Zinc(II) Terpyridine Complexes: Substituent Effect on Photoluminescence, Antiproliferative Activity, and DNA Interaction
Abstract
:1. Introduction
2. Results and discussion
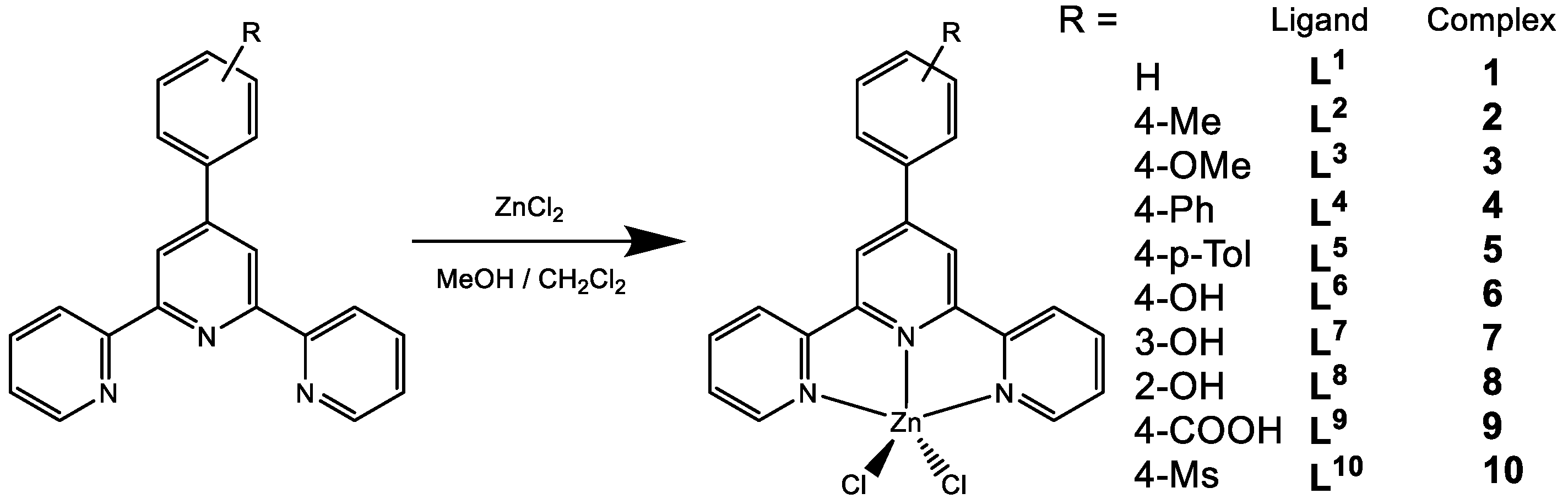
2.1. Synthesis and Characterization
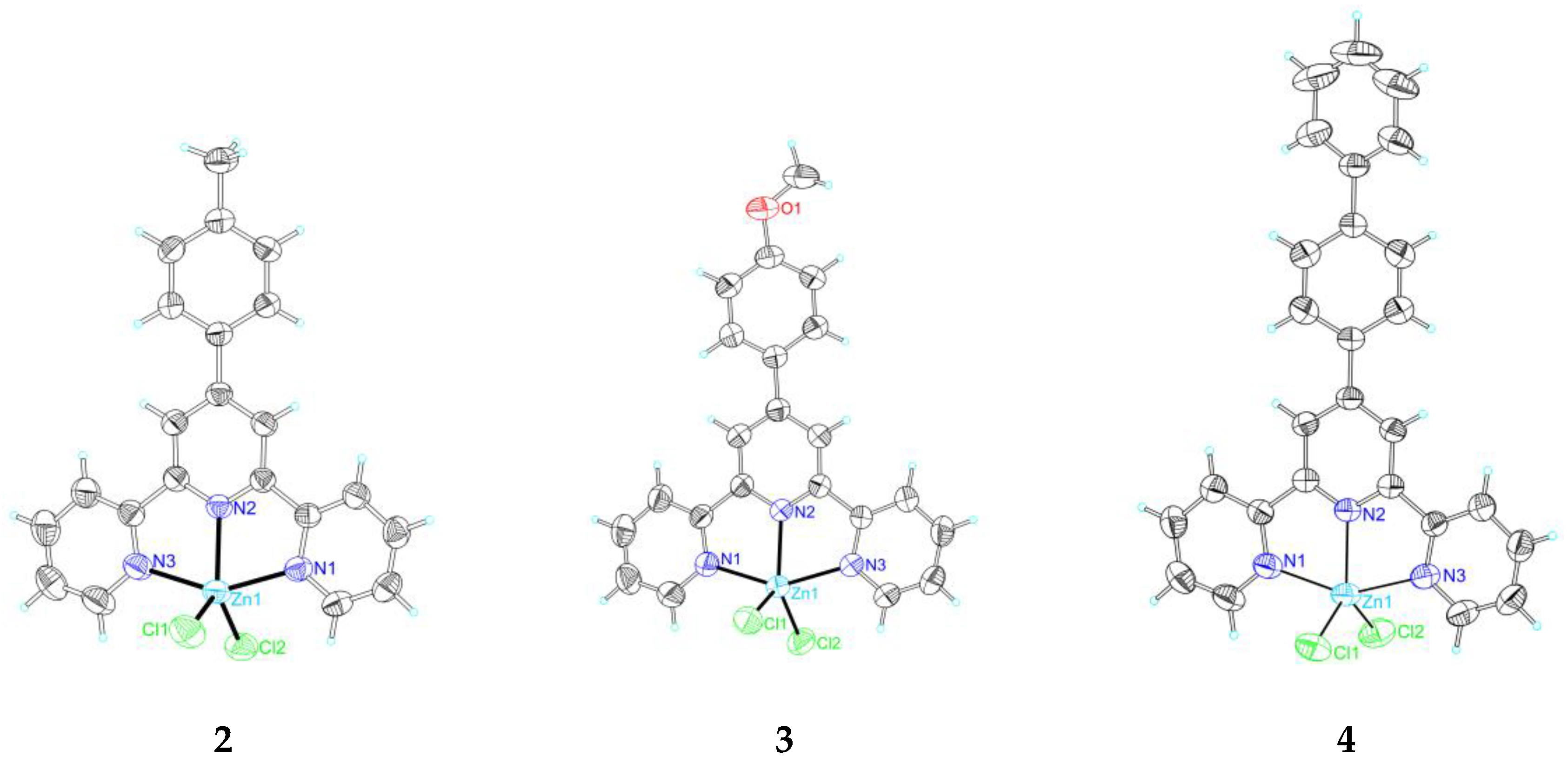
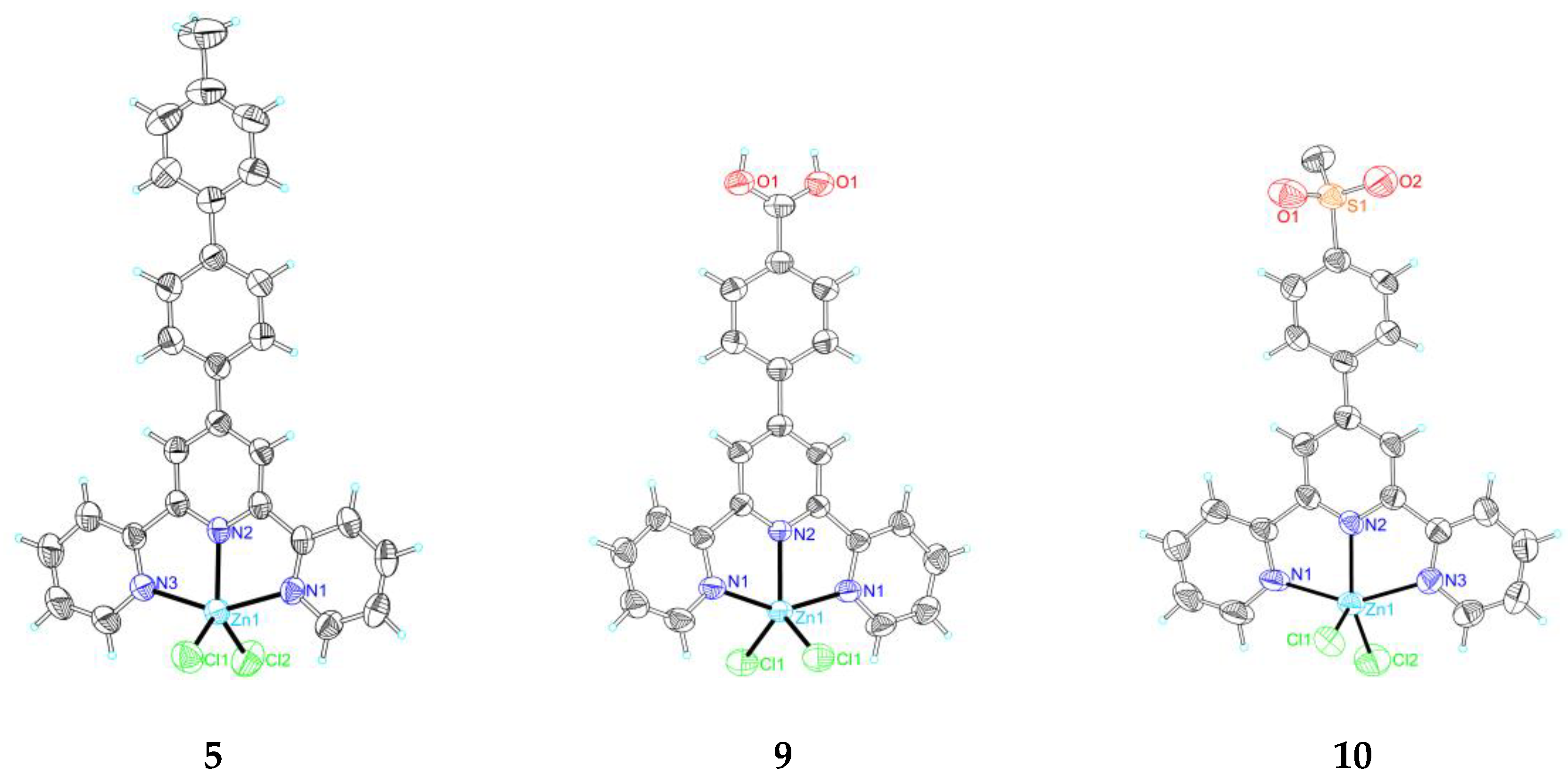
2.2. Single Crystal X-Ray Crystallography
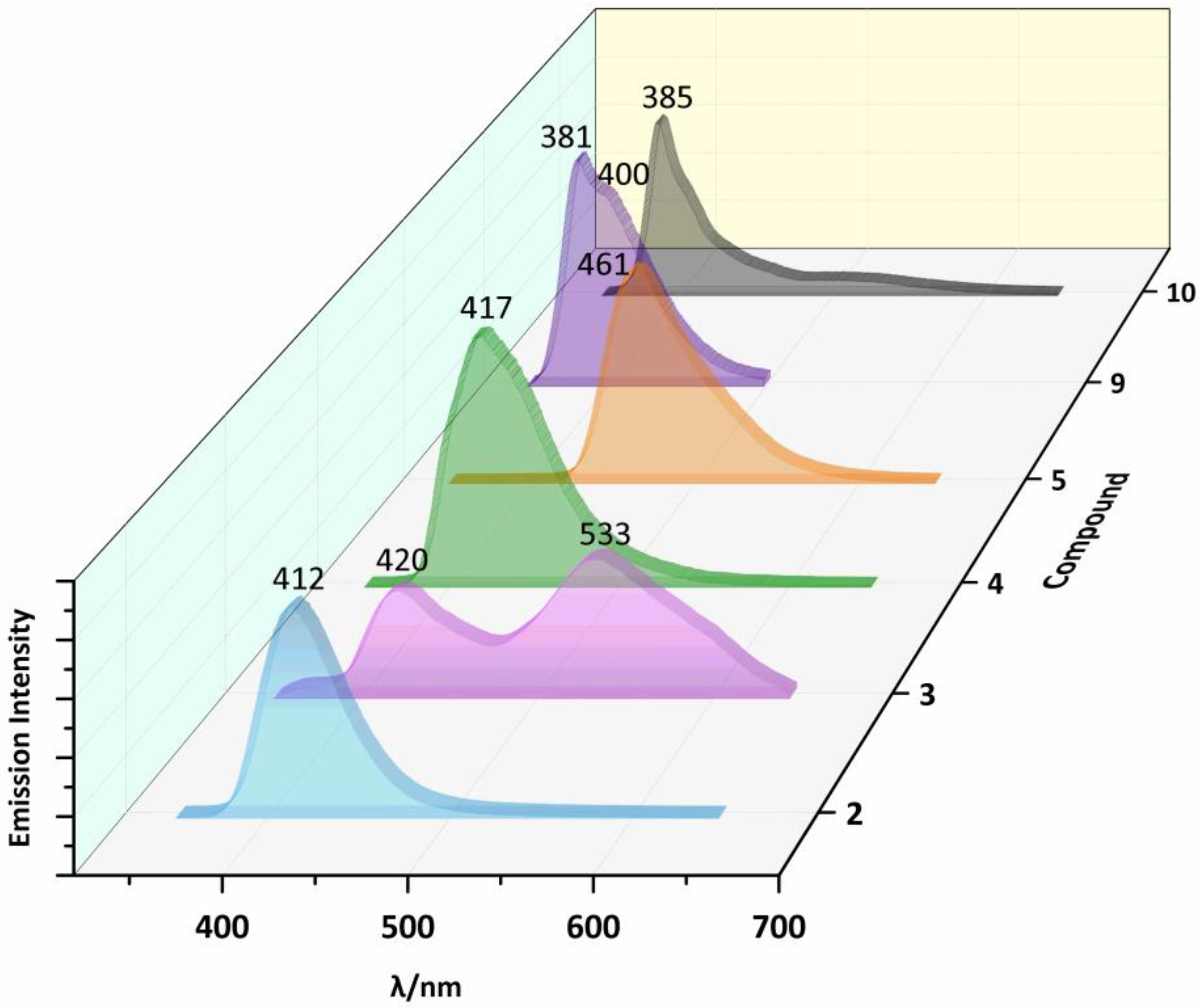
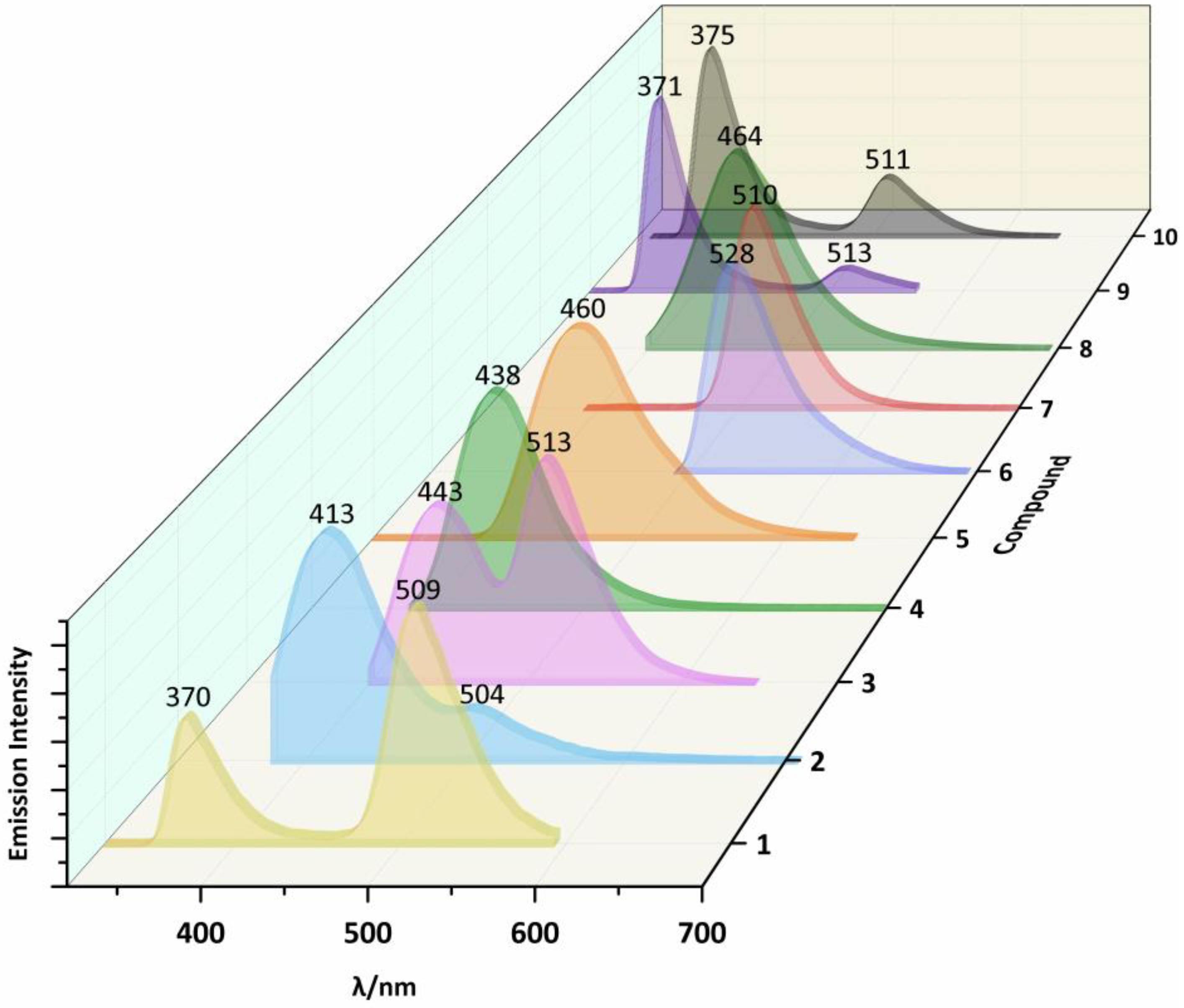
2.3. Photoluminescent Properties
2.4. Solution Stability
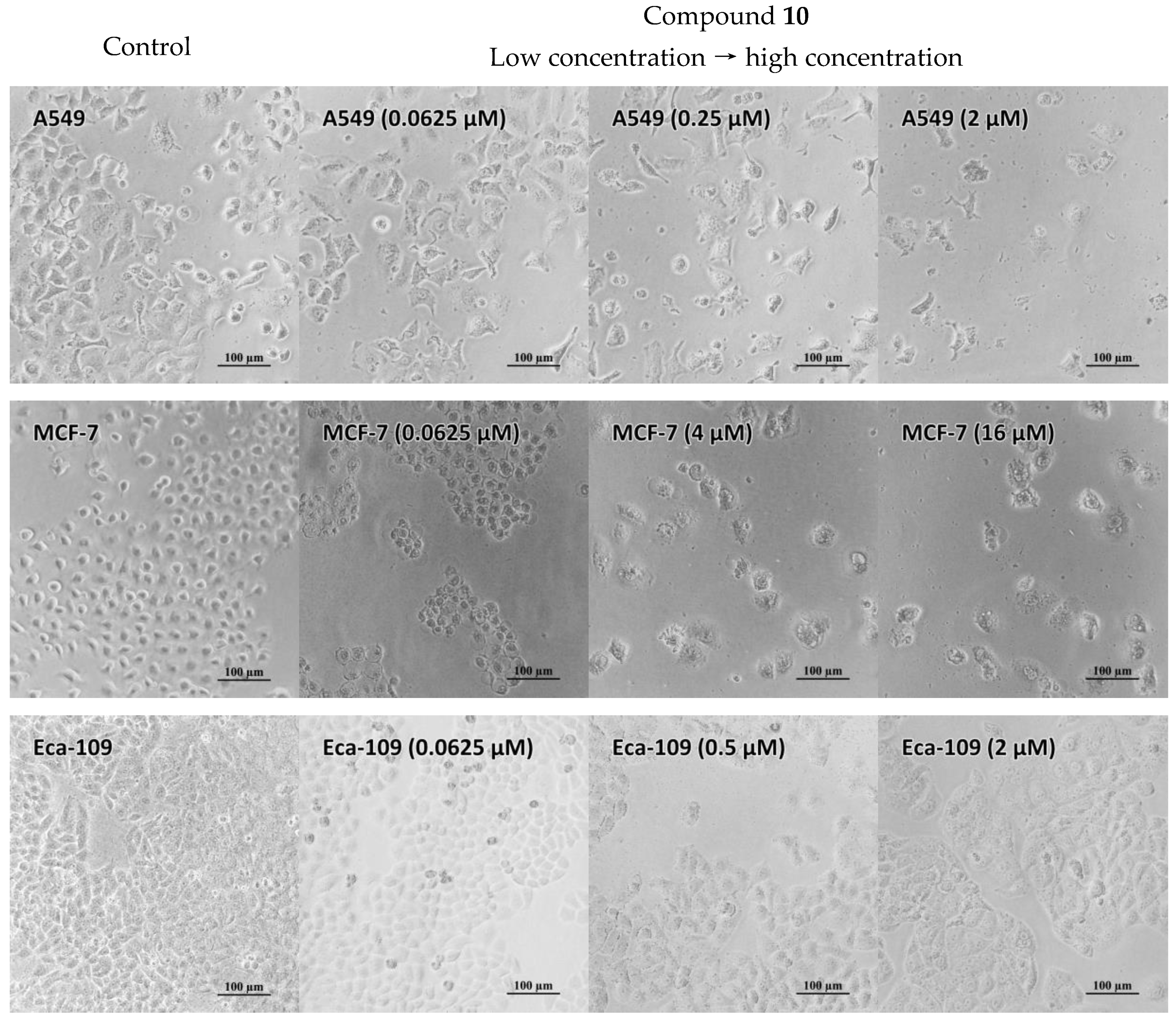
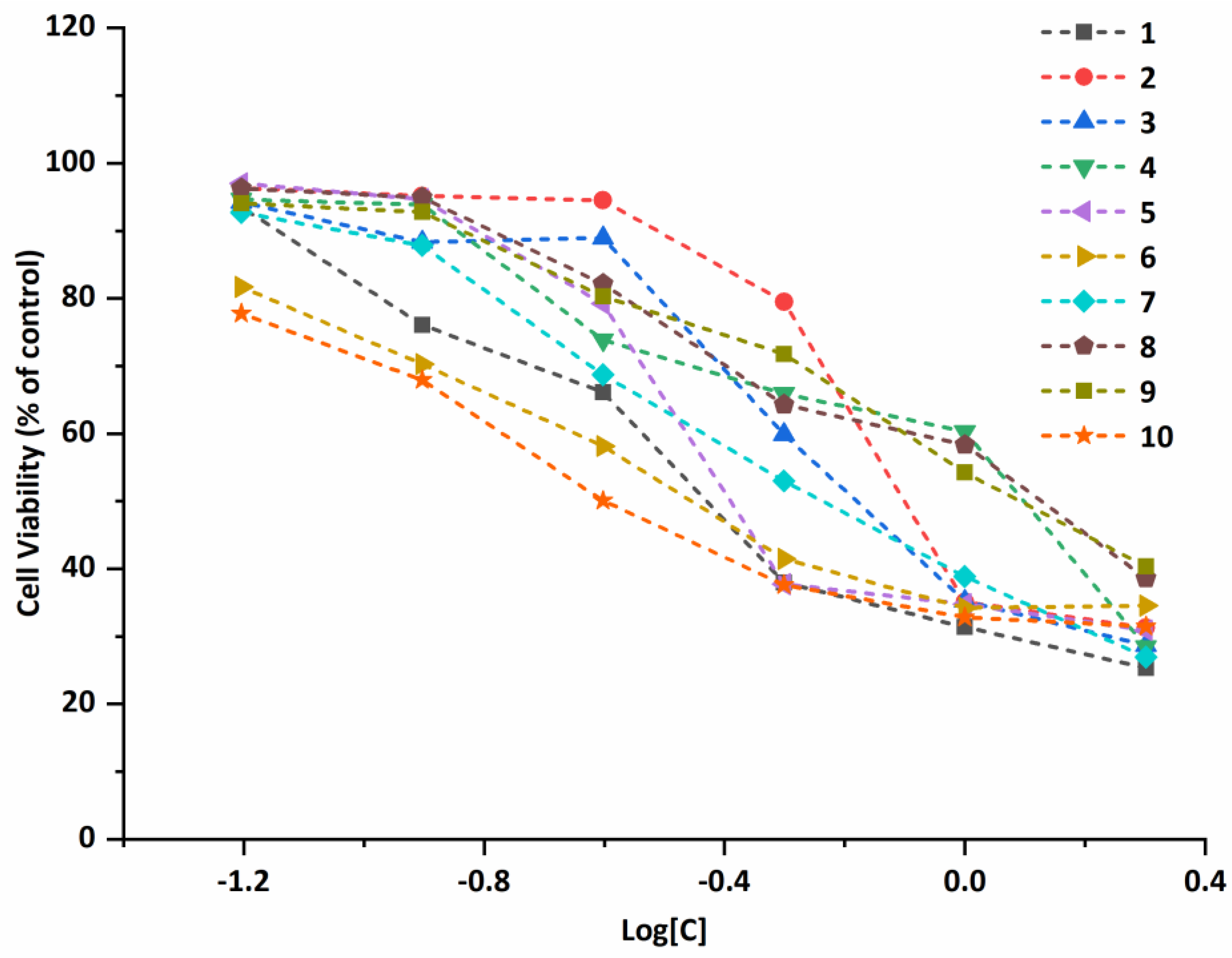
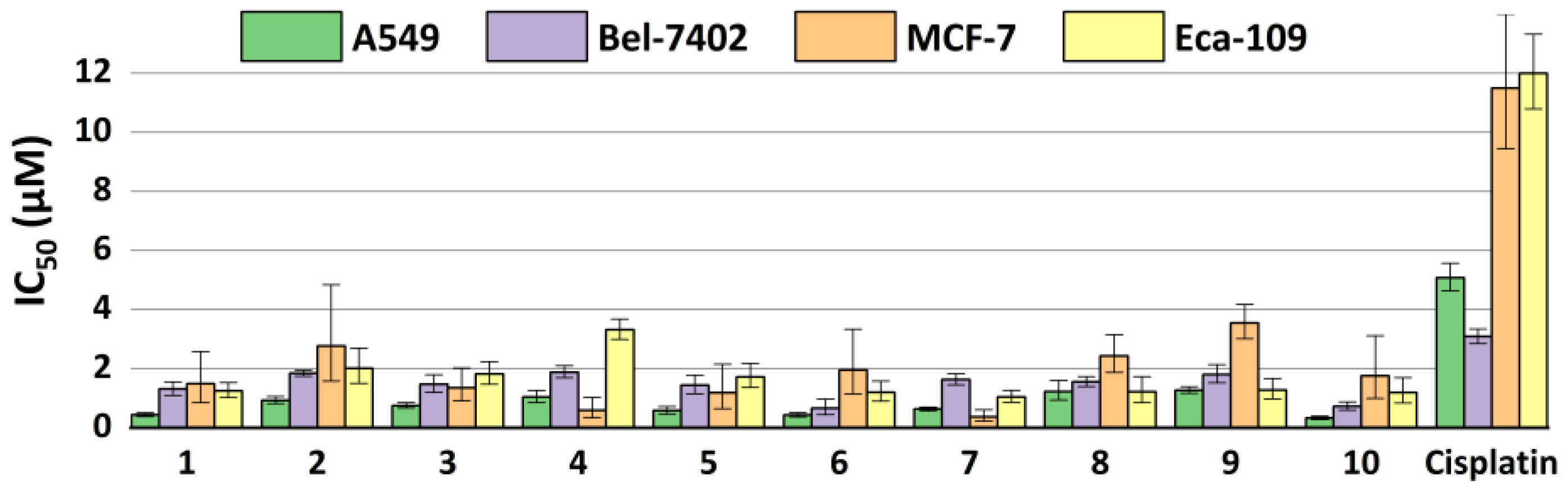
2.5. Antiproliferative Properties
2.6. Interaction of the Compounds with DNA.
2.6.1. UV-vis Absorption Spectroscopy
2.6.2. Fluorescence Titration
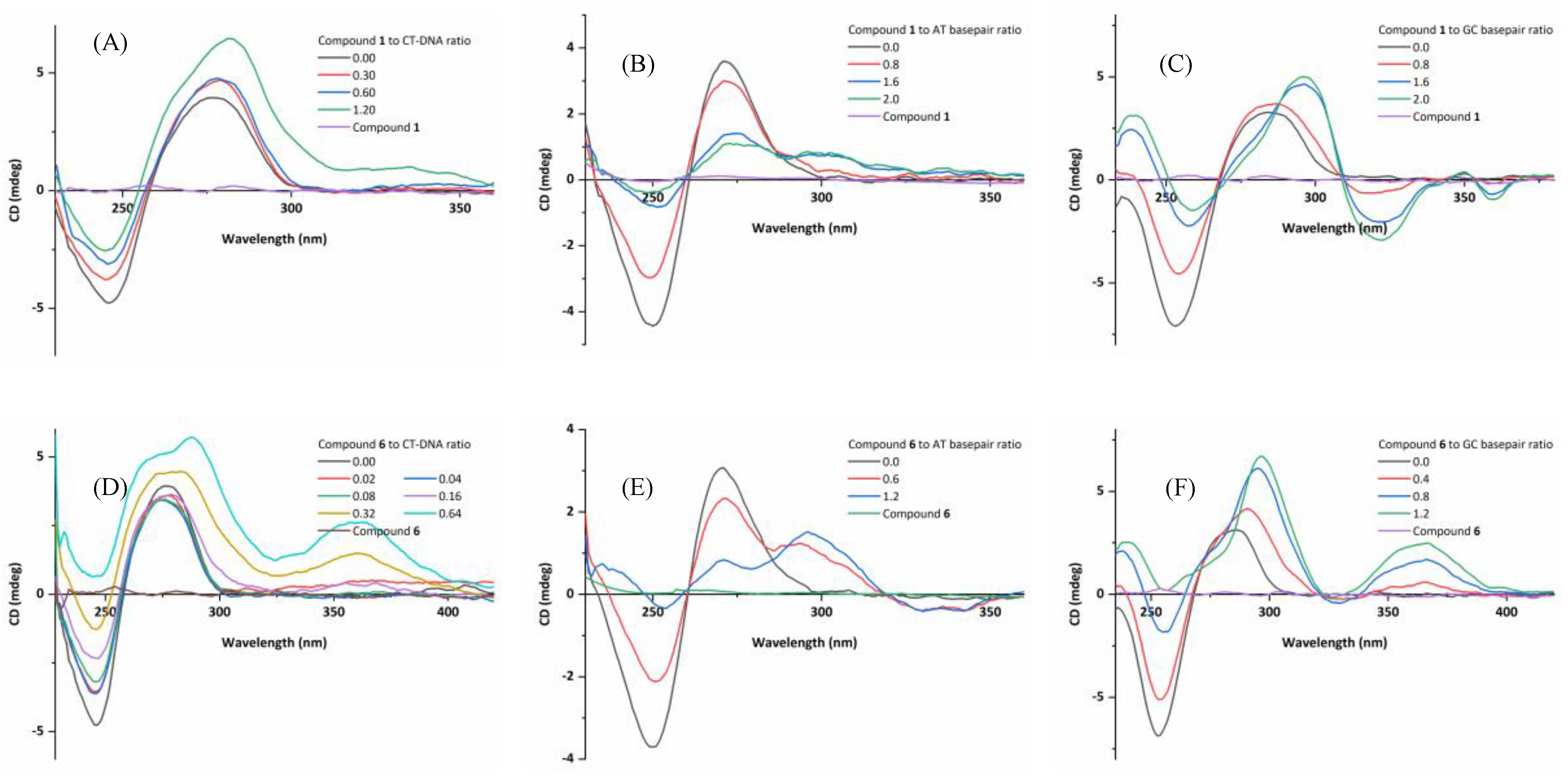
2.6.3. Circular Dichroism (CD) Spectroscopic Studies
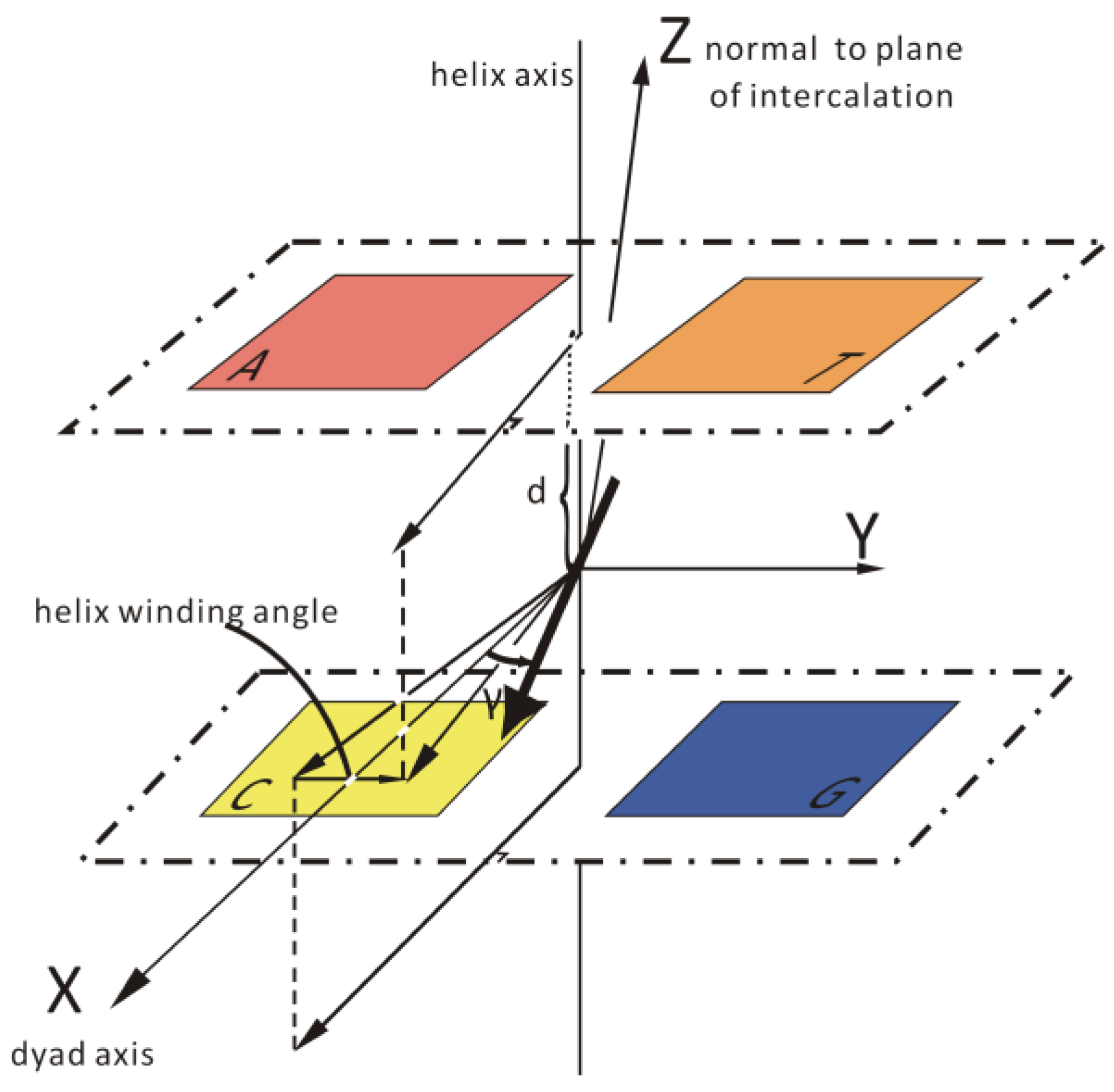
2.6.4. Induced CD (ICD) Characteristics
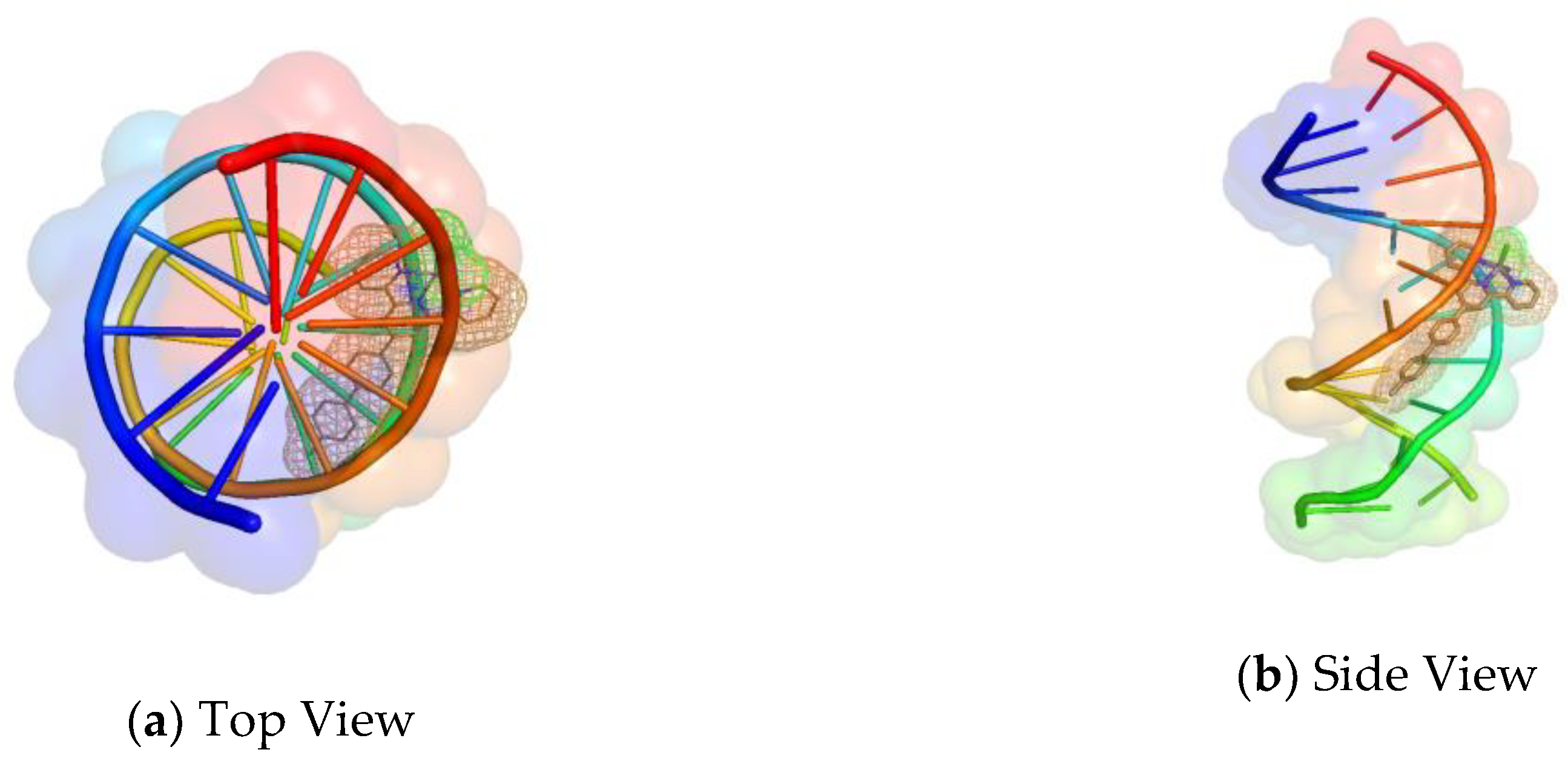
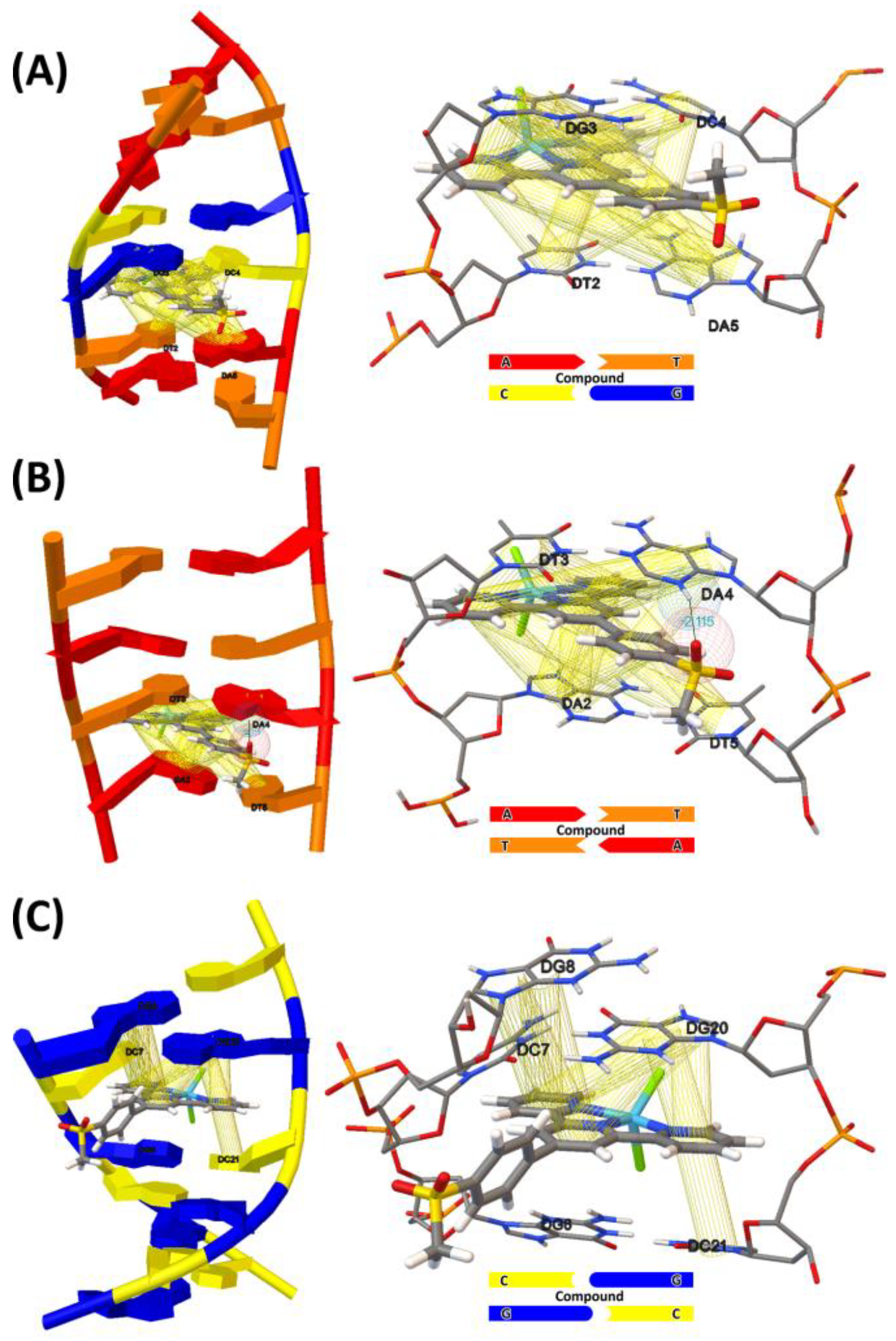
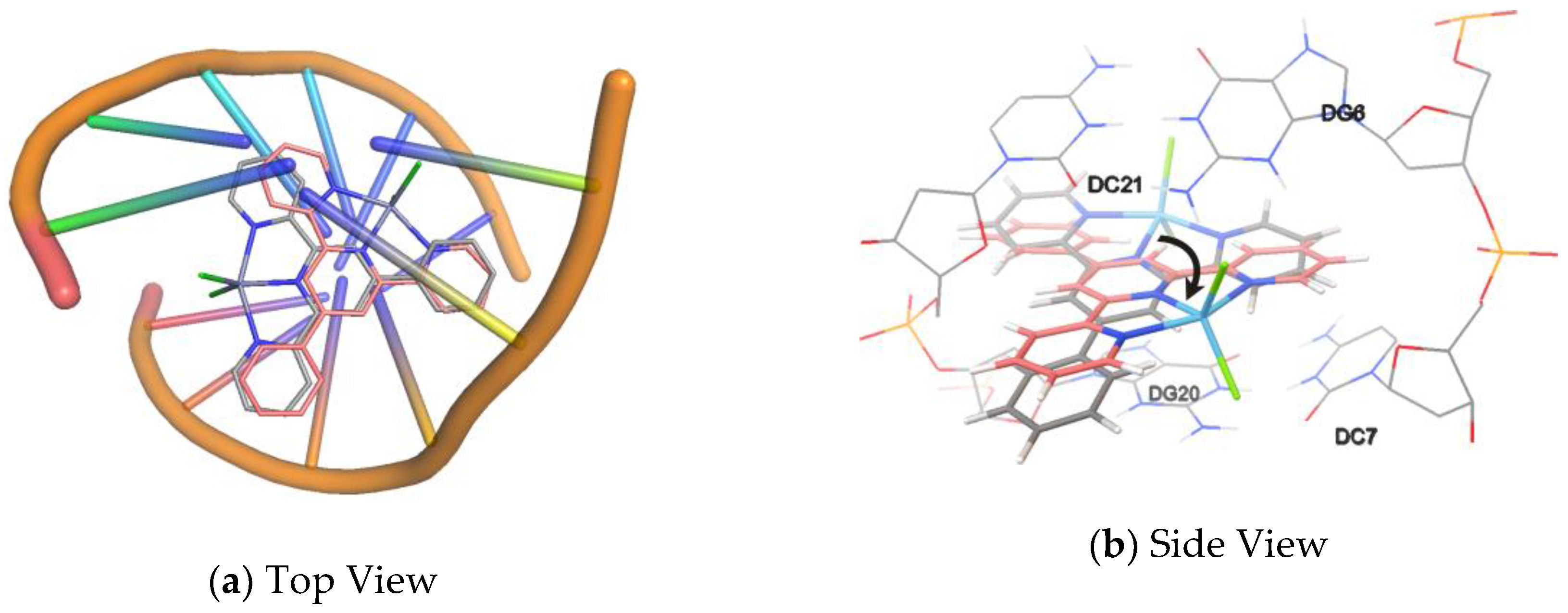
2.7. Molecular Docking Studies
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Physical Measurements
3.3. Synthesis of Ligands and the Complexes
3.4. Crystallography
3.5. Solution Chemistry
3.6. Antiproliferative Activity In Vitro
3.7. DNA Binding Studies
3.7.1. UV-vis Absorption Studies
3.7.2. Fluorescence Studies
3.7.3. Circular Dichroism (CD) Spectropolarimetry
3.8. Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Guo, Z.; Sadler, P.J. Metals in medicine. Angew. Chem. Int. Ed. 1999, 38, 1512–1531. [Google Scholar] [CrossRef]
- Wong, E.; Giandomenico, C.M. Current status of platinum-based antitumor drugs. Chem. Rev. 1999, 99, 2451–2466. [Google Scholar] [CrossRef] [PubMed]
- Jamieson, E.R.; Lippard, S.J. Structure, recognition, and processing of cisplatin−DNA adducts. Chem. Rev. 1999, 99, 2467–2498. [Google Scholar] [CrossRef] [PubMed]
- Reedijk, J. Why does cisplatin reach guanine-N7 with competing S-donor ligands available in the cell? Chem. Rev. 1999, 99, 2499–2510. [Google Scholar] [CrossRef]
- Roos, W.P.; Kaina, B. DNA damage-induced cell death by apoptosis. Trends Mol. Med. 2006, 12, 440–450. [Google Scholar] [CrossRef]
- Zuber, G.; Quada, J.C.; Hecht, S.M. Sequence selective cleavage of a DNA octanucleotide by chlorinated bithiazoles and bleomycins. J. Am. Chem. Soc. 1998, 120, 9368–9369. [Google Scholar] [CrossRef]
- Hecht, S.M. Bleomycin: New perspectives on the mechanism of action. J. Nat. Prod. 2000, 63, 158–168. [Google Scholar] [CrossRef]
- Metcalfe, C.; Thomas, J.A. Kinetically inert transition metal complexes that reversibly bind to DNA. Chem. Soc. Rev. 2003, 32, 215–224. [Google Scholar] [CrossRef]
- Silvestri, A.; Barone, G.; Ruisi, G.; Giudice, M.T.L.; Tumminello, S. The interaction of native DNA with iron (III)-N, N′-ethylene-bis (salicylideneiminato)-chloride. J. Inorg. Biochem. 2004, 98, 589–594. [Google Scholar] [CrossRef] [Green Version]
- Navarro, M.; Cisneros-Fajardo, E.J.; Sierralta, A.; Fernandez-Mestre, M.; Silva, P.; Arrieche, D.; Marchán, E. Design of copper DNA intercalators with leishmanicidal activity. JBIC J. Biol. Inorg. Chem. 2003, 8, 401–408. [Google Scholar] [CrossRef]
- Sigman, D.S.; Mazumder, A.; Perrin, D.M. Chemical nucleases. Chem. Rev. 1993, 93, 2295–2316. [Google Scholar] [CrossRef]
- Wolkenberg, S.E.; Boger, D.L. Mechanisms of in situ activation for DNA-targeting antitumor agents. Chem. Rev. 2002, 102, 2477–2496. [Google Scholar] [CrossRef] [PubMed]
- Erkkila, K.E.; Odom, D.T.; Barton, J.K. Recognition and reaction of metallointercalators with DNA. Chem. Rev. 1999, 99, 2777–2796. [Google Scholar] [CrossRef] [PubMed]
- Bruijnincx, P.C.; Sadler, P.J. New trends for metal complexes with anticancer activity. Curr. Opin. Chem. Biol. 2008, 12, 197–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, H.; Van Lier, J.E. Metal complexes as photo-and radiosensitizers. Chem. Rev. 1999, 99, 2379–2450. [Google Scholar] [CrossRef]
- Seng, H.L.; Tan, K.W.; Maah, M.J.; Tan, W.T.; Hamada, H.; Chikira, M.; Ng, C.H. Copper (II) complexes of methylated glycine derivatives: Effect of methyl substituent on their DNA binding and nucleolytic property. Polyhedron 2009, 28, 2219–2227. [Google Scholar] [CrossRef]
- Zeglis, B.M.; Pierre, V.C.; Barton, J.K. Metallo-intercalators and metallo-insertors. Chem. Commun. 2007, 4565–4579. [Google Scholar] [CrossRef] [Green Version]
- Siegel, R.; Naishadham, D.; Jemal, A. Cancer statistics for hispanics/latinos, 2012. CA. Cancer J. Clin. 2012, 62, 283–298. [Google Scholar] [CrossRef]
- Liu, L.; Fu, L.; Jing, T.; Ruan, Z.; Yan, L. pH-triggered polypeptides nanoparticles for efficient BODIPY imaging-guided near infrared photodynamic therapy. ACS Appl. Mater. Interfaces 2016, 8, 8980–8990. [Google Scholar] [CrossRef]
- Song, Y.; Wu, Q.; Yang, P.; Luan, N.; Wang, L.; Liu, Y. DNA Binding and cleavage activity of Ni (II) complex with all-trans retinoic acid. J. Inorg. Biochem. 2006, 100, 1685–1691. [Google Scholar] [CrossRef]
- Tan, C.; Liu, J.; Chen, L.; Shi, S.; Ji, L. Synthesis, structural characteristics, DNA binding properties and cytotoxicity studies of a series of Ru (III) complexes. J. Inorg. Biochem. 2008, 102, 1644–1653. [Google Scholar] [CrossRef] [PubMed]
- Schubert, U.S.; Hofmeier, H.; Newkome, G.R. Modern Terpyridine Chemistry; John Wiley & Sons: Hoboken, NJ, USA, 2006. [Google Scholar]
- Ma, Z.; Lu, W.; Liang, B.; Pombeiro, A.J. Synthesis, characterization, photoluminescent and thermal properties of zinc (II) 4′-phenyl-terpyridine compounds. New J. Chem. 2013, 37, 1529–1537. [Google Scholar] [CrossRef]
- Ma, Z.; Wei, L.; Alegria, E.C.; Martins, L.M.; da Silva, M.F.C.G.; Pombeiro, A.J. Synthesis and characterization of copper (II) 4′-phenyl-terpyridine compounds and catalytic application for aerobic oxidation of benzylic alcohols. Dalton Trans. 2014, 43, 4048–4058. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Zhang, B.; Da Silva, M.F.C.G.; Silva, J.; Mendo, A.S.; Baptista, P.V.; Fernandes, A.R.; Pombeiro, A.J. Synthesis, Characterization, thermal properties and antiproliferative potential of Copper (II) 4′-phenyl-terpyridine compounds. Dalton Trans. 2016, 45, 5339–5355. [Google Scholar] [CrossRef] [PubMed]
- Winter, A.; Gottschaldt, M.; Newkome, G.R.; Schubert, U.S. Terpyridines and their complexes with first row transition metal ions: Cytotoxicity, nuclease activity and self-assembly of biomacromolecules. Curr. Top. Med. Chem. 2012, 12, 158–175. [Google Scholar] [CrossRef]
- Schubert, U.S.; Winter, A.; Newkome, G.R. Terpyridine-Based Materials: For Catalytic, Optoelectronic and Life Science Applications; John Wiley & Sons: Hoboken, NJ, USA, 2012. [Google Scholar]
- Huang, Y.; Wang, Q.; Ma, Z. A Neutral Dinuclear Silver (I) 4′-Phenyl-Terpyridine Compound. J. Struct. Chem. 2018, 59, 182–187. [Google Scholar] [CrossRef]
- Liang, X.; Jiang, J.; Xue, X.; Huang, L.; Ding, X.; Nong, D.; Chen, H.; Pan, L.; Ma, Z. Synthesis, characterization, photoluminescence, anti-tumor activity, DFT calculations and molecular docking with proteins of zinc (II) halogen substituted terpyridine compounds. Dalton Trans. 2019, 48, 10488. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, P.; Liang, B.; Huang, L.; Zhou, Y.; Ma, Z. Effects of counterions of colorful sandwich-type zinc (II) 4′-phenyl-terpyridine compounds on photoluminescent and thermal properties. J. Mol. Struct. 2017, 1146, 504–511. [Google Scholar] [CrossRef]
- Medlycott, E.A.; Hanan, G.S. Designing tridentate ligands for ruthenium (II) complexes with prolonged room temperature luminescence lifetimes. Chem. Soc. Rev. 2005, 34, 133–142. [Google Scholar] [CrossRef]
- Xue, X.; Wang, Q.; Mai, F.; Liang, X.; Huang, Y.; Li, J.; Zhou, Y.; Yang, D.; Ma, Z. Study on the Photoluminescent and Thermal Properties of Zinc Complexes with a N6O4 Macrocyclic Ligand. Molecules 2018, 23, 1735. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Liu, C.; Bu, X.; Yang, M. Synthesis, crystal structure, cytotoxic activity and DNA-binding properties of the copper (II) and zinc (II) complexes with 1-[3-(2-pyridyl) pyrazol-1-ylmethyl] naphthalene. J. Inorg. Biochem. 2005, 99, 1119–1125. [Google Scholar] [CrossRef] [PubMed]
- Sheng, X.; Guo, X.; Lu, X.; Lu, G.; Shao, Y.; Liu, F.; Xu, Q. DNA binding, cleavage, and cytotoxic activity of the preorganized dinuclear zinc (II) complex of triazacyclononane derivatives. Bioconjug. Chem. 2008, 19, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Ng, C.H.; Kong, K.C.; Von, S.T.; Balraj, P.; Jensen, P.; Thirthagiri, E.; Hamada, H.; Chikira, M. Synthesis, characterization, DNA-binding study and anticancer properties of ternary metal (II) complexes of edda and an intercalating ligand. Dalton Trans. 2008, 4, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Stehbens, W.E. Oxidative stress, toxic hepatitis, and antioxidants with particular emphasis on zinc. Exp. Mol. Pathol. 2003, 75, 265–276. [Google Scholar] [CrossRef]
- Vaidyanathan, V.G.; Nair, B.U. Nucleobase oxidation of DNA by (terpyridyl) chromium (III) derivatives. Eur. J. Inorg. Chem. 2004, 2004, 1840–1846. [Google Scholar] [CrossRef]
- Indumathy, R.; Radhika, S.; Kanthimathi, M.; Weyhermuller, T.; Nair, B.U. Cobalt complexes of terpyridine ligand: Crystal structure and photocleavage of DNA. J. Inorg. Biochem. 2007, 101, 434–443. [Google Scholar] [CrossRef]
- Messori, L.; Marcon, G.; Innocenti, A.; Gallori, E.; Franchi, M.; Orioli, P. Molecular recognition of metal complexes by DNA: A comparative study of the interactions of the parent complexes [PtCl (TERPY)] Cl and [AuCl (TERPY)] Cl 2 with double stranded DNA. Bioinorg. Chem. Appl. 2005, 3, 239–253. [Google Scholar] [CrossRef] [Green Version]
- Constable, E.C.; Lewis, J.; Liptrot, M.C.; Raithby, P.R. The coordination chemistry of 4′-phenyl-2,2′: 6′, 2″-terpyridine; the synthesis, crystal and molecular structures of 4′-phenyl-2,2′: 6′, 2″-terpyridine and bis (4′-phenyl-2,2′: 6′,2″-terpyridine) nickel (II) chloride decahydrate. Inorg. Chim. Acta 1990, 178, 47–54. [Google Scholar] [CrossRef]
- Ma, Z.; Cao, Y.; Li, Q.; da Silva, M.F.C.G.; da Silva, J.J.F.; Pombeiro, A.J. Synthesis, characterization, solid-state photo-luminescence and anti-tumor activity of zinc (II) 4′-phenyl-terpyridine compounds. J. Inorg. Biochem. 2010, 104, 704–711. [Google Scholar] [CrossRef]
- Zhou, P.; Huang, L.; Zhang, Y.; Xue, X.; Zhou, Y.; Ma, Z. Synthesis, characterization and photoluminescence of substituted terpyridine compounds and their molecular docking studies with bovine hemoglobin. J. Photochem. Photobiol. A Chem. 2018, 358, 17–25. [Google Scholar] [CrossRef]
- Jeitler, J.R.; Turnbull, M.M.; Wikaira, J.L. Synthesis, characterization and structure of transition metal complexes of 4′-methylthio-2,2′: 6′,2″-terpyridyl. Inorg. Chim. Acta 2003, 351, 331–344. [Google Scholar] [CrossRef]
- Hou, L.; Li, D. A novel photoluminescent Ag–terpyridyl complex: One-dimensional linear metal string with double-helical structure. Inorg. Chem. Commun. 2005, 8, 128–130. [Google Scholar] [CrossRef]
- Hasegawa, Y.; Nakagawa, T.; Kawai, T. Recent progress of luminescent metal complexes with photochromic units. Coord. Chem. Rev. 2010, 254, 2643–2651. [Google Scholar] [CrossRef]
- Di, L.; Kerns, E.H.; Chen, H.; Petusky, S.L. Development and application of an automated solution stability assay for drug discovery. J. Biomol. Screen. 2006, 11, 40–47. [Google Scholar] [CrossRef] [Green Version]
- Reedijk, J. Platinum anticancer coordination compounds: Study of DNA binding inspires new drug design. Eur. J. Inorg. Chem. 2009, 10, 1303–1312. [Google Scholar] [CrossRef]
- Dasari, S.; Tchounwou, P.B. Cisplatin in cancer therapy: Molecular mechanisms of action. Eur. J. Pharmacol. 2014, 740, 364–378. [Google Scholar] [CrossRef] [Green Version]
- Brabec, V.; Kasparkova, J. Modifications of DNA by platinum complexes: Relation to resistance of tumors to platinum antitumor drugs. Drug Resist. Updat. 2005, 8, 131–146. [Google Scholar] [CrossRef]
- Pyle, A.; Rehmann, J.; Meshoyrer, R.; Kumar, C.; Turro, N.; Barton, J.K. Mixed-ligand complexes of ruthenium (II): Factors governing binding to DNA. J. Am. Chem. Soc. 1989, 111, 3051–3058. [Google Scholar] [CrossRef]
- Shahabadi, N.; Fatahi, P. DNA Interaction Studies of a Copper (II) Complex Containing an Antiviral Drug, Valacyclovir: The Effect of Metal Center on the Mode of Binding. DNA Cell Biol. 2012, 31, 1328–1334. [Google Scholar] [CrossRef]
- Long, E.C.; Barton, J.K. On demonstrating DNA intercalation. Acc. Chem. Res. 1990, 23, 271–273. [Google Scholar] [CrossRef]
- Fiel, R.; Howard, J.; Mark, E.; Gupta, N.D. Interaction of DNA with a porphyrin ligand: Evidence for intercalation. Nucleic Acids Res. 1979, 6, 3093–3118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terenzi, A.; Barone, G.; Silvestri, A.; Giuliani, A.M.; Ruggirello, A.; Liveri, V.T. Interaction of DNA with a porphyrin ligand: Evidence for intercalation. J. Inorg. Biochem. 2009, 103, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darabi, F.; Hadadzadeh, H.; Ebrahimi, M.; Khayamian, T.; Rudbari, H.A. The piroxicam complex of cobalt (II): Synthesis in two different ionic liquids, structure, DNA-and BSA interaction and molecular modeling. Inorg. Chim. Acta 2014, 409, 379–389. [Google Scholar] [CrