Quantitative Analysis of Cold Stress Inducing Lipidomic Changes in Shewanella putrefaciens Using UHPLC-ESI-MS/MS
Abstract
:1. Introduction
2. Materials and Methods
2.1. Pretreatment of Samples
2.2. Lipids Separation by UHPLC
2.3. Lipids Quantification by Mass Spectrometric Analysis
2.4. External Calibration Method
2.5. Lipidomic Data Processing
3. Results and Discussion
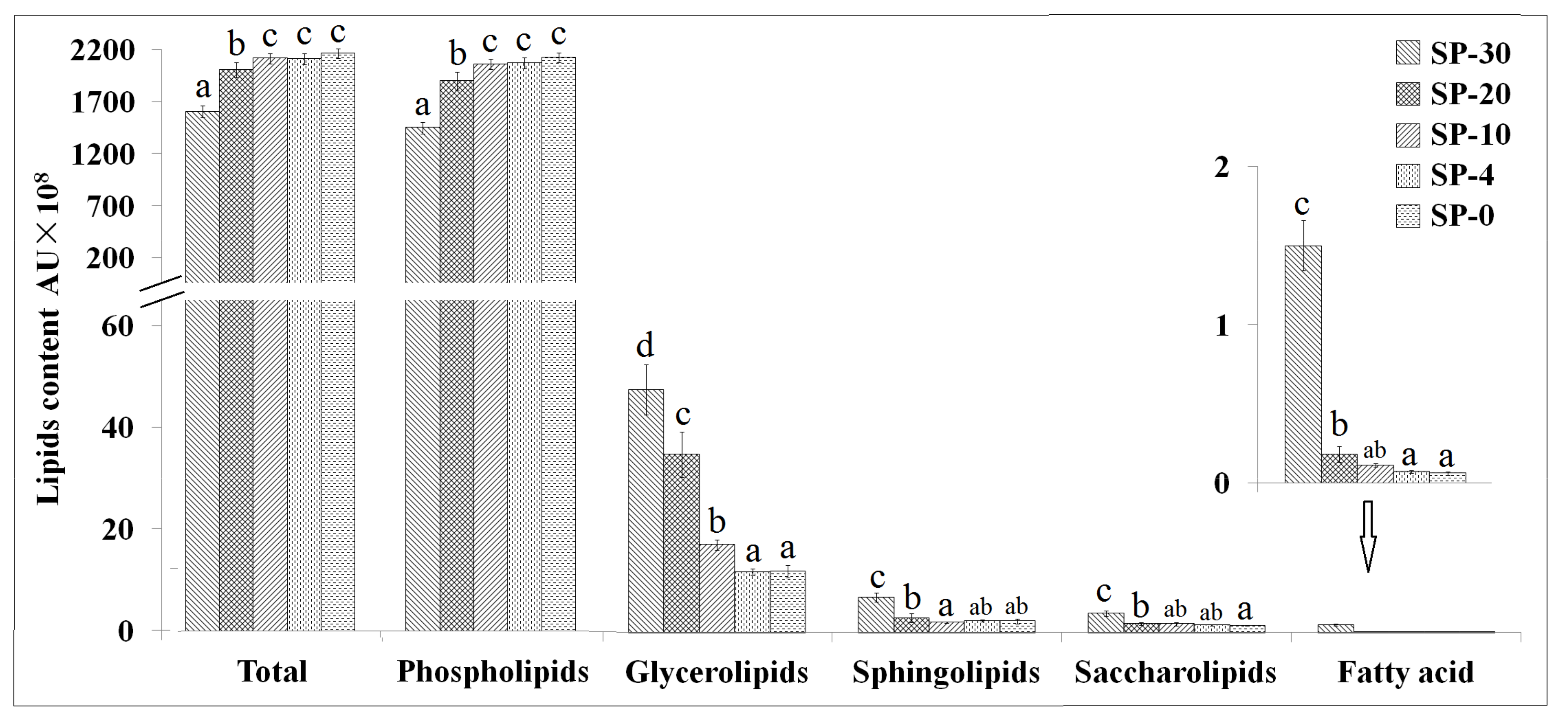
3.1. Changes in Lipids Content
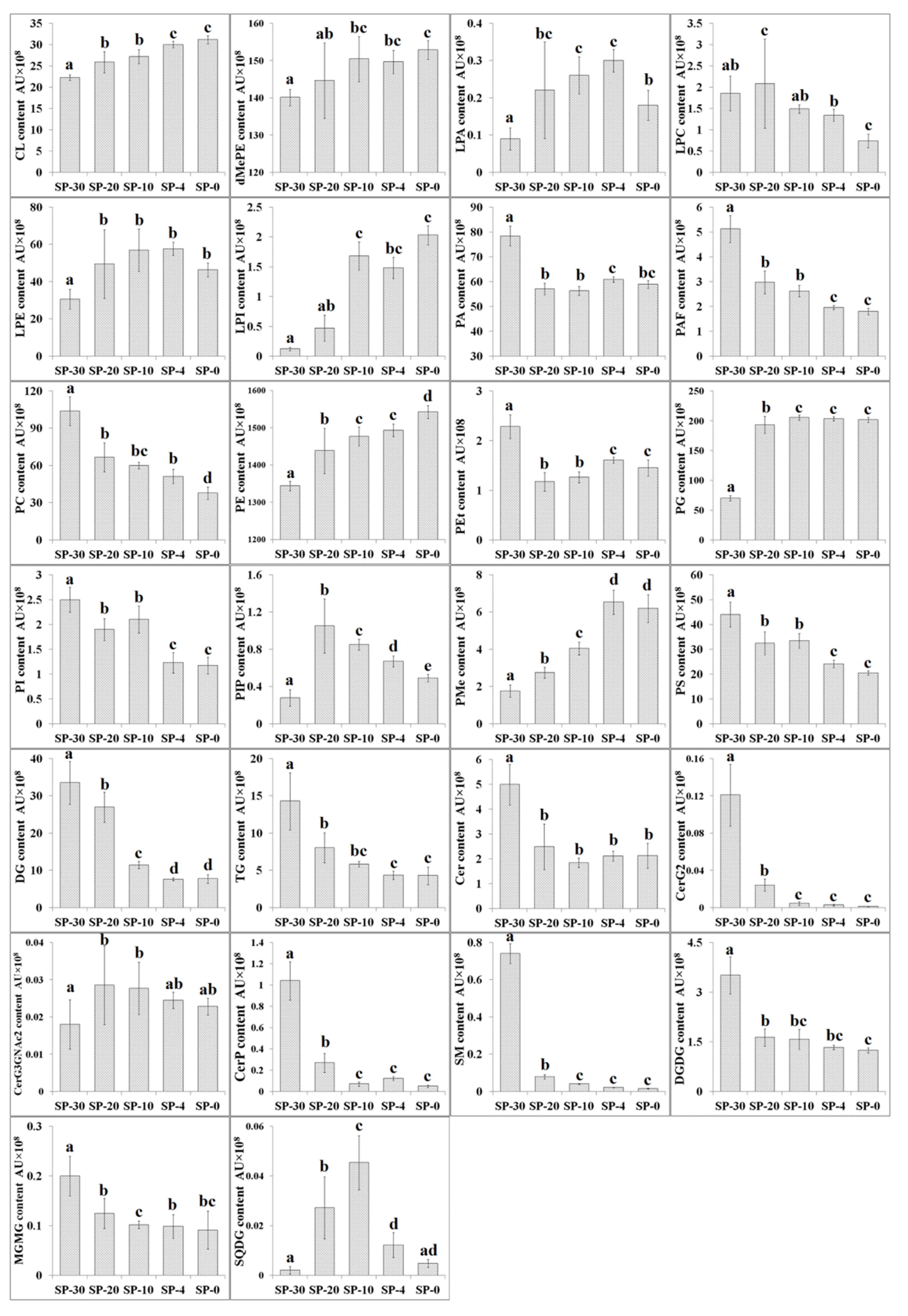
3.2. Changes in Content of Different Compositional Lipid Species
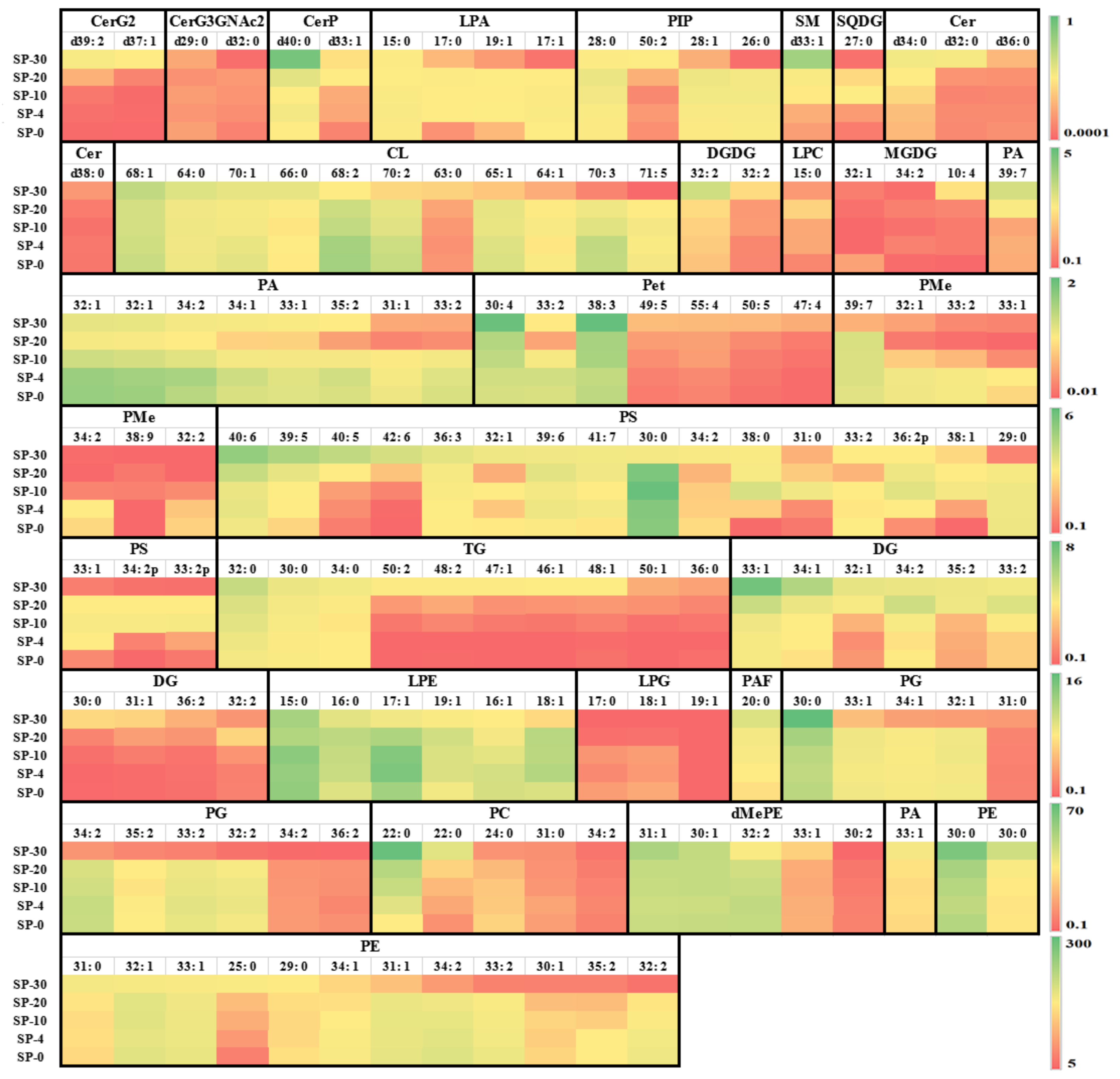
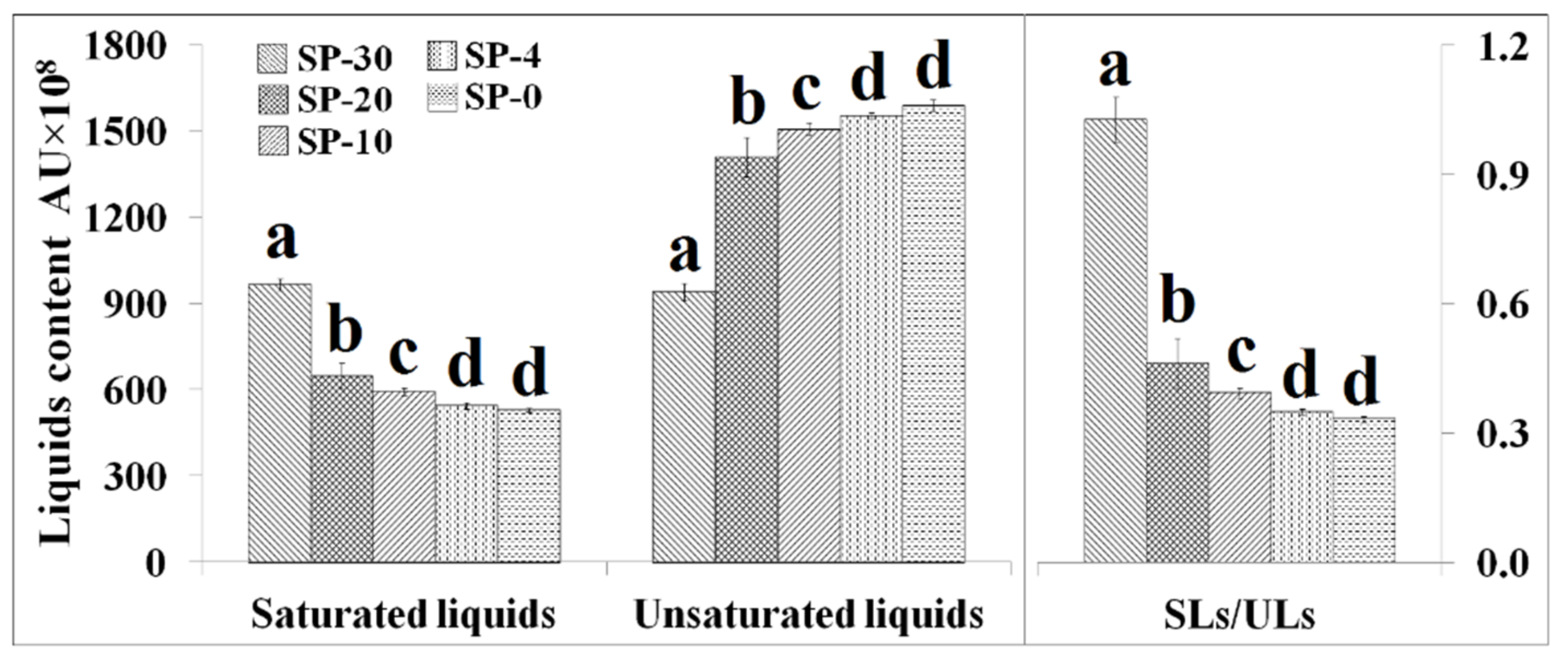
3.3. Changes in the Unsaturation Level of Compositional Lipid Species in Response to Cold Stress
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sohlenkamp, C.; Geiger, O. Bacterial membrane lipids: Diversity in structures and pathways. Fems Microbiol. Rev. 2016, 40, 133–159. [Google Scholar] [CrossRef] [Green Version]
- Rowlett, V.W.; Vkps, M.; Karlstaedt, A.; Dowhan, W.; Taegtmeyer, H.; Margolin, W.; Vitrac, H. The impact of membrane phospholipid alterations in Escherichia coli on cellular function and bacterial stress adaptation. J. Bacteriol. 2017, 199, e00849-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yong-Mei, Z.; Rock, C.O. Membrane lipid homeostasis in bacteria. Nat. Rev. Microbiol. 2008, 6, 222–233. [Google Scholar]
- Montanari, C.; Kamdem, S.L.S.; Serrazanetti, D.I.; Etoa, F.X.; Guerzoni, M.E. Synthesis of cyclopropane fatty acids in Lactobacillus helveticus and Lactobacillus sanfranciscensis and their cellular fatty acids changes following short term acid and cold stresses. Food Microbiol. 2010, 27, 493–502. [Google Scholar] [CrossRef] [PubMed]
- Velly, H.; Bouix, M.; Passot, S.; Penicaud, C.; Beinsteiner, H.; Ghorbal, S.; Lieben, P.; Fonseca, F. Cyclopropanation of unsaturated fatty acids and membrane rigidification improve the freeze-drying resistance of Lactococcus lactis subsp. lactis TOMSC161. Appl. Microbiol. Biotechnol. 2015, 99, 907–918. [Google Scholar] [CrossRef]
- Lopes, C.; Barbosa, J.; Maciel, E.; da Costa, E.; Alves, E.; Domingues, P.; Mendo, S.; Domingues, M.R.M. Lipidomic signature of Bacillus licheniformis I89 during the different growth phases unravelled by high-resolution liquid chromatography-mass spectrometry. Arch. Biochem. Biophys. 2019, 663, 83–94. [Google Scholar] [CrossRef]
- Kishimoto, K.; Urade, R.; Ogawa, T.; Moriyama, T. Nondestructive quantification of neutral lipids by thin-layer chromatography and laser-fluorescent scanning: Suitable methods for “lipidome” analysis. Biochem. Biophys. Res. Commun. 2001, 281, 657–662. [Google Scholar] [CrossRef]
- Xianlin, H.; Gross, R.W. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: A bridge to lipidomics. J. Lipid Res. 2010, 24, 1071–1079. [Google Scholar]
- Laudicella, V.A.; Whitfield, P.D.; Carboni, S.; Doherty, M.K.; Hughes, A.D. Application of lipidomics in bivalve aquaculture, a review. Rev. Aquac. 2019, 1–25. [Google Scholar] [CrossRef]
- Fuchs, B.; Süß, R.; Teuber, K.; Eibisch, M.; Schiller, J. Lipid analysis by thin-layer chromatography-A review of the current state. J. Chromatogr. A 2011, 1218, 2754–2774. [Google Scholar] [CrossRef]
- Crompton, M.J.; Dunstan, R.H. Evaluation of in-situ fatty acid extraction protocols for the analysis of staphylococcal cell membrane associated fatty acids by gas chromatography. J. Chromatogr. B 2018, 1084, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Lather, P.; Mohanty, A.K.; Jha, P.; Garsa, A.K.; Sood, S.K. Changes associated with cell membrane composition of Staphylococcus aureus on acquisition of resistance against class IIa bacteriocin and its in vitro substantiation. Eur. Food Res. Technol. 2015, 240, 101–107. [Google Scholar] [CrossRef]
- Warschawski, D.E.; Arnold, A.A.; Marcotte, I. A new method of assessing lipid mixtures by 31 P magic-angle spinning NMR. Biophys. J. 2018, 114, 1368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bibow, S.; Hiller, S. A guide to quantifying membrane protein dynamics in lipids and other native-like environments by solution-state NMR spectroscopy. Febs J. 2019, 286, 1610–1623. [Google Scholar] [CrossRef] [Green Version]
- Kondakova, T.; Merlet-Machour, N.; Chapelle, M.; Preterre, D.; Dionnet, F.; Feuilloley, M.; Orange, N.; Poc, C.D. A new study of the bacterial lipidome: HPTLC-MALDI-TOF imaging enlightening the presence of phosphatidylcholine in airborne Pseudomonas fluorescens MFAF76a. Res. Microbiol. 2015, 166, 1–8. [Google Scholar] [CrossRef]
- Calvano, C.D.; Ventura, G.; Sardanelli, A.M.M.; Savino, L.; Losito, I.; De Michele, G.; Palmisano, F.; Cataldi, T.R.I. Searching for potential lipid biomarkers of Parkinson’s disease in Parkin-mutant human skin fibroblasts by HILIC-ESI-MS/MS: Preliminary findings. Int. J. Mol. Sci. 2019, 20, 18. [Google Scholar] [CrossRef] [Green Version]
- Blevins, M.S.; Klein, D.R.; Brodbelt, J.S. Localization of cyclopropane modifications in bacterial lipids via 213 nm ultraviolet photodissociation mass spectrometry. Anal. Chem. 2019, 91, 6820–6828. [Google Scholar] [CrossRef]
