Functional Gene Network of Prenyltransferases in Arabidopsis thaliana
Abstract
:1. Introduction
2. Prenyltransferases in Arabidopsis thaliana
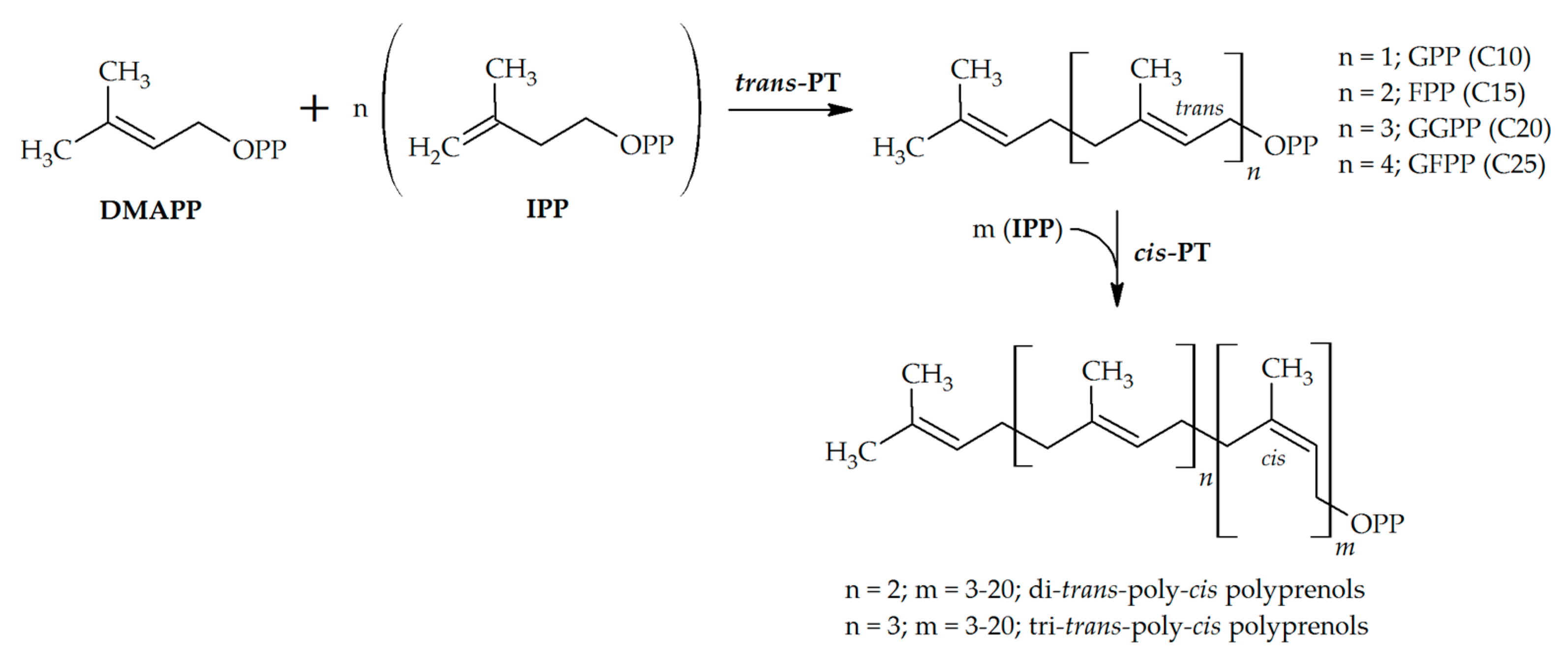
2.1. Trans-Prenyltransferases
2.2. Cis-Prenyltransferases
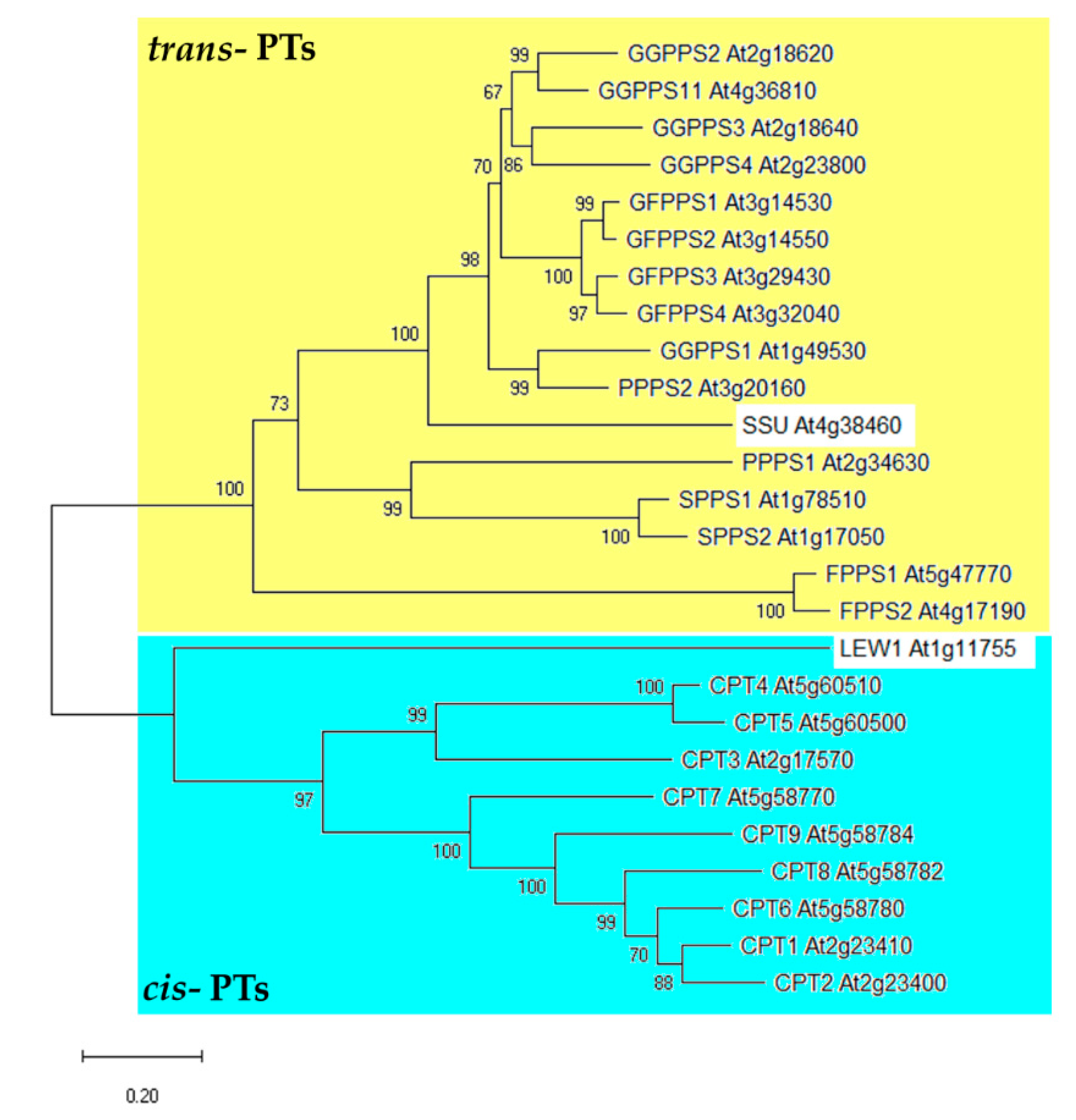
3. Primary Structure of PTs Can Be Indicative for Determination of Their Function
4. Prenyltransferase Mutants: A Source of Information about Their Function and Topology
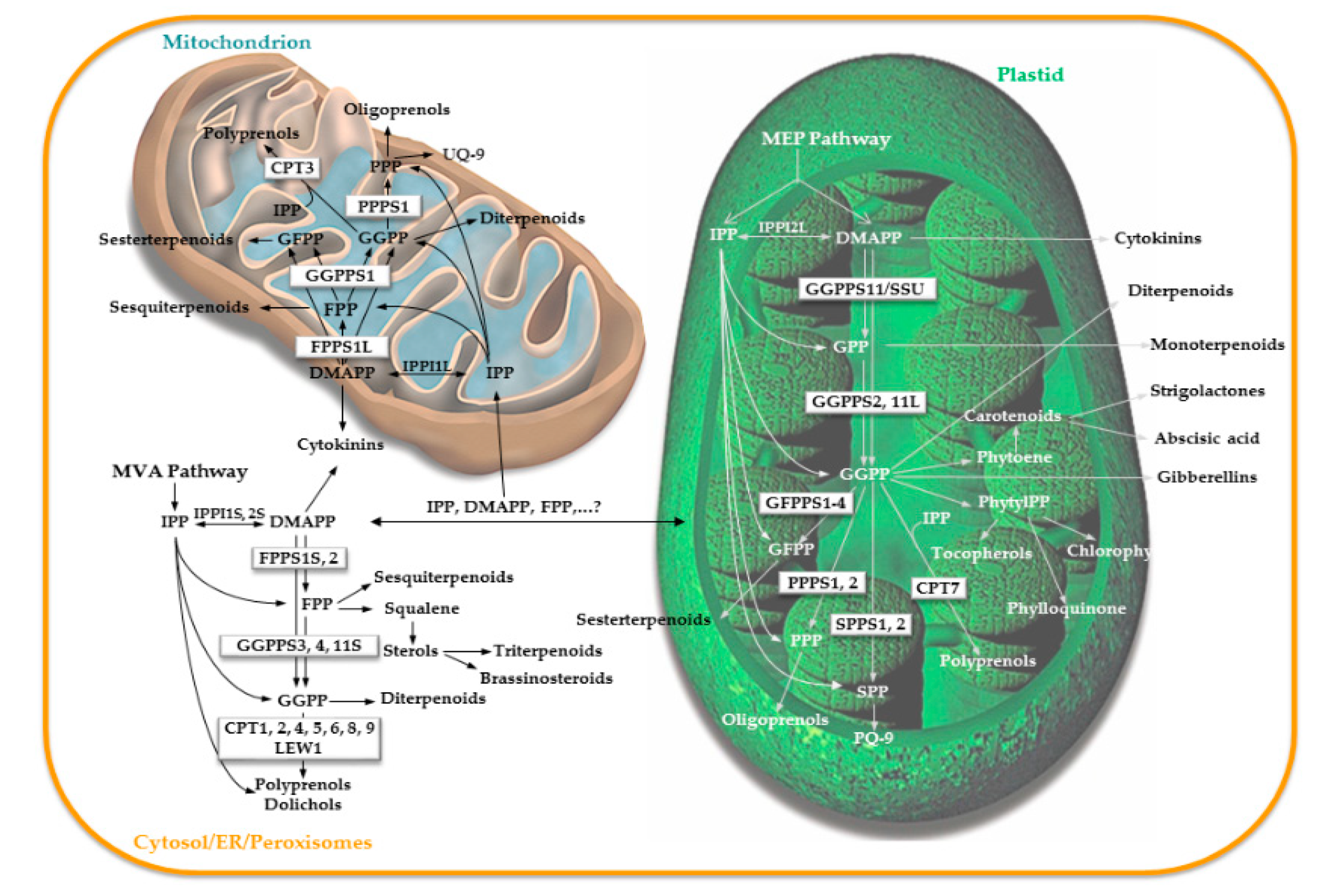
4.1. Prenyltransferase Mutants in the Cytosolic/Mitochondrial Branch of the Isoprenoid Pathway
4.2. Prenyltransferase Mutants in the Plastidial Branch of the Isoprenoid Pathway
5. Co-Expression Analysis and Prediction of Isoprenoid Fluxes
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wallrapp, F.H.; Pan, J.-J.; Ramamoorthy, G.; Almonacid, D.E.; Hillerich, B.S.; Seidel, R.; Patskovsky, Y.; Babbitt, P.C.; Almo, S.C.; Jacobson, M.P.; et al. Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily. Proc. Natl. Acad. Sci. USA 2013, 110, E1196–E1202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vranová, E.; Coman, D.; Gruissem, W. Network Analysis of the MVA and MEP Pathways for Isoprenoid Synthesis. Annu. Rev. Plant. Biol. 2013, 64, 665–700. [Google Scholar] [CrossRef] [PubMed]
- Liang, P.-H.; Ko, T.-P.; Wang, A.H.-J. Structure, mechanism and function of prenyltransferases. Eur. J. Biochem. 2002, 269, 3339–3354. [Google Scholar] [CrossRef] [PubMed]
- Winkelblech, J.; Shu-Ming Li, A.-F. Prenyltransferases as key enzymes in primary and secondary metabolism. Appl. Microbiol. Biotech. 2015, 99, 7379. [Google Scholar] [CrossRef]
- Nagel, R.; Bernholz, C.; Vranová, E.; Košuth, J.; Bergau, N.; Ludwig, S.; Wessjohann, L.; Gershenzon, J.; Tissier, A.; Schmidt, A. Arabidopsis thaliana isoprenyl diphosphate synthases produce the C25 intermediate geranylfarnesyl diphosphate. Plant. J. 2015, 84, 847–859. [Google Scholar] [CrossRef]
- Koyama, T.; Ogura, K. Isopentenyl Diphosphate Isomerase and Prenyltransferases. In Isoprenoids Including Carotenoids and Steroids; Cane, D.E., Ed.; Elsevier: Amsterdam, The Netherlands, 1999; Volume 2, pp. 69–95. [Google Scholar]
- Vandermoten, S.; Haubruge, É.; Cusson, M. New insights into short-chain prenyltransferases: Structural features, evolutionary history and potential for selective inhibition. Cell Mol. Life Sci. 2009, 66, 3685–3695. [Google Scholar] [CrossRef]
- Chen, Q.; Fan, D.; Wang, G. Heteromeric Geranyl(geranyl) Diphosphate Synthase Is Involved in Monoterpene Biosynthesis in Arabidopsis Flowers. Mol. Plant. 2015, 8, 1434–1437. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Dixon, R.A. Heterodimeric geranyl(geranyl)diphosphate synthase from hop Humulus lupulus and the evolution of monoterpene biosynthesis. Proc. Natl. Acad. Sci. USA 2009, 106, 9914–9919. [Google Scholar] [CrossRef] [Green Version]
- Kwon, M.; Kwon, E.J.G.; Ro, D.K. Chapter Six - cis-Prenyltransferase and Polymer Analysis from a Natural Rubber Perspective. In Methods in Enzymology; O’Connor, S.E., Ed.; Elsevier: Amsterdam, The Netherlands, 2016; Volume 576, pp. 121–145. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Campbell, M.; Hahn, F.M.; Poulter, C.D.; Leustek, T. Analysis of the isopentenyl diphosphate isomerase gene family from Arabidopsis thaliana. Plant. Mol. Biol 1998, 36, 323–328. [Google Scholar] [CrossRef]
- Okada, K.; Kasahara, H.; Yamaguchi, S.; Kawaide, H.; Kamiya, Y.; Nojiri, H.; Yamane, H. Genetic evidence for the role of isopentenyl diphosphate isomerases in the mevalonate pathway and plant development in Arabidopsis. Plant. Cell Physiol. 2008, 49, 604–616. [Google Scholar] [CrossRef] [PubMed]
- Phillips, M.A.; D’Auria, J.C.; Gershenzon, J.; Pichersky, E. The Arabidopsis thaliana Type I Isopentenyl Diphosphate Isomerases Are Targeted to Multiple Subcellular Compartments and Have Overlapping Functions in Isoprenoid Biosynthesis. Plant. Cell 2008, 20, 677–696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sapir-Mir, M.; Mett, A.; Belausov, E.; Tal-Meshulam, S.; Frydman, A.; Gidoni, D.; Eyal, Y. Peroxisomal Localization of Arabidopsis Isopentenyl Diphosphate Isomerases Suggests That Part of the Plant Isoprenoid Mevalonic Acid Pathway Is Compartmentalized to Peroxisomes. Plant. Physiol. 2008, 148, 1219–1228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vranová, E.; Hirsch-Hoffmann, M.; Gruissem, W. AtIPD: A Curated Database of Arabidopsis Isoprenoid Pathway Models and Genes for Isoprenoid Network Analysis. Plant. Physiol. 2011, 156, 1655–1660. [Google Scholar] [CrossRef] [Green Version]
