Antibacterial Isoquinoline Alkaloids from the Fungus Penicillium Spathulatum Em19
Abstract
:1. Introduction
2. Results and Discussion
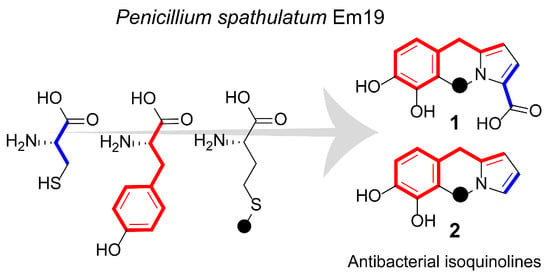
2.1. Isolation of Compounds
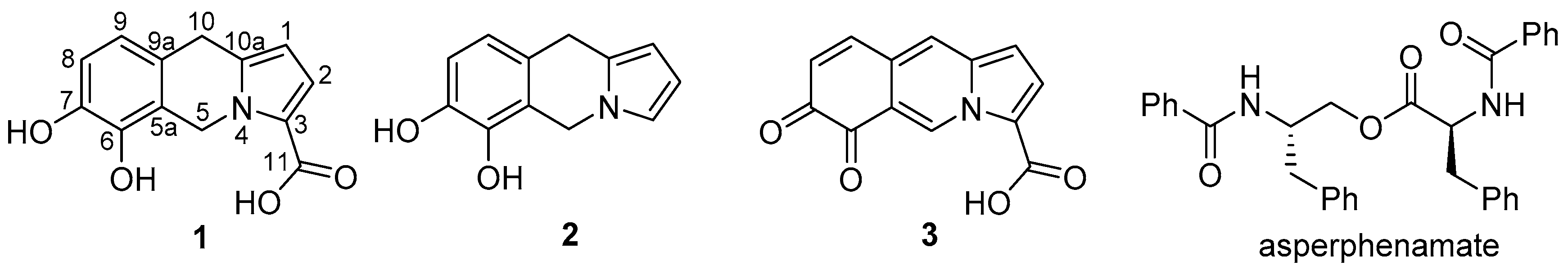
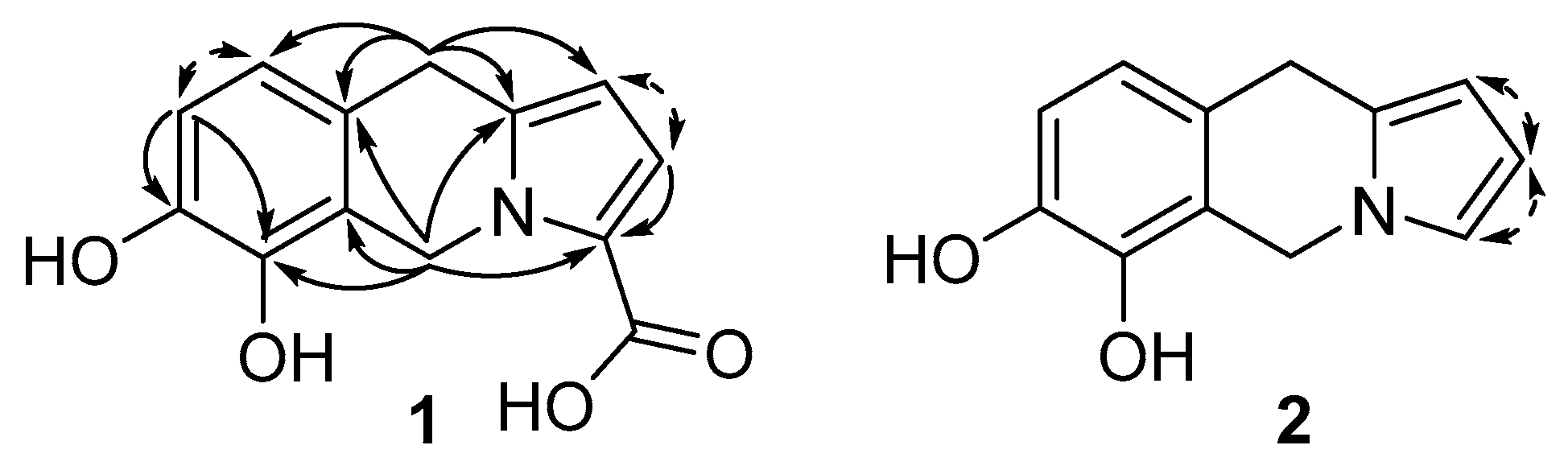
2.2. Compound Identification
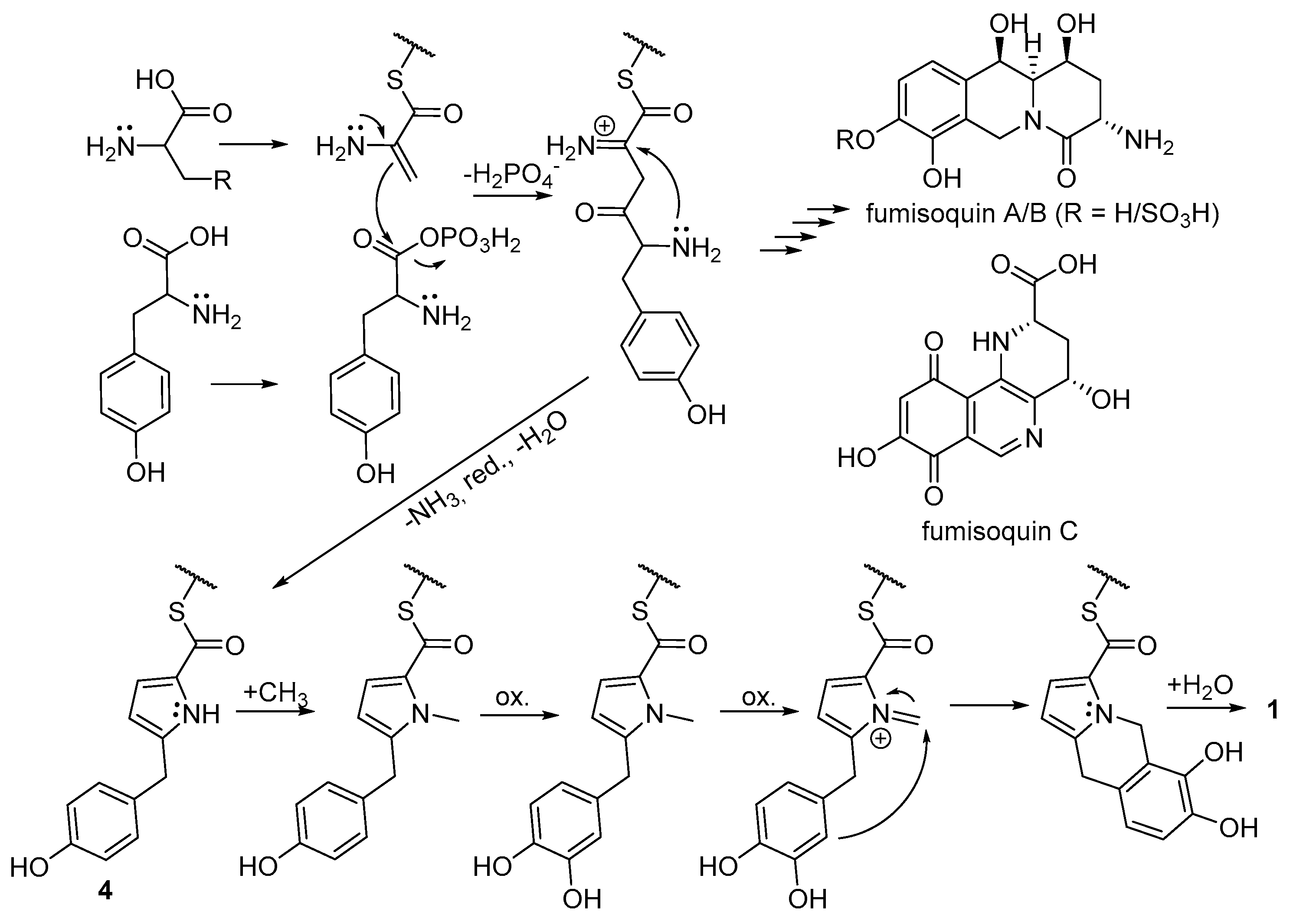
2.3. Biosynthetic Considerations
2.4. Biological Activity of Compounds 1 and 2
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Strain Origin, Identity and Maintenance
3.3. Culture Conditions and Metabolite Sampling
3.4. Isolation of Compounds 1 and 2
3.5. In Vitro Bioassay
3.6. Analysis by HPLC-MS and UHPLC-MS
3.7. MIC Determination
3.8. Determination of Toxicity
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Global Antimicrobial Resistance Surveillance System (GLASS) Report: Early Implementation 2017-2018. World Health Organization, 2018. Licence: CC BY-NC-SA 3.0 IGO. Available online: https://apps.who.int/iris/bitstream/handle/10665/279656/9789241515061-eng.pdf (accessed on 11 September 2019).
- Laxminarayan, R.; Duse, A.; Wattal, C.; Zaidi, A.K.M.; Wertheim, H.F.L.; Sumpradit, N.; Vlieghe, E.; Hara, G.L.; Gould, I.M.; Goossens, H.; et al. Antibiotic resistance—the need for global solutions. Lancet. Infect. Dis. 2013, 13, 1057–1098. [Google Scholar] [CrossRef] [Green Version]
- Infectious Diseases Society of America. The 10 × ‘20 Initiative: Pursuing a Global Commitment to Develop 10 New Antibacterial Drugs by 2020. Clin. Infect. Dis. 2010, 50, 1081–1083. [Google Scholar] [CrossRef] [Green Version]
- Drugs@FDA: FDA Approved Drug Products. Available online: www.fda.gov/drugsatfda (accessed on 24 September 2019).
- Srivastava, A.; Talaue, M.; Liu, S.; Degen, D.; Ebright, R.Y.; Sineva, E.; Chakraborty, A.; Druzhinin, S.Y.; Chatterjee, S.; Mukhopadhyay, J.; et al. New target for inhibition of bacterial RNA polymerase: ‘switch region’. Curr. Opin. Microbiol. 2011, 14, 532–543. [Google Scholar] [CrossRef] [Green Version]
- Theuretzbacher, U.; Gottwalt, S.; Beyer, P.; Butler, M.; Czaplewski, L.; Lienhardt, C.; Moja, L.; Paul, M.; Paulin, S.; Rex, J.; et al. Analysis of the clinical antibacterial and antituberculosis pipeline. Lancet. Infect. Dis. 2019, 19, E40–E50. [Google Scholar] [CrossRef]
- Dictionary of Natural Products. Available online: http://dnp.chemnetbase.com/faces/chemical/ChemicalSearch.xhtml (accessed on 24 September 2019).