- Granafei, S.; Losito, I.; Trotta, M.; Italiano, F.; Leo, V.D.; Agostiano, A.; Palmisano, F.; Cataldi, T.R.I. Profiling of ornithine lipids in bacterial extracts of Rhodobacter sphaeroides by reversed-phase liquid chromatography with electrospray ionization and multistage mass spectrometry (RPLC-ESI-MS n ). Anal. Chim. Acta 2016, 903, 110–120. [Google Scholar] [CrossRef]
- Luo, Y.; Javed, M.A.; Deneer, H. Comparative study on nutrient depletion-induced lipidome adaptations in Staphylococcus haemolyticus and Staphylococcus epidermidis. Sci. Rep. 2018, 8, 2356. [Google Scholar] [CrossRef]
- Jeon, J.; Park, S.C.; Jin, H.; Lee, J.W.; Ban, C. Comparative lipidomic profiling of the human commensal bacterium Propionibacterium acnes and its extracellular vesicles. Rsc Adv. 2018, 8, 15241–15247. [Google Scholar] [CrossRef] [Green Version]
- Jørgensen, B.R.; Huss, H.H. Growth and activity of Shewanella putrefaciens isolated from spoiling fish. Int. J. Food Microbiol. 1989, 9, 51–62. [Google Scholar] [CrossRef]
- Mu, H.; Guo, Q.Y.; Wei, S.; Li, B.G.; Zhang, G.W. Inhibitory effects of chitosan combined with nisin on Shewanella spp. isolated from Pseudosciaena crocea. Food Control. 2017, 79, 349–355. [Google Scholar]
- Zhu, J.; Zhao, A.; Feng, L.; Gao, H. Quorum sensing signals affect spoilage of refrigerated large yellow croaker (Pseudosciaena crocea) by Shewanella baltica. Int. J. Food Microbiol. 2016, 217, 146–155. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Lin, H.; Sui, J.; Cao, L. Effects of specific egg yolk antibody (IgY) on the quality and shelf life of refrigerated Paralichthys olivaceus. J. Sci. Food Agric. 2012, 92, 1267–1272. [Google Scholar] [CrossRef] [PubMed]
- Briones, L.S.; Reyes, J.E.; Tabilo-Munizaga, G.E.; Pérez-Won, M.O. Microbial shelf-life extension of chilled Coho salmon (Oncorhynchus kisutch) and abalone (Haliotis rufescens) by high hydrostatic pressure treatment. Food Control. 2010, 21, 1530–1535. [Google Scholar] [CrossRef]
- Parlapani, F.F.; Meziti, A.; Kormas, K.A.; Boziaris, I.S. Indigenous and spoilage microbiota of farmed sea bream stored in ice identified by phenotypic and 16S rRNA gene analysis. Food Microbiol. 2013, 33, 85–89. [Google Scholar] [CrossRef] [PubMed]
- Jääskeläinen, E.; Jakobsen, L.M.A.; Hultman, J.; Eggers, N.; Bertram, H.C.; Björkroth, J. Metabolomics and bacterial diversity of packaged yellowfin tuna (Thunnus albacares) and salmon (Salmo salar) show fish species-specific spoilage development during chilled storage. Int. J. Food Microbiol. 2019, 293, 44–52. [Google Scholar] [CrossRef]
- Remya, S.; Mohan, C.O.; Venkateshwarlu, G.; Sivaraman, G.K. Combined effect of O2 scavenger and antimicrobial film on shelf life of fresh cobia (Rachycentron canadum) fish steaks stored at 2 °C. Food Control. 2017, 71, 71–78. [Google Scholar] [CrossRef]
- Tsogas, A.; Vatavali, K.; Dimitriou, E.; Badeka, A.; Kontakos, S.; Kontominas, M.G. Combined effect of light salting and vacuum packaging on the microbiological, chemical, and sensory attributes of mullet fillets (Mugil cephalus) during refrigerated and frozen/refrigerated storage. J. Food Process. Pres. 2019, 43, e14009. [Google Scholar] [CrossRef]
- Kuuliala, L.; Abatih, E.; Ioannidis, A.G.; Vanderroost, M.; De Meulenaer, B.; Ragaert, P.; Devlieghere, F. Multivariate statistical analysis for the identification of potential seafood spoilage indicators. Food Control. 2018, 84, 49–60. [Google Scholar] [CrossRef]
- Olatunde, O.O.; Benjakul, S. Natural preservatives for extending the shelf-life of seafood: A revisit. Compr. Rev. Food Sci. Food Saf. 2018, 17, 1595–1612. [Google Scholar] [CrossRef] [Green Version]
- Esposito, G.; Sciuto, S.; Acutis, P.L. Quantification of TMA in fishery products by direct sample analysis with high resolution mass spectrometry. Food Control 2018, 94, 162–166. [Google Scholar] [CrossRef]
- Tribelli, M.P.; López, I.N. Reporting key features in cold-adapted bacteria. Life 2018, 8, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Descalzo, L.; Alcazar, A.; Baquero, F.; Cid, C. Biotechnological applications of cold-adapted bacteria. In Extremophiles: Sustainable Resources and Biotechnological Implications; Singh, O.V., Ed.; Wiley: Hoboken, NJ, USA, 2012; pp. 159–174. [Google Scholar]
- Lily, T.; Williams, T.J.; Cowley, M.J.; Lauro, F.M.; Michael, G.; Raftery, M.J.; Ricardo, C. Cold adaptation in the marine bacterium, Sphingopyxis alaskensis, assessed using quantitative proteomics. Env. Microbiol. 2010, 12, 2658–2676. [Google Scholar]
- Baraúna, R.; Freitas, D.; Pinheiro, J.; Folador, A.; Silva, A. A proteomic perspective on the bacterial adaptation to cold: Integrating OMICs data of the Psychrotrophic bacterium Exiguobacterium antarcticum B7. Proteomes 2017, 5, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barria, C.; Malecki, M.; Arraiano, C.M. Bacterial adaptation to cold. Microbiology 2013, 159, 2437–2443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Budiman, C.; Koga, Y.; Takano, K.; Kanaya, S. FK506-binding protein 22 from a psychrophilic bacterium, a cold shock-inducible peptidyl prolyl isomerase with the ability to assist in protein folding. Int. J. Mol. Sci. 2011, 12, 5261–5284. [Google Scholar] [CrossRef] [Green Version]
- Bird, S.S.; Marur, V.R.; Sniatynski, M.J.; Greenberg, H.K.; Kristal, B.S. Serum lipidomics profiling using LC-MS and high-energy collisional dissociation fragmentation: Focus on triglyceride detection and characterization. Anal. Chem. 2011, 83, 6648–6657. [Google Scholar] [CrossRef] [Green Version]
- Hu, C.; Van Dommelen, J.; Rob, V.D.H.; Spijksma, G.; Reijmers, T.H.; Wang, M.; Slee, E.; Lu, X.; Xu, G.; Greef, J.; et al. Rplc-ion-trap-ftms method for lipid profiling of plasma: Method validation and application to p53 mutant mouse model. J. Proteome Res. 2008, 7, 4982–4991. [Google Scholar] [CrossRef]
- Cajka, T.; Smilowitz, J.T.; Fiehn, O. Validating quantitative untargeted lipidomics across nine liquid chromatography-high-resolution mass spectrometry platforms. Anal. Chem. 2017, 89, 12360–12368. [Google Scholar] [CrossRef]
- Davydova, L.; Bakholdina, S.; Barkina, M.; Velansky, P.; Bogdanov, M.; Sanina, N. Effects of elevated growth temperature and heat shock on the lipid composition of the inner and outer membranes of Yersinia pseudotuberculosis. Biochimie 2016, 123, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Benforte, F.C.; Colonnella, M.A.; Ricardi, M.M.; Solar Venero, E.C.; Lizarraga, L.; López, N.I.; Tribelli, P.M. Novel role of the LPS core glycosyltransferase WapH for cold adaptation in the Antarctic bacterium Pseudomonas extremaustralis. PLoS ONE 2018, 13, e0192559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bale, N.J.; Rijpstra, W.I.C.; Sahonero-Canavesi, D.X.; Oshkin, I.Y.; Belova, S.E.; Dedysh, S.N.; Sinninghe Damsté, J.S. Fatty acid and hopanoid adaption to cold in the methanotroph Methylovulum psychrotolerans. Front. Microbiol. 2019, 10, 589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dowhan, W. A retrospective: Use of Escherichia coli as a vehicle to study phospholipid synthesis and function. Biochim. Biophys. Acta 2013, 1831, 471–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niemi, A.R.; Rilfors, L.; Lindblom, G. Influence of monoglucosyldiacylglycerol and monoacylmonoglucosyldiacylglycerol on the lipid bilayer of the membrane from Acholeplasma laidlawii strain A-EF22. Bba Biomembr. 1995, 1239, 186. [Google Scholar] [CrossRef] [Green Version]
- Jorasch, P.; Wolter, F.U.; Heinz, E. A UDP glucosyltransferase from Bacillus subtilis successively transfers up to four glucose residues to 1,2-diacylglycerol: Expression of ypfP in Escherichia coli and structural analysis of its reaction products. Mol. Microbiol. 2010, 29, 419–430. [Google Scholar] [CrossRef]
- Jesper, L.; Tuulia, R.M.; Klement, M.L.R.; Elsa, B.W.; Epand, R.M.; Epand, R.F.; Lena, M.L.; Ake, W. High cationic charge and bilayer interface-binding helices in a regulatory lipid glycosyltransferase. Biochemistry 2007, 46, 5664–5677. [Google Scholar]
- Romain, V.; Kattria, V.D.P.; David, R. Membrane lipid saturation activates endoplasmic reticulum unfolded protein response transducers through their transmembrane domains. PNAS 2013, 110, 4628–4633. [Google Scholar]
- Falkenburger, B.H.; Jensen, J.B.; Dickson, E.J.; Suh, B.-C.; Hille, B. Phosphoinositides: Lipid regulators of membrane proteins. J. Physiol. 2010, 588, 3179–3185. [Google Scholar] [CrossRef]
- Wang, X.; Devaiah, S.P.; Zhang, W.; Welti, R. Signaling functions of phosphatidic acid. Prog. Lipid Res. 2006, 45, 250–278. [Google Scholar] [CrossRef]
- Hoyo, J.; Torrent-Burgués, J.; Tzanov, T. Physical states and thermodynamic properties of model Gram-negative bacterial inner membranes. Chem. Phys. Lipids 2019, 218, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Nichols, D.S.; Miller, M.R.; Davies, N.W.; Amber, G.; Mark, R.; Ricardo, C. Cold adaptation in the Antarctic Archaeon Methanococcoides burtonii involves membrane lipid unsaturation. J. Bacteriol. 2004, 186, 8508–8515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mastronicolis, S.K.; Arvanitis, N.; Karaliota, A.; Magiatis, P.; Heropoulos, G.; Litos, C.; Moustaka, H.; Tsakirakis, A.; Paramera, E.; Papastavrou, P. Coordinated regulation of cold-induced changes in fatty acids with cardiolipin and phosphatidylglycerol composition among phospholipid species for the food pathogen Listeria monocytogenes. Appl. Environ. Microbiol. 2008, 74, 4543–4549. [Google Scholar] [CrossRef] [Green Version]
- Athenstaedt, K.; Daum, G. Phosphatidic acid, a key intermediate in lipid metabolism. Febs J. 2010, 266, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Rock, C.O. Exogenous fatty acid metabolism in bacteria. Biochimie 2017, 141, 30. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Su, Y.; Wang, X. Phosphatidic acid-mediated signaling. In Lipid-Mediated Protein Signaling; Capelluto, D.G.S., Ed.; Springer: Dordrecht, The Netherlands, 2013; pp. 159–176. [Google Scholar]
- Redón, M.; Guillamón, J.M.; Mas, A.; Rozès, N. Effect of growth temperature on yeast lipid composition and alcoholic fermentation at low temperature. Eur. Food Res. Technol. 2011, 232, 517–527. [Google Scholar] [CrossRef]
- Klose, C.; Surma, M.A.; Gerl, M.J.; Meyenhofer, F.; Shevchenko, A.; Kai, S. Flexibility of a eukaryotic lipidome-insights from yeast lipidomics. PLoS ONE 2012, 7, e35063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Řezanka, T.; Kolouchová, I.; Sigler, K. Lipidomic analysis of psychrophilic yeasts cultivated at different temperatures. Biochim. Biophys. Acta 2016, 1861, 1634–1642. [Google Scholar] [CrossRef]
- Torija, M.J.; Beltran, G.; Novo, M.; Poblet, M.; Guillamón, J.M.; Mas, A.; Rozès, N. Effects of fermentation temperature and Saccharomyces species on the cell fatty acid composition and presence of volatile compounds in wine. Int. J. Food Microbiol. 2003, 85, 127–136. [Google Scholar] [CrossRef]
- Sohlenkamp, C.; Lopez, L.I.O. Biosynthesis of phosphatidylcholine in bacteria. Prog. Lipid Res. 2003, 42, 115–162. [Google Scholar] [CrossRef]
- Uttlová, P.; Pinkas, D.; Bechyňková, O.; Fišer, R.; Svobodová, J.; Seydlová, G. Bacillus subtilis alters the proportion of major membrane phospholipids in response to surfactin exposure. Bba Biomembr. 2016, 1858, 2965–2971. [Google Scholar]
- Zheng, L.; Lin, Y.; Lu, S.; Zhang, J.; Bogdanov, M. Biogenesis, transport and remodeling of lysophospholipids in Gram-negative bacteria. Bba Mol. Cell Biol. Lipids 2016, 1862, 1404–1413. [Google Scholar] [CrossRef] [PubMed]
- Paola, D.A.; Stefano, S. Synthesis of lysophospholipids. Molecules 2010, 15, 1354. [Google Scholar]
- Piñeiro, R.; Falasca, M. Lysophosphatidylinositol signalling: New wine from an old bottle. Bba Mol. Cell Biol. Lipids 2012, 1821, 694–705. [Google Scholar]
- Sanina, N.; Davydova, L.; Bakholdina, S.; Novikova, O.; Pornyagina, O.; Solov’Eva, T.; Shnyrov, V.; Bogdanov, M.; Sanina, N.; Davydova, L. Effect of phenol-induced changes in lipid composition on conformation of OmpF-like porin of Yersinia pseudotuberculosis. Febs Lett. 2013, 587, 2260–2265. [Google Scholar]
- Connell, L.B.; Rodriguez, R.R.; Redman, R.S.; Dalluge, J.J. Cold-Adapted Yeasts in Antarctic Deserts. In Cold-Adapted Yeasts; Buzzini, P., Margesin, R., Eds.; Springer: New York, NY, USA, 2014; pp. 75–98. [Google Scholar]
- Weiqi, L.; Maoyin, L.; Wenhua, Z.; Ruth, W.; Xuemin, W. The plasma membrane-bound phospholipase Ddelta enhances freezing tolerance in Arabidopsis thaliana. Nat. Biotechnol. 2004, 22, 427–433. [Google Scholar]
- Dalluge, J.J.; Connell, L.B. On the potential of mass spectrometry-based metabolite profiling approaches to the study of biochemical adaptation in psychrophilic yeast. Extremophiles 2013, 17, 953–961. [Google Scholar] [CrossRef]
- Altabe, S.G.; Mansilla, M.C.; de Mendoza, D. Remodeling of Membrane Phospholipids by Bacterial Desaturases. In Stearoyl-CoA Desaturase Genes in Lipid Metabolism; Ntambi, M.J., Ed.; Springer: New York, NY, USA, 2013; pp. 209–231. [Google Scholar]
- Yoon, Y.; Lee, H.; Lee, S.; Kim, S.; Choi, K.H. Membrane fluidity-related adaptive response mechanisms of foodborne bacterial pathogens under environmental stresses. Food Res. Int. 2015, 72, 25–36. [Google Scholar] [CrossRef]
- Kim, S.-S.; Lee, J.-I.; Kang, D.-H. Resistance of Escherichia coli O157:H7 ATCC 35150 to ohmic heating as influenced by growth temperature and sodium chloride concentration in salsa. Food Control. 2019, 103, 119–125. [Google Scholar] [CrossRef]
- Russell, N.J. Mechanisms of thermal adaptation in bacteria: Blueprints for survival. Trends Biochem. Sci. 1984, 9, 108–112. [Google Scholar] [CrossRef]
- Mansilla, M.C.; Cybulski, L.E.; Daniela, A.; Diego, D.M. Control of membrane lipid fluidity by molecular thermosensors. J. Bacteriol. 2004, 186, 6681–6688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.I.; Talaty, N.; Costa, A.B.; Xia, Y.; Tao, W.A.; Bell, R.; Callahan, J.H.; Cooks, R.G. Rapid direct lipid profiling of bacteria using desorption electrospray ionization mass spectrometry. Int. J. Mass Spectrom. 2011, 301, 37–44. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, X.; Liu, W.; Mei, J.; Xie, J. Quantitative Analysis of Cold Stress Inducing Lipidomic Changes in Shewanella putrefaciens Using UHPLC-ESI-MS/MS. Molecules 2019, 24, 4609. https://doi.org/10.3390/molecules24244609
Gao X, Liu W, Mei J, Xie J. Quantitative Analysis of Cold Stress Inducing Lipidomic Changes in Shewanella putrefaciens Using UHPLC-ESI-MS/MS. Molecules. 2019; 24(24):4609. https://doi.org/10.3390/molecules24244609
Chicago/Turabian StyleGao, Xin, Wenru Liu, Jun Mei, and Jing Xie. 2019. "Quantitative Analysis of Cold Stress Inducing Lipidomic Changes in Shewanella putrefaciens Using UHPLC-ESI-MS/MS" Molecules 24, no. 24: 4609. https://doi.org/10.3390/molecules24244609
APA StyleGao, X., Liu, W., Mei, J., & Xie, J. (2019). Quantitative Analysis of Cold Stress Inducing Lipidomic Changes in Shewanella putrefaciens Using UHPLC-ESI-MS/MS. Molecules, 24(24), 4609. https://doi.org/10.3390/molecules24244609