- Beck, G.; Coman, D.; Herren, E.; Ruiz-Sola, M.Á.; Rodríguez-Concepción, M.; Gruissem, W.; Vranová, E. Characterization of the GGPP synthase gene family in Arabidopsis thaliana. Plant. Mol. Biol. 2013, 82, 393–416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouvier, F.; Suire, C.; d’Harlingue, A.; Backhaus, R.A.; Camara, B. Molecular cloning of geranyl diphosphate synthase and compartmentation of monoterpene synthesis in plant cells. Plant. J. 2000, 24, 241–252. [Google Scholar] [CrossRef]
- Joly, A.; Edwards, P.A. Effect of site-directed mutagenesis of conserved aspartate and arginine residues upon farnesyl diphosphate synthase activity. J. Biol. Chem. 1993, 268, 26983–26989. [Google Scholar]
- Song, L.; Poulter, C.D. Yeast farnesyl-diphosphate synthase: Site-directed mutagenesis of residues in highly conserved prenyltransferase domains I and II. Proc. Natl. Acad. Sci. USA 1994, 91, 3044–3048. [Google Scholar] [CrossRef] [Green Version]
- Armenteros, J.J.A.; Salvatore, M.; Winther, O.; Emanuelsson, O.; von Heijne, G.; Elofsson, A.; Nielsen, H. Detecting Sequence Signals in Targeting Peptides Using Deep Learning. Life Sci. Alliance 2019, 2, e201900429. [Google Scholar] [CrossRef] [Green Version]
- Hosfield, D.J.; Zhang, Y.; Dougan, D.R.; Broun, A.; Tari, L.W.; Swanson, R.V.; Finn, J. Structural Basis for Bisphosphonate-mediated Inhibition of Isoprenoid Biosynthesis. J. Biol. Chem. 2004, 279, 8526–8529. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Chen, Q.; Fan, D.; Li, J.; Wang, G.; Zhang, P. Structural Analyses of Short-Chain Prenyltransferases Identify an Evolutionarily Conserved GFPPS Clade in Brassicaceae Plants. Mol. Plant. 2016, 9, 195–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lange, B.M.; Ghassemian, M. Genome organization in Arabidopsis thaliana: A survey for genes involved in isoprenoid and chlorophyll metabolism. Plant. Mol. Biol. 2003, 51, 925–948. [Google Scholar] [CrossRef] [PubMed]
- Keim, V.; Manzano, D.; Fernández, F.J.; Closa, M.; Andrade, P.; Caudepón, D.; Bortolotti, C.; Vega, M.C.; Arró, M.; Ferrer, A. Characterization of Arabidopsis FPS Isozymes and FPS Gene Expression Analysis Provide Insight into the Biosynthesis of Isoprenoid Precursors in Seeds. PLoS ONE 2012, 7, 49109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Running, M.P. The role of lipid post-translational modification in plant developmental processes. Front. Plant. Sci. 2014, 5, 50. [Google Scholar] [CrossRef] [Green Version]
- Delourme, D.; Lacroute, F.; Karst, F. Cloning of an Arabidopsis thaliana cDNA coding for farnesyl diphosphate synthase by functional complementation in yeast. Plant. Mol. Biol. 1994, 26, 1867–1873. [Google Scholar] [CrossRef]
- Cunillera, N.; Arró, M.; Delourme, D.; Karst, F.; Boronat, A.; Ferrer, A. Arabidopsis thaliana Contains Two Differentially Expressed Farnesyl-Diphosphate Synthase Genes. J. Biol. Chem. 1996, 271, 7774–7780. [Google Scholar] [CrossRef] [Green Version]
- Masferrer, A.; Arro, M.; Manzano, D.; Schaller, H.; Fernandez-Busquets, X.; Moncalean, P.; Fernandez, B.; Cunillera, N.; Boronat, A.; Ferrer, A. Overexpression of Arabidopsis thaliana farnesyl diphosphate synthase (FPS1S) in transgenic Arabidopsis induces a cell death/senescence-like response and reduced cytokinin levels. Plant. J. 2002, 30, 123–132. [Google Scholar] [CrossRef]
- Manzano, D.; Fernández-Busquets, X.; Schaller, H.; González, V.; Boronat, A.; Arró, M.; Ferrer, A. The metabolic imbalance underlying lesion formation in Arabidopsis thaliana overexpressing farnesyl diphosphate synthase (isoform 1S) leads to oxidative stress and is triggered by the developmental decline of endogenous HMGR activity. Planta 2004, 219, 982–992. [Google Scholar] [CrossRef]
- Closa, M.; Vranová, E.; Bortolotti, C.; Bigler, L.; Arró, M.; Ferrer, A.; Gruissem, W. The Arabidopsis thaliana FPP synthase isozymes have overlapping and specific functions in isoprenoid biosynthesis, and complete loss of FPP synthase activity causes early developmental arrest. Plant. J. 2010, 63, 512–525. [Google Scholar] [CrossRef]
- Manzano, D.; Andrade, P.; Caudepón, D.; Altabella, T.; Arró, M.; Ferrer, A. Suppressing Farnesyl Diphosphate Synthase Alters Chloroplast Development and Triggers Sterol-Dependent Induction of Jasmonate- and Fe-Related Responses. Plant. Physiol. 2016, 172, 93–117. [Google Scholar] [CrossRef]
- Bhatia, V.; Maisnam, J.; Jain, A.; Sharma, K.K.; Bhattacharya, R. Aphid-repellent pheromone E-β-farnesene is generated in transgenic Arabidopsis thaliana over-expressing farnesyl diphosphate synthase2. Ann. Bot 2015, 115, 581–591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okada, K.; Saito, T.; Nakagawa, T.; Kawamukai, M.; Kamiya, Y. Five geranylgeranyl diphosphate synthases expressed in different organs are localized into three subcellular compartments in Arabidopsis. Plant. Physiol. 2000, 122, 1045–1056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakakibara, H.; Kasahara, H.; Ueda, N.; Kojima, M.; Takei, K.; Hishiyama, S.; Asami, T.; Okada, K.; Kamiya, Y.; Yamaya, T.; et al. Agrobacterium tumefaciens increases cytokinin production in plastids by modifying the biosynthetic pathway in the host plant. Proc. Natl. Acad. Sci. USA 2005, 102, 9972–9977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz-Sola, M.Á.; Barja, M.V.; Manzano, D.; Llorente, B.; Schipper, B.; Beekwilder, J.; Rodriguez-Concepcion, M. A Single Arabidopsis Gene Encodes Two Differentially Targeted Geranylgeranyl Diphosphate Synthase Isoforms. Plant. Phys. 2016, 172, 1393–1402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cunillera, N.; Boronat, A.; Ferrer, A. The Arabidopsis thaliana FPS1 Gene Generates a Novel mRNA That Encodes a Mitochondrial Farnesyl-diphosphate Synthase Isoform. J. Biol. Chem. 1997, 272, 15381–15388. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Suzuki, K.; Saito, T.; Okada, K.; Tanaka, K.; Nakagawa, T.; Matsuda, H.; Kawamukai, M. Geranylgeranyl pyrophosphate synthase encoded by the newly isolated gene GGPS6 from Arabidopsis thaliana is localized in mitochondria. Plant. Mol. Biol. 1997, 35, 331–341. [Google Scholar] [CrossRef]
- Lee, J.; Lee, H.; Kim, J.; Lee, S.; Kim, D.H.; Kim, S.; Hwang, I. Both the Hydrophobicity and a Positively Charged Region Flanking the C-Terminal Region of the Transmembrane Domain of Signal-Anchored Proteins Play Critical Roles in Determining Their Targeting Specificity to the Endoplasmic Reticulum or Endosymbiotic Organelles in Arabidopsis Cells. Plant. Cell 2011, 23, 1588. [Google Scholar] [CrossRef] [Green Version]
- Vranová, E.; Kopcsayová, D.; Košuth, J.; Colinas, M. Mutant-Based Model of Two Independent Pathways for Carotenoid-Mediated Chloroplast Biogenesis in Arabidopsis Embryos. Front. Plant. Sci. 2019, 10, 1034. [Google Scholar] [CrossRef]
- Hsieh, F.-L.; Chang, T.-H.; Ko, T.-P.; Wang, A.H.J. Structure and Mechanism of an Arabidopsis Medium/Long-Chain-Length Prenyl Pyrophosphate Synthase. Plant. Phys. 2011, 155, 1079–1090. [Google Scholar] [CrossRef] [Green Version]
- Block, A.; Fristedt, R.; Rogers, S.; Kumar, J.; Barnes, B.; Barnes, J.; Elowsky, C.G.; Wamboldt, Y.; Mackenzie, S.A.; Redding, K.; et al. Functional Modeling Identifies Paralogous Solanesyl-diphosphate Synthases That Assemble the Side Chain of Plastoquinone-9 in Plastids. J. Biol. Chem. 2013, 288, 27594–27606. [Google Scholar] [CrossRef] [Green Version]
- Ducluzeau, A.-L.; Wamboldt, Y.; Elowsky, C.G.; Mackenzie, S.A.; Schuurink, R.C.; Basset, G.J.C. Gene network reconstruction identifies the authentic trans-prenyl diphosphate synthase that makes the solanesyl moiety of ubiquinone-9 in Arabidopsis. Plant. J. 2012, 69, 366–375. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.-H.; Lee, Y.; Kim, H.U. Fibrillin 5 Is Essential for Plastoquinone-9 Biosynthesis by Binding to Solanesyl Diphosphate Synthases in Arabidopsis. Plant. Cell 2015, 27, 2956–2971. [Google Scholar] [CrossRef] [PubMed]
- Jun, L.; Saiki, R.; Tatsumi, K.; Nakagawa, T.; Kawamukai, M. Identification and Subcellular Localization of Two Solanesyl Diphosphate Synthases from Arabidopsis thaliana. Plant. Cell Physiol. 2004, 45, 1882–1888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirooka, K.; Izumi, Y.; An, C.-I.; Nakazawa, Y.; Fukusaki, E.; Kobayashi, A. Functional analysis of two solanesyl diphosphate synthases from Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2005, 69, 592–601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Surmacz, L.; Swiezewska, E. Polyisoprenoids—Secondary metabolites or physiologically important superlipids? Biochem. Bioph. Res. Co 2011, 407, 627–632. [Google Scholar] [CrossRef] [PubMed]
- Kera, K.; Takahashi, S.; Sutoh, T.; Koyama, T.; Nakayama, T. Identification and characterization of a cis,trans-mixed heptaprenyl diphosphate synthase from Arabidopsis thaliana. FEBS J. 2012, 279, 3813–3827. [Google Scholar] [CrossRef]
- Surowiecki, P.; Onysk, A.; Manko, K.; Swiezewska, E.; Surmacz, L. Long-Chain Polyisoprenoids Are Synthesized by AtCPT1 in Arabidopsis thaliana. Molecules 2019, 24, 2789. [Google Scholar] [CrossRef] [Green Version]
- Surmacz, L.; Plochocka, D.; Kania, M.; Danikiewicz, W.; Swiezewska, E. cis-Prenyltransferase AtCPT6 produces a family of very short-chain polyisoprenoids in planta. Biochim. Biophys. Acta 2014, 1841, 240–250. [Google Scholar] [CrossRef]
- Akhtar, T.A.; Surowiecki, P.; Siekierska, H.; Kania, M.; Van Gelder, K.; Rea, K.A.; Virta, L.K.A.; Vatta, M.; Gawarecka, K.; Wojcik, J.; et al. Polyprenols Are Synthesized by a Plastidial cis-Prenyltransferase and Influence Photosynthetic Performance. Plant. Cell 2017, 29, 1709–1725. [Google Scholar] [CrossRef] [Green Version]
