu, L.; Chen, J. Manipulation of NADH metabolism in industrial strains. Sheng Wu Gong Cheng Xue Bao 2009, 25, 161–169. [Google Scholar] [PubMed]
- Imlay, J.A. Pathways of oxidative damage. Ann. Rev. Microbiol. 2003, 57, 395–418. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Sun, Z.; Yin, J.; Qiu, J. Enhanced production of polymyxin E in Paenibacillus polymyxa by replacement of glucose by starch. Biomed Res. Int. 2018, 2018, 1934309. [Google Scholar] [CrossRef]
- Yu, Z.; Zhang, L.; Qin, W.; Yin, J.; Qiu, J. Exogenous catalase stimulates the polymyxin E-induced rapid killing of Paenibacillus polymyxa. Int. J. Pept. Res. Ther. 2017, 1–8. [Google Scholar] [CrossRef]
- Brochmann, R.P.; Toft, A.; Ciofu, O.; Briales, A.; Kolpen, M.; Hempel, C.; Bjarnsholt, T.; Høiby, N.; Jensen, P.Ø. Bactericidal effect of colistin on planktonic Pseudomonas aeruginosa is independent of hydroxyl radical formation. Int. J. Antimicrob. Agents 2014, 43, 140–147. [Google Scholar] [CrossRef]
- Zusterzeel, P.L.; Mulder, T.P.; Peters, W.H.; Wiseman, S.A.; Steegers, E.A. Plasma protein carbonyls in nonpregnant, healthy pregnant and preeclamptic women. Free Radic. Res. 2000, 33, 471–476. [Google Scholar] [CrossRef]
- Yu, Z.; Yang, Y.; Wang, Y.; Yin, J.; Qiu, J. Reactive oxygen species-scavenging system is involved in L-amino acid oxidase accumulation in Pseudoalteromonas sp. B3. 3 Biotech 2017, 7, 326. [Google Scholar] [CrossRef]
- Yu, Z.; Wang, J.; Lin, J.; Zhao, M.; Qiu, J. Exploring regulation genes involved in the expression of L-amino acid oxidase in Pseudoalteromonas sp. Rf-1. PLoS ONE 2015, 10, e0122741. [Google Scholar] [CrossRef]
- Yu, Z.; Ding, Y.; Yin, J.; Yu, D.; Zhang, J.; Zhang, M.; Ding, M.; Zhong, W.; Qiu, J.; Li, J. Dissemination of genetic acquisition/loss provides a variety of quorum sensing regulatory properties in Pseudoalteromonas. Int. J. Mol. Sci. 2018, 19, 3636. [Google Scholar] [CrossRef] [PubMed]
- Hinc, K.; Iwanicki, A.; Seror, S.; Obuchowski, M. Mapping of a transcription promoter located inside the priA gene of the Bacillus subtilis chromosome. Acta Biochim. Pol. 2006, 53, 497–505. [Google Scholar] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, N.; Isshiki, R.; Kawai, Y.; Tanaka, D.; Sekiguchi, T.; Matsumoto, S.; Tsuneda, S. Stochastic expression of lactate dehydrogenase A induces Escherichia coli persister formation. J. Biosci. Bioeng. 2018, 126, 30–37. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are available from the authors. |





| Genes | Nucleotide Sequences (5′–3′) | Product Size (bp) | |
|---|---|---|---|
| Forward | Reverse | ||
| icdA | TCGCAACGACATCAAACT | CAACCCGATTATCCCATT | 860 |
| sucB | ATTCTTCCGCTTGAGTGC | GACGCCTGAATGTCTTGG | 1465 |
| mdh | GGTAGATGCCTTCATAGCC | AACCCGACAAAGGGAAAA | 685 |
| ndh | GCTGGTTATGGCGGAGTT | CAGTTGGCGGGTATGGAC | 904 |
| cydB | TACACGAGGGAATAGGGA | AGAACGCAGAGTGCTGAT | 712 |
| Genes | Nucleotide Sequences (5′–3′) | Product Size (bp) | |
|---|---|---|---|
| Forward | Reverse | ||
| icdA | GTTGGAAGCCATCCGTGAGT | TGACGCCGGACAGAATTACG | 866 |
| sucB | CACAGGAACGCAAGTCGTTG | TACTCGAAGCCGAGGACTGA | 639 |
| mdh | CACCCAGATCATGGGCACTT | GTAGGCACACCGAGGAACAA | 1462 |
| ndh | GTGGATCGGATGCCCTTTCA | GTGGGCAATCTGCTCTCCTT | 780 |
| cydB | ACTGAGACGCTTATGGAAA | AAAGGGTGGACAGGAACG | 666 |
| Genes | Nucleotide Sequences (5′–3′) | Product Size (bp) | |
|---|---|---|---|
| Forward | Reverse | ||
| icdA | TCTTGTCTGAGCGCTACGTT | GTGGCTCCCTGCTGAAACAT | 117 |
| sucB | TCCATAGCGTCTGTACCCGA | GTGAATGCGGACGATCCTGA | 114 |
| mdh | ACCATATCGTCACCGTGTCC | AGGTGTGCTTGATACGGCAA | 108 |
| ndh | CACGCGTGACAGTGGTAAAC | TGGCATTGCAACCGCTTTTT | 101 |
| cydB | AACAGCGAGCGGAATGGTAA | CCGGAAAATGGCGCAAAAGA | 124 |
| sigA | GCCTGTCTGATCCACCACGTAGC | CGGTATGTCGGACGCGGTATG | 137 |
| Genes | Nucleotide Sequences (5′–3′) | Product Size (bp) | |
|---|---|---|---|
| Forward | Reverse | ||
| icdA | CCTATTGGCGGTGGTATCCG | TACTGGAGATGGGACACCGT | 108 |
| sucB | GCTTGTCCCTGTACTCGACG | TGCCAATACATTCAGACGGC | 103 |
| mdh | GGTACCGCTCGTACGCTATT | CACCCACTCGTGTACGTTGT | 101 |
| ndh | TCATCGCGCTAGGTTGTACC | CAAGCGCAGATACGTTTGGC | 107 |
| cydB | GCACTGTTGATCGAGCTCAC | GAAGGATGGGCTTTCGGGAT | 104 |
| 16S rRNA | GAGAAGAAAGCCCCGGCTAA | ACCAGACTTAAAGAGCCGCC | 116 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, Z.; Zhu, Y.; Fu, J.; Qiu, J.; Yin, J. Enhanced NADH Metabolism Involves Colistin-Induced Killing of Bacillus subtilis and Paenibacillus polymyxa. Molecules 2019, 24, 387. https://doi.org/10.3390/molecules24030387
Yu Z, Zhu Y, Fu J, Qiu J, Yin J. Enhanced NADH Metabolism Involves Colistin-Induced Killing of Bacillus subtilis and Paenibacillus polymyxa. Molecules. 2019; 24(3):387. https://doi.org/10.3390/molecules24030387
Chicago/Turabian StyleYu, Zhiliang, Yuyi Zhu, Jianv Fu, Juanping Qiu, and Jianhua Yin. 2019. "Enhanced NADH Metabolism Involves Colistin-Induced Killing of Bacillus subtilis and Paenibacillus polymyxa" Molecules 24, no. 3: 387. https://doi.org/10.3390/molecules24030387
APA StyleYu, Z., Zhu, Y., Fu, J., Qiu, J., & Yin, J. (2019). Enhanced NADH Metabolism Involves Colistin-Induced Killing of Bacillus subtilis and Paenibacillus polymyxa. Molecules, 24(3), 387. https://doi.org/10.3390/molecules24030387