- Fleming, A. On the antibacterial action of cultures of a Penicillium, with special reference to their use in the isolation of B. influenzae. Br. J. Exp. Pathol. 1929, 10, 226–236. [Google Scholar] [CrossRef]
- Birch, A.J.; Wright, J.J. Studies in relation to biosynthesis—XLII: The structural elucidation and some aspects of the biosynthesis of the brevianamides-A and –E. Tetrahedron 1970, 26, 2329–2344. [Google Scholar] [CrossRef]
- Oxford, A.E.; Raistrick, H.; Simonart, P. Studies in the biochemistry of micro-organisms. Biochem. J. 1939, 33, 240–248. [Google Scholar] [CrossRef]
- Samson, R.A.; Stolk, A.C.; Hadlok, R. Revision of the subsection Fasciculata of Penicillium and some allied species. Stud. Mycol. Baarn. 1976, 11, 1–47. [Google Scholar]
- MacMillan, J.; Vanstone, A.E.; Yeboah, S.K. Structure of Wortmannin a Steroidal Fungal Metabolite. Chem. Commun. 1968, 11, 613–614. [Google Scholar] [CrossRef]
- Dunn, G.E.; Lee, G.K.L. Kinetics and Mechanism of the Decarboxylation of Pyrrole-2-carboxylic Acid in Aqueous Solution. Can. J. Chem. 1971, 49, 1032–1035. [Google Scholar] [CrossRef]
- Clark, A.M.; Hufford, C.D.; Robertson, L.W. Two metabolites from Aspergillus flavipes. Lloydia 1977, 40, 146–151. [Google Scholar]
- Frisvad, J.C.; Houbraken, J.; Popma, S.; Samson, R.A. Two new Penicillium species Penicillium buchwaldii and Penicillium spathulatum, producing the anticancer compound asperphenamate. FEMS Microbiol. Lett. 2013, 339, 77–92. [Google Scholar] [CrossRef] [Green Version]
- Del Valle, P.; Martinez, A.L.; Figueroa, M.; Raja, H.A.; Mata, R. Alkaloids from the Fungus Penicillium spathulatum as α-Glucosidase Inhibitors. Planta Medica 2016, 82, 1286–1294. [Google Scholar] [CrossRef]
- Yao, G.M.; Sebisubi, F.M.; Voo, L.Y.C.; Ho, C.C.; Tan, G.T.; Chang, L.C. Citrinin Derivatives from the Soil Filamentous Fungus Penicillium sp. H9318. J. Braz. Chem. Soc. 2011, 22, 1125–1129. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.L.X.; Lin, Y.; Yuan, J.; Lu, Y.; Zhu, X.; Li, J.; Li, M.; Lin, Y.; He, J.; Liu, L. Meroterpenes and azaphilones from marine mangrove endophytic fungus Penicillium 303#. Fitoterapia 2014, 97, 241–246. [Google Scholar]
- Trisuwan, K.; Rukachaisirikul, V.; Sukpondma, Y.; Phongpaichit, S.; Preedanon, S.; Sakayaroj, J. Furo[3,2-h]isochroman,furo[3,2-h]isoquinoline, isochroman, phenol, pyranone, and pyrone derivatives from the seafan-derived fungus Penicillium sp. PSU-F40. Tetrahedron 2010, 66, 4484–4489. [Google Scholar] [CrossRef]
- Ogihara, J.; Kato, J.; Oishi, K.; Fujimoto, Y. PP-R, 7-(2-hydroxyethyl)-monascorubramine, a red pigment produced in the mycelia of Penicillium sp AZ. J. Biosci. Bioengin. 2001, 91, 44–47. [Google Scholar] [CrossRef]
- Ogihara, J.; Kato, J.; Oishi, K.; Fujimoto, Y.; Eguchi, T. Production and structural analysis of PP-V, a homologue of monascorubramine, produced by a new isolate of Penicillium sp. J. Biosci. Bioengin. 2000, 90, 549–554. [Google Scholar] [CrossRef]
- Evidente, A.; Andolfi, A.; Abou-Donia, A.H.; Touema, S.M.; Hammoda, H.M.; Shawky, E.; Motta, A. (−)-Amarbellisine, a lycorine-type alkaloid from Amaryllis belladonna L. growing in Egypt. Phytochemistry 2004, 65, 2113–2118. [Google Scholar] [CrossRef]
- Subramaniam, G.; Ang, K.K.H.; Ng, S.; Buss, A.D.; Butler, M.S. A benzopyrroloisoquinoline alkaloid from Ficus fistulosa. Phytochem. Lett. 2009, 2, 88–90. [Google Scholar] [CrossRef]
- Baccile, J.A.; Spraker, J.E.; Le, H.H.; Brandenburger, E.; Gomez, C.; Bok, J.W.; Macheleidt, J.; Brakhage, A.A.; Hoffmeister, D.; Keller, N.P.; et al. Plant-like biosynthesis of isoquinoline alkaloids in Aspergillus fumigatus. Nat. Chem. Biol. 2016, 12, 419–424. [Google Scholar] [CrossRef] [Green Version]
- Pascard, C.; Ducruix, A.; Lunel, J.; Prange, T. Highly modified cysteine-containing antibiotics. Chemical structure and configuration of nosiheptide. J. Am. Chem. Soc. 1977, 99, 6418–6423. [Google Scholar] [CrossRef]
- Gross, E.; Morell, J.L. Presence of dehydroalanine in the antibiotic nisin and its relation to activity. J. Am. Chem. Soc. 1967, 89, 2791–2792. [Google Scholar] [CrossRef]
- Houck, D.R.; Chen, L.C.; Keller, P.J.; Beale, J.M.; Floss, H.G. Biosynthesis of the modified peptide antibiotic nosiheptide in Streptomyces actuosus. J. Am. Chem. Soc. 1988, 110, 5800–5806. [Google Scholar] [CrossRef]
- Weil, H.-P.; Beck-Sickinger, A.G.; Metzger, J.; Stevanovic, S.; Jung, G.; Josten, M.; Sahl, H.-G. Biosynthesis of the lantibiotic Pep5. Isolation and characterization of a prepeptide containing dehydroamino acids. Eur. J. Biochem. 1990, 194, 217–223. [Google Scholar] [CrossRef]
- Siegers, K.; Heinzmann, S.; Entian, K.-D. Biosynthesis of Lantibiotic Nisin. Posttranslational modification of its prepeptide occurs at a multimeric membrane-associated lanthionine synthetase complex. J. Biol. Chem. 1996, 271, 12294–12301. [Google Scholar] [CrossRef] [Green Version]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA Genes for phylogenetics. In PCR-Protocols and Applications—A Laboratory Manual; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: Boston, MA, USA, 1990; pp. 315–324. [Google Scholar]
- Levenfors, J.J.; Hedman, R.; Thaning, C.; Gerhardson, B.; Welch, C.J. Broad-spectrum antifungal metabolites produced by the soil bacterium Serratia plymuthica A 153. Soil. Biol. Biochem. 2004, 36, 677–685. [Google Scholar] [CrossRef]
- Stanier, R.Y.; Palleroni, N.J.; Doudoroff, M. The aerobic pseudomonads: A taxonomic study. J. Gen. Microbiol. 1966, 43, 159–271. [Google Scholar] [CrossRef] [Green Version]
- Pohanka, A.; Broberg, A.; Johansson, M.; Kenne, L.; Levenfors, J. Antimicrobial Dialkylresorcinols from Pseudomonas sp. Ki19. J. Nat. Prod. 2005, 68, 1380–1385. [Google Scholar] [CrossRef]
- Laiho, O. Paxillus involutus as a mycorrhizal symbiont of forest trees. Acta Forest. Fenn. 1970, 106, 1–72. [Google Scholar] [CrossRef] [Green Version]
- Thaning, C.; Welch, C.J.; Borowicz, J.J.; Hedman, R.; Gerhardson, B. Suppression of Sclerotinia sclerotiorum apothecial formation by the soil bacterium Serratia plymuthica: Identification of a chlorinated macrolide as one of the causal agents. Soil Biol. Biochem. 2001, 33, 1817–1826. [Google Scholar] [CrossRef]
- Shirling, E.B.; Gottlieb, D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 1966, 16, 313–340. [Google Scholar] [CrossRef] [Green Version]
- Weislow, O.S.; Kiser, R.; Fine, D.L.; Bader, J.; Shoemaker, R.H.; Boyd, M.R. New soluble-formazan assay for HIV-1 cytopathic effects: Application to high-flux screening of synthetic and natural products for AIDS-antiviral activity. J. Natl. Cancer Inst. 1989, 81, 577–586, [Published erratum in J. Natl. Cancer Inst. 1989, 81, 963]. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |
| 1 | 2 | 3 | ||||
|---|---|---|---|---|---|---|
| Pos. | δC | δH, mult. (J in Hz) | δC | δH, mult. (J in Hz) | δC | δH, mult. (J in Hz) |
| 1 | 105.5 | 6.02, d (4.0) | 103.7 | 5.83, m | 108.8 | 6.92, d (4.3) |
| 2 | 119.0 | 6.97, d (4.0) | 108.6 | 6.04, dd (3.4, 2.7) | 125.8 | 7.64, d (4.3) |
| 3 | 121.6 | - | 118.9 | 6.76, dd (2.7, 1.7) | 120.8 | - |
| 5 | 43.4 | 5.54, s | 43.4 | 5.05, br s | 134.2 | 10.16, s |
| 5a | 120.6 | - | 121.0 | - | 119.6 | - |
| 6 | 142.7 | - | 142.6 | - | 179.1 | - |
| 7 | 143.5 | - | 143.3 | - | 181.4 | - |
| 8 | 115.0 | 6.81, d (8.1) | 114.9 | 6.77, d (8.0) | 129.3 | 6.48, d (10.0) |
| 9 | 119.2 | 6.67, d (8.1) | 119.4 | 6.65, d (8.0) | 144.5 | 7.71, d (10.0) |
| 9a | 124.7 | - | 126.3 | - | 126.7 | - |
| 10 | 28.4 | 3.97, s | 28.1 | 3.87, br s | 121.4 | 7.85, s |
| 10a | 136.9 | - | 128.2 | - | 137.8 | - |
| 11 | 162.4 | - | - | - | n.d.a | - |
| MIC (μg/mL) | ||||
|---|---|---|---|---|
| Test Organisms | 1 | 2 | MEM | CIP |
| Escherichia coli LMG15862 | 15 | 5 | 0.5 | 0.06 |
| Acinetobacter baumannii LMG1041T | 15 | 5 | 2 | 1 |
| Enterobacter cloacae LMG2783T | 15 | 5 | 1 | 0.03 |
| Klebsiella pneumonia LMG20218 | 64 | 32 | 1 | 0.5 |
| Pseudomonas aeruginosa LMG6395 | >64 | 64 | 1 | 0.25 |
| Staphylococcus aureus LMG15975 | 4 | 1 | 0.5 | 0.25 |
| Candida albicans H-29 | >64 | >64 | n.t.a | n.t. |
| Aspergillus fumigatus J7 | >64 | >64 | n.t. | n.t. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nord, C.; Levenfors, J.J.; Bjerketorp, J.; Sahlberg, C.; Guss, B.; Öberg, B.; Broberg, A. Antibacterial Isoquinoline Alkaloids from the Fungus Penicillium Spathulatum Em19. Molecules 2019, 24, 4616. https://doi.org/10.3390/molecules24244616
Nord C, Levenfors JJ, Bjerketorp J, Sahlberg C, Guss B, Öberg B, Broberg A. Antibacterial Isoquinoline Alkaloids from the Fungus Penicillium Spathulatum Em19. Molecules. 2019; 24(24):4616. https://doi.org/10.3390/molecules24244616
Chicago/Turabian StyleNord, Christina, Jolanta J. Levenfors, Joakim Bjerketorp, Christer Sahlberg, Bengt Guss, Bo Öberg, and Anders Broberg. 2019. "Antibacterial Isoquinoline Alkaloids from the Fungus Penicillium Spathulatum Em19" Molecules 24, no. 24: 4616. https://doi.org/10.3390/molecules24244616