- Hooper, C.M.; Castleden, I.R.; Tanz, S.K.; Aryamanesh, N.; Millar, A.H. SUBA4: The interactive data analysis centre for Arabidopsis subcellular protein locations. Nucleic Acids Res. 2016, 45, 1064–1074. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Suzuki, K.; Okada, K.; Tanaka, K.; Nakagawa, T.; Kawamukai, M.; Matsuda, K. Cloning and functional expression of a novel geranylgeranyl pyrophosphate synthase gene from Arabidopsis thaliana in Escherichia coli. Plant. Cell Physiol. 1997, 38, 357–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz-Sola, M.Á.; Coman, D.; Beck, G.; Barja, M.V.; Colinas, M.; Graf, A.; Welsch, R.; Rütimann, P.; Bühlmann, P.; Bigler, L.; et al. Arabidopsis Geranylgeranyl Diphosphate Synthase 11 is a hub isozyme required for the production of most photosynthesis-related isoprenoids. New Phytol. 2016, 209, 252–264. [Google Scholar] [CrossRef] [PubMed]
- Evidente, A.; Kornienko, A.; Lefranc, F.; Cimmino, A.; Dasari, R.; Evidente, M.; Mathieu, V.; Kiss, R. Sesterterpenoids with Anticancer Activity. Curr. Med. Chem. 2015, 22, 3502–3522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, A.C.; Kautsar, S.A.; Hong, Y.J.; Medema, M.H.; Bond, A.D.; Tantillo, D.J.; Osbourn, A. Unearthing a sesterterpene biosynthetic repertoire in the Brassicaceae through genome mining reveals convergent evolution. Proc. Natl. Acad. Sci. USA 2017, 114, 6005–6014. [Google Scholar] [CrossRef] [Green Version]
- Shao, J.; Chen, Q.-W.; Lv, H.-J.; He, J.; Liu, Z.-F.; Lu, Y.-N.; Liu, H.-L.; Wang, G.-D.; Wang, Y. (+)-Thalianatriene and (−)-Retigeranin B Catalyzed by Sesterterpene Synthases from Arabidopsis thaliana. Org. Lett. 2017, 19, 1816–1819. [Google Scholar] [CrossRef]
- Nowicka, B.; Kruk, J. Occurrence, biosynthesis and function of isoprenoid quinones. Biochim. Biophys. Acta 2010, 1797, 1587–1605. [Google Scholar] [CrossRef] [Green Version]
- Hirooka, K.; Bamba, T.; Fukusaki, E.; Kobayashi, A. Cloning and kinetic characterization of Arabidopsis thaliana solanesyl diphosphate synthase. Biochem. J. 2003, 370, 679–686. [Google Scholar] [CrossRef]
- Mackenzie, S.A. Plant organellar protein targeting: A traffic plan still under construction. Trends Cell. Biol. 2005, 15, 548–554. [Google Scholar] [CrossRef] [Green Version]
- Liang, P.-H. Reaction Kinetics, Catalytic Mechanisms, Conformational Changes, and Inhibitor Design for Prenyltransferases. Biochemistry 2009, 48, 6562–6570. [Google Scholar] [CrossRef]
- Swiezewska, E.; Danikiewicz, W. Polyisoprenoids: Structure, biosynthesis and function. Prog. Lipid. Res. 2005, 44, 235–258. [Google Scholar] [CrossRef]
- Oh, S.K.; Han, K.H.; Ryu, S.B.; Kang, H. Molecular Cloning, Expression, and Functional Analysis of a cis-Prenyltransferase from Arabidopsis thaliana. Implications in Rubber Biosynthesis. J. Biol. Chem. 2000, 275, 18482–18488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cunillera, N.; Arro, M.; Fores, O.; Manzano, D.; Ferrer, A. Characterization of dehydrodolichyl diphosphate synthase of Arabidopsis thaliana, a key enzyme in dolichol biosynthesis. FEBS Lett. 2000, 477, 170–174. [Google Scholar] [CrossRef] [Green Version]
- Grabińska, K.A.; Edani, B.H.; Park, E.J.; Kraehling, J.R.; Sessa, W.C. A conserved C-terminal RXG motif in the NgBR subunit of cis-prenyltransferase is critical for prenyltransferase activity. J. Biol. Chem. 2017, 292, 17351–17361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, Y.; Chakrabarty, R.; Tran, H.T.; Kwon, E.-J.G.; Kwon, M.; Nguyen, T.-D.; Ro, D.-K. A lettuce (Lactuca sativa) homolog of human Nogo-B receptor interacts with cis-prenyltransferase and is necessary for natural rubber biosynthesis. J. Biol. Chem. 2015, 290, 1898–1914. [Google Scholar] [CrossRef] [Green Version]
- Epping, J.; van Deenen, N.; Niephaus, E.; Stolze, A.; Fricke, J.; Huber, C.; Eisenreich, W.; Twyman, R.M.; Prüfer, D.; Schulze Gronover, C. A rubber transferase activator is necessary for natural rubber biosynthesis in dandelion. Nat. Plants 2015, 1, 15048. [Google Scholar] [CrossRef]
- Brasher, M.I.; Surmacz, L.; Leong, B.; Pitcher, J.; Swiezewska, E.; Pichersky, E.; Akhtar, T.A. A two-component enzyme complex is required for dolichol biosynthesis in tomato. Plant. J. 2015, 82, 903–914. [Google Scholar] [CrossRef] [Green Version]
- Addou, S.; Rentzsch, R.; Lee, D.; Orengo, C.A. Domain-Based and Family-Specific Sequence Identity Thresholds Increase the Levels of Reliable Protein Function Transfer. J. Mol. Biol. 2009, 387, 416–430. [Google Scholar] [CrossRef]
- Chen, A.; Kroon, P.A.; Poulter, C.D. Isoprenyl diphosphate synthases: Protein sequence comparisons, a phylogenetic tree, and predictions of secondary structure. Protein Sci. 1994, 3, 600–607. [Google Scholar] [CrossRef] [Green Version]
- Tarshis, L.C.; Proteau, P.J.; Kellogg, B.A.; Sacchettini, J.C.; Poulter, C.D. Regulation of product chain length by isoprenyl diphosphate synthases. Proc. Natl. Acad. Sci. USA 1996, 93, 15018–15023. [Google Scholar] [CrossRef] [Green Version]
- Ohnuma, S.-i.; Hirooka, K.; Tsuruoka, N.; Yano, M.; Ohto, C.; Nakane, H.; Nishino, T. A Pathway Where Polyprenyl Diphosphate Elongates in Prenyltransferase: insight into a common mechanism of chain length determination of prenyltransferases. J. Biol. Chem. 1998, 273, 26705–26713. [Google Scholar] [CrossRef] [Green Version]
- Guo, R.-T.; Kuo, C.-J.; Chou, C.-C.; Ko, T.-P.; Shr, H.-L.; Liang, P.-H.; Wang, A.H.J. Crystal Structure of Octaprenyl Pyrophosphate Synthase from Hyperthermophilic Thermotoga maritima and Mechanism of Product Chain Length Determination. J. Biol. Chem. 2004, 279, 4903–4912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, T.-H.; Guo, R.-T.; Ko, T.-P.; Wang, A.H.J.; Liang, P.-H. Crystal Structure of Type-III Geranylgeranyl Pyrophosphate Synthase from Saccharomyces cerevisiae and the Mechanism of Product Chain Length Determination. J. Biol. Chem. 2006, 281, 14991–15000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gabelli, S.B.; McLellan, J.S.; Montalvetti, A.; Oldfield, E.; Docampo, R.; Amzel, L.M. Structure and mechanism of the farnesyl diphosphate synthase from Trypanosoma cruzi: Implications for drug design. Proteins 2006, 62, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Kavanagh, K.L.; Dunford, J.E.; Bunkoczi, G.; Russell, R.G.G.; Oppermann, U. The Crystal Structure of Human Geranylgeranyl Pyrophosphate Synthase Reveals a Novel Hexameric Arrangement and Inhibitory Product Binding. J. Biol. Chem. 2006, 281, 22004–22012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulmanov, M.; Khan, M.A.; Hoehndorf, R.; Wren, J. DeepGO: Predicting protein functions from sequence and interactions using a deep ontology-aware classifier. Bioinformatics 2018, 34, 660–668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bepler, T.; Berger, B. Learning Protein Sequence Embeddings Using Information from Structure. In Proceedings of the International Conference on Learning Representations, New Orleans, LA, USA, 6–9 May 2019. [Google Scholar]
- Jin, H.; Song, Z.; Nikolau, B.J. Reverse genetic characterization of two paralogous acetoacetyl CoA thiolase genes in Arabidopsis reveals their importance in plant growth and development. Plant. J. 2012, 70, 1015–1032. [Google Scholar] [CrossRef]
- Okada, K.; Ohara, K.; Yazaki, K.; Nozaki, K.; Uchida, N.; Kawamukai, M.; Nojiri, H.; Yamane, H. The AtPPT1 gene encoding 4-hydroxybenzoate polyprenyl diphosphate transferase in ubiquinone biosynthesis is required for embryo development in Arabidopsis thaliana. Plant. Mol. Biol 2004, 55, 567–577. [Google Scholar] [CrossRef]
- Babiychuk, E.; Bouvier-Navé, P.; Compagnon, V.; Suzuki, M.; Muranaka, T.; Van Montagu, M.; Kushnir, S.; Schaller, H. Allelic mutant series reveal distinct functions for Arabidopsis cycloartenol synthase 1 in cell viability and plastid biogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 3163–3168. [Google Scholar] [CrossRef] [Green Version]
- Gutkowska, M.; Wnuk, M.; Nowakowska, J.; Lichocka, M.; Stronkowski, M.M.; Swiezewska, E. Rab geranylgeranyl transferase β subunit is essential for male fertility and tip growth in Arabidopsis. J. Exp. Bot 2015, 66, 213–224. [Google Scholar] [CrossRef]
- Jozwiak, A.; Gutkowska, M.; Gawarecka, K.; Surmacz, L.; Buczkowska, A.; Lichocka, M.; Nowakowska, J.; Swiezewska, E. POLYPRENOL REDUCTASE2 Deficiency Is Lethal in Arabidopsis Due to Male Sterility. Plant. Cell 2015, 27, 3336–3353. [Google Scholar] [CrossRef]
- Motohashi, R.; Ito, T.; Kobayashi, M.; Taji, T.; Nagata, N.; Asami, T.; Yoshida, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Functional analysis of the 37 kDa inner envelope membrane polypeptide in chloroplast biogenesis using a Ds-tagged Arabidopsis pale-green mutant. Plant. J. 2003, 34, 719–731. [Google Scholar] [CrossRef] [PubMed]
- Yamasato, A.; Tanaka, R.; Tanaka, A. Loss of the N-terminal domain of chlorophyllide a oxygenase induces photodamage during greening of Arabidopsis seedlings. BMC Plant. Biol. 2008, 8, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruppel, N.J.; Kropp, K.N.; Davis, P.A.; Martin, A.E.; Luesse, D.R. Mutations in Geranylgeranyl Diphosphate Synthase 1 affect chloroplast development in Arabidopsis thaliana (Brassicaceae) Am. J. Bot 2013, 100, 2074–2084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Schie, C.C.N.; Ament, K.; Schmidt, A.; Lange, T.; Haring, M.A.; Schuurink, R.C. Geranyl diphosphate synthase is required for biosynthesis of gibberellins. Plant. J. 2007, 52, 752–762. [Google Scholar] [CrossRef]
- Zhang, H.; Ohyama, K.; Boudet, J.; Chen, Z.; Yang, J.; Zhang, M.; Muranaka, T.; Maurel, C.; Zhu, J.-K.; Gong, Z. Dolichol Biosynthesis and Its Effects on the Unfolded Protein Response and Abiotic Stress Resistance in Arabidopsis. Plant. Cell 2008, 20, 1879–1898. [Google Scholar] [CrossRef] [Green Version]
- Wille, A.; Zimmermann, P.; Vranova, E.; Furholz, A.; Laule, O.; Bleuler, S.; Hennig, L.; Prelic, A.; von Rohr, P.; Thiele, L.; et al. Sparse graphical Gaussian modeling of the isoprenoid gene network in Arabidopsis thaliana. Genome Biol. 2004, 5, 92. [Google Scholar] [CrossRef] [Green Version]
- Ghassemian, M.; Lutes, J.; Tepperman, J.M.; Chang, H.-S.; Zhu, T.; Wang, X.; Quail, P.H.; Markus Lange, B. Integrative analysis of transcript and metabolite profiling data sets to evaluate the regulation of biochemical pathways during photomorphogenesis. Arch. Biochem. Biophys. 2006, 448, 45–59. [Google Scholar] [CrossRef]
- Meier, S.; Tzfadia, O.; Vallabhaneni, R.; Gehring, C.; Wurtzel, E. A transcriptional analysis of carotenoid, chlorophyll and plastidial isoprenoid biosynthesis genes during development and osmotic stress responses in Arabidopsis thaliana. BMC Syst. Biol. 2011, 5, 77. [Google Scholar] [CrossRef] [Green Version]
- Nützmann, H.-W.; Scazzocchio, C.; Osbourn, A. Metabolic Gene Clusters in Eukaryotes. Annu. Rev. Genet. 2018, 52, 159–183. [Google Scholar] [CrossRef]


| ENZYME | GENE | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Acronym | Name | EC Number | AGI | Gene Model | References | Alternative Names | References | Localization b | References |
| GPPS | Geranyl diphosphate synthase c | EC 2.5.1.1 | At4g36810/At4g38460 | GGPPS11La/SSU | IDS11; GGPPS11; GGPS1/GGPPS12 | [5,24,34]/[24] | Pl/Pl | [17,34,35,36]/[8] | |
| FPPS | Farnesyl diphosphate synthase | EC 2.5.1.10 | At5g47770 | FPPS1Sa | [24,37] | C | [25] | ||
| FPPS1La | [24,37] | Mt | [37] | ||||||
| At4g17190 | FPPS2 | [24] | C | [25] | |||||
| GGPPS | Geranylgeranyl diphosphate synthase | EC 2.5.1.29 | At1g49530 | GGPPS1 | [24] | IDS1; GGPS6 | [5,34] | Mt | [5,17,34,35,38] |
| At2g18620 | GGPPS2 | [24] | IDS2 | [5] | Pl | [17] | |||
| At2g18640 | GGPPS3 | [24] | IDS3; GGPS4 | [5,34] | ER | [17,34] | |||
| At2g23800 | GGPPS4 | [24] | IDS4; GGPS2 | [5,34] | ER | [17,34,39] | |||
| At4g36810 | GGPPS11Sa | [36] | C | [36,40] | |||||
| At4g36810 | GGPPS11La | IDS11; GGPPS11; GGPS1 | [5,24,34] | Pl | [17,34,35] | ||||
| GFPPS | Geranylfarnesyl diphosphate synthase | EC 2.5.1.81 | At3g14530 | GFPPS1 | [23] | IDS6; GGPPS6 | [5,24] | Pl | [5,17] |
| At3g14550 | GFPPS2 | [23] | IDS7; GGPPS7; GGPS3 | [5,24,34] | Pl | [5,17,34] | |||
| At3g29430 | GFPPS3 | [23] | IDS9; GGPPS9 | [5,24] | Pl | [5,17] | |||
| At3g32040 | GFPPS4 | [23] | IDS10; GGPPS10 | [5,24] | Pl | [5,17] | |||
| PPPS | Polyprenyl diphosphate synthase | EC 2.5.1.91 | At2g34630 | PPPS1 | [41] | GPPS; SPPS3 | [24,42] | Pl/Mt | [18,43] |
| At3g20160 | PPPS2 | [23] | IDS8; GGPPS8 | [5,24] | Pl | [17] | |||
| SPPS | Solanesyl diphosphate synthase | EC 2.5.1.85 | At1g78510 | SPPS1 | [42] | Pl | [42,44] | ||
| At1g17050 | SPPS2 | [42] | Pl | [44,45,46] | |||||
| CPT | cis-Prenyltransferase | EC 2.5.1.87 | At1g11755 | LEW1 | [47] | PMd | |||
| At2g23410 | CPT1 | [47] | AtcPT3 | [48] | ER | [48,49] | |||
| At2g23400 | CPT2 | [47] | AtcPT2 | [48] | Cd | ||||
| At2g17570 | CPT3 | [47] | AtcPT1 | [48] | Mtd | ||||
| At5g60510 | CPT4 | [47] | AtcPT9 | [48] | Cd | ||||
| At5g60500 | CPT5 | [47] | AtcPT8 | [48] | Cd | ||||
| At5g58780 | CPT6 | [47] | AtcPT5 | [48] | ER | [48,50] | |||
| At5g58770 | CPT7 | [47] | AtcPT4/AtHEPS | [48,51] | Pl | [48,51] | |||
| At5g58782 | CPT8 | [47] | AtcPT6 | [48] | ER | [47] | |||
| At5g58784 | CPT9 | [47] | AtcPT7 | [48] | ERd | ||||
| Gene | AGI | Mutagen | Allele | Mutant Line | Phenotype | References |
|---|---|---|---|---|---|---|
| FPPS1 | At5g47770 | T-DNA | fpps1-1 | SAIL_310-D07 | Wild-type phenotype with slightly reduced levels of sterols and UQ-9 | [31] |
| T-DNA | fpps1-2 | SALK_073576 | ||||
| FPPS2 | At4g17190 | T-DNA | fpps2-1 | SAIL_328_G06 | Wild-type phenotype with slightly reduced levels of sterols and UQ-9 | [31] |
| Ds | fpps2-2 | GT7041 | ||||
| FPPS1/FPPS2 | At5g47770/At4g17190 | T-DNA/Ds | fpps1/fpps2 | SAIL_310_D07; SALK_073576/SAIL_328_G06; GT7041 | Embryo lethal at the globular stage; slightly impaired pollen tube elongation | [31] |
| amiRNA | fpps1/fpps2 | amiFPS1 (21%/26% mRNA), amiFPS2 (16%/35% mRNA) | Impaired growth and development; chlorosis; reduces level of chlorophylls, carotenoids, sterols and UQ-9; higher level of UQ-10 | [32] | ||
| SSU | At4g38460 | T-DNA | ssu-1 (ggpps12-1) | pst11416 | Wild-type phenotype with reduced level of monoterpenoids, and wild-type level of carotenoids, chlorophylls, sesquiterpenoids | [8] |
| T-DNA | ssu (ggpps12) | pst01798 (20% mRNA) | ||||
| GGPPS1 | At1g49530 | T-DNA | ggpps1-1 | SAIL_559_G01 | Wild-type phenotype with wild-type level of UQ-9, carotenoids, tocopherols, chlorophylls, PQ-9, phylloquinones, plastochromanol-8 | [36] |
| GGPPS2 | At2g18620 | T-DNA | ggpps2-1 | FLAG_134_B10 | Wild-type phenotype with wild-type levels of carotenoids, chlorophylls, phylloquinones | [54] |
| GGPPS3 | At2g18640 | - | - | - | - | - |
| GGPPS4 | At2g23800 | - | - | - | - | - |
| GGPPS11 | At4g36810 | EMS mutagenesis | ggpps11-1 | D163R point mutation | Variegated phenotype; germination delayed on the inhibitor of gibberellin biosynthesis; reduced level of chlorophylls and carotenoids | [54,86] |
| T-DNA in chloroplast targeting sequence | ggpps11-2 | SALK_015098 | Albino seedling | [54,86] | ||
| T-DNA | ggpps11-3 | SALK_085914 | Embryo lethal at the heart stage | [54,86] | ||
| T-DNA | ggpps11-4 | SAIL_712_D06 | [54] | |||
| T-DNA in 5′-UTR | ggpps11-5 | SALK_140601 | Pale green phenotype with reduced level of monoterpenoids, carotenoids, tocopherols, chlorophylls, PQ-9, phylloquinones, plastochromanol-8 | [8,36,54] | ||
| GFPPS1 | At3g14530 | T-DNA | gfpps1-1 (ggpps6-1) | SAIL_1148_A03 | Wild-type phenotype with wild-type level of carotenoids, chlorophylls, phylloquinones | [54] |
| GFPPS2 | At3g14550 | T-DNA | gfpps2-1 (ggpps7-1) | SALK_119280 | Wild-type phenotype with wild-type level of carotenoids, chlorophylls, phylloquinones | [54] |
| GFPPS3 | At3g29430 | RNAi | gfpps3 (ggpps9) | RNAi GGPPS9-1 (21% mRNA); RNAi GGPPS9-6 (16% mRNA) | Wild-type phenotype with wild-type level of carotenoids, chlorophylls, phylloquinones | [54] |
| GFPPS4 | At3g32040 | T-DNA | gfpps4-1 (ggpps10-1) | SM_3_32015 | Wild-type phenotypes with wild-type level of carotenoids, chlorophylls, phylloquinones | [54] |
| PPPS1 | At2g34630 | T-DNA | ppps1-1 (gps1-1) | GABI 097_G02 | Embryo lethal | [87] |
| RNAi | ppps1 | RNAi PPPS1-1-6 (10% mRNA) | Growth reduction; dwarfed plants with delayed flowering | [87] | ||
| RNAi | ppps1 | RNAi PPPS1-1-3 (20% mRNA) | Reduced level of UQ-9; wild-type level of PQ | [43] | ||
| RNAi | ppps1 | GPPS RNAi-2, GPPS RNAi-4 (10% mRNA) | Wild-type phenotype with wild-type level of monoterpenoids and sesquiterpenoids | [8] | ||
| PPPS2 | At3g20160 | T-DNA | ppps2-1 (ggpps8-1) | FLAG_470_E09 | Higher level of carotenoids; reduced level of chlorophyll b | [54] |
| SPPS1 | At1g78510 | T-DNA | spps1 | SALK_126948 | Wild-type phenotype with reduced level of PQ and plastochromanol, and wild-type level of tocopherol and UQ | [42] |
| SPPS2 | At1g17050 | T-DNA | spps2 | SALK_064292 | Developmental delay; stunted phenotype and pale yellowish leaves at the high light; reduced level of PQ; no plastochromanol; higher level of tocopherol, and wild-type level of UQ | [42] |
| SPPS1/SPPS2 | At1g78510/ | T-DNA | spps1/spps2 | SALK_126948/SALK_064292 | Seedling lethal, albino phenotype; no PQ and plastochromanol; wild-type level of UQ and tocopherol | [42] |
| At1g17050 | ||||||
| LEW1 | At1g11755 | EMS mutagenesis | lew1-1 | G159A point mutation | Leaf-wilting phenotype, increased drought resistance, impaired plasma membrane integrity, impaired protein N-glycosylation, reduced the total plant content of main dolichols C75, C80 by 85% and protein glycosylation defects | [88] |
| T-DNA | lew1-2 | SALK_032276 | Lethal | [88] | ||
| CPT1 | At2g23410 | T-DNA | cpt1-1 | SALK_038151 | Extremely stunted growth and shorter root length; reduced level from Dol-18 to -23 (Dol-21 dominating) | [49] |
| T-DNA | cpt1-2 | SALK_032276 | ||||
| T-DNA | cpt1-3 | SALK_100795 | ||||
| CPT2 | At2g23400 | - | - | - | - | - |
| CPT3 | At2g17570 | - | - | - | - | - |
| CPT4 | At5g60510 | - | - | - | - | - |
| CPT5 | At5g60500 | - | - | - | - | - |
| CPT6 | At5g58780 | T-DNA | cpt6-1 | SALK_071255 | Reduced level of Dol-7 and short-chain dolichols (Dol-13 dominating) | [50] |
| cpt6-2 | SALK_064499 | |||||
| CPT7 | At5g58770 | T-DNA | cpt7-1 | SALK_022111 | Decreased thylakoid membrane fluidity and photosynthetic performance; no polyprenols 9, 10, 11 (Pol-10 dominating); wild-type level of tocopherols, phylloquinone, carotenoids, PQ, and chlorophylls | [51] |
| RNAi | cpt7 | RNAi-23; RNAi-24 and RNAi-31 (15% mRNA) | ||||
| CPT8 | At5g58782 | - | - | - | - | - |
| CPT9 | At5g58784 | - | - | - | - | - |
| CYTOSOL AND MITOCHONDRIA | ||||||||||
| Pathway | MVA | Sterol | Triterpenoids | Sesquiterpenoids/Diterpenoids | Protein Prenylation | Ubiquinone | ||||
| Total No. of Genes per Pathway | 9 | 24 | 13 | 13 | 7 | 4 | ||||
| FPPS1 | At5g47770 | 6 | 13 | 2 | 1 | 4 | 0 | |||
| FPPS2 | At4g17190 | 7 | 14 | 0 | 1 | 3 | 1 | |||
| GGPPS1 | At1g49530 | 0 | 0 | 2 | 0 | 0 | 3 | |||
| GGPPS3 | At2g18640 | 0 | 4 | 3 | 6 | 1 | 2 | |||
| GGPPS11 | At4g36810 | 0 | 11 | 2 | 0 | 1 | 0 | |||
| GGPPS4 | At2g23800 | 2 | 2 | 3 | 1 | 0 | 2 | |||
| PPPS1 | At2g34630 | 2 | 3 | 0 | 2 | 1 | 4 | |||
| PLASTIDS | ||||||||||
| Pathway | MEP | Plastoquinone | Chlorophyll | Carotenoid | Phylloquinone | Gibberellins | Monoterpenoids | Diterpenoids/Sesquiterpenoids | Sesterterpenoids | |
| Total No. of Genes per Pathway | 7 | 6 | 37 | 32 | 7 | 23 | 6 | 13 | 3 | |
| SSU | At4g38460 | 5 | 5 | 25 | 22 | 0 | 5 | 2 | 1 | 0 |
| GGPPS11 | At4g36810 | 7 | 3 | 29 | 18 | 1 | 7 | 1 | 0 | 0 |
| GGPPS2 | At2g18620 | 0 | 0 | 3 | 1 | 5 | 2 | 2 | 7 | 1 |
| GFPPS1 | At3g14530 | 0 | 0 | 1 | 0 | 3 | 2 | 0 | 5 | 2 |
| GGPPS3 | At3g29430 | 0 | 0 | 4 | 0 | 4 | 2 | 2 | 8 | 1 |
| GGPPS4 | At3g32040 | 0 | 0 | 3 | 0 | 5 | 7 | 2 | 7 | 1 |
| PPPS2 | At3g20160 | 0 | 0 | 4 | 0 | 4 | 4 | 2 | 8 | 1 |
| PPPS1 | At2g34630 | 0 | 0 | 0 | 2 | 4 | 2 | 4 | 2 | 1 |
| SPPS1 | At1g78510 | 6 | 6 | 29 | 20 | 0 | 4 | 2 | 1 | 0 |
| SPPS2 | At1g17050 | 7 | 4 | 29 | 22 | 0 | 7 | 2 | 1 | 0 |
 Table 3 shows number of pathway genes that are significantly co-expressed with the trans-PTs. The intensity of the highlighted background is directly proportional to the relative number of co-expressed genes/total number of pathway genes. Data for co-expression analysis were obtained from BAR/Expression Browser (http://bar.utoronto.ca/affydb/cgi-bin/affy_db_exprss_browser_in.cgi) and log2 transformed (see Table S2 for log2 transformed expression data). In general, AtGeneExpress_plus-Extended Tissue Series data/Average of replicate treatments were downloaded. Genes that are missing were not present on the microarrays, and therefore, they are not included in the analysis. Pearson correlation coefficients and the corresponding false discovery rate (FDR) p-values were calculated. The threshold for significance is p-value ≤ 0.05. A detailed description of the analysis can be found in [54]. Genes in categories mono-, di-, sesqui-, sester- and triterpenoids contain genes encoding only terpene cyclases. Genes in the category diterpenoids/sesquiterpenoids and sesquiterpenoids/diterpenoids partly overlap. In both categories were kept the genes that have no experimentally proven one or other activity, and plastidial genes were considered diterpenoids and kept only in the category diterpenoids/sesquiterpenoids.
Table 3 shows number of pathway genes that are significantly co-expressed with the trans-PTs. The intensity of the highlighted background is directly proportional to the relative number of co-expressed genes/total number of pathway genes. Data for co-expression analysis were obtained from BAR/Expression Browser (http://bar.utoronto.ca/affydb/cgi-bin/affy_db_exprss_browser_in.cgi) and log2 transformed (see Table S2 for log2 transformed expression data). In general, AtGeneExpress_plus-Extended Tissue Series data/Average of replicate treatments were downloaded. Genes that are missing were not present on the microarrays, and therefore, they are not included in the analysis. Pearson correlation coefficients and the corresponding false discovery rate (FDR) p-values were calculated. The threshold for significance is p-value ≤ 0.05. A detailed description of the analysis can be found in [54]. Genes in categories mono-, di-, sesqui-, sester- and triterpenoids contain genes encoding only terpene cyclases. Genes in the category diterpenoids/sesquiterpenoids and sesquiterpenoids/diterpenoids partly overlap. In both categories were kept the genes that have no experimentally proven one or other activity, and plastidial genes were considered diterpenoids and kept only in the category diterpenoids/sesquiterpenoids.© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kopcsayová, D.; Vranová, E. Functional Gene Network of Prenyltransferases in Arabidopsis thaliana. Molecules 2019, 24, 4556. https://doi.org/10.3390/molecules24244556
Kopcsayová D, Vranová E. Functional Gene Network of Prenyltransferases in Arabidopsis thaliana. Molecules. 2019; 24(24):4556. https://doi.org/10.3390/molecules24244556
Chicago/Turabian StyleKopcsayová, Diana, and Eva Vranová. 2019. "Functional Gene Network of Prenyltransferases in Arabidopsis thaliana" Molecules 24, no. 24: 4556. https://doi.org/10.3390/molecules24244556
APA StyleKopcsayová, D., & Vranová, E. (2019). Functional Gene Network of Prenyltransferases in Arabidopsis thaliana. Molecules, 24(24), 4556. https://doi.org/10.3390/molecules24244556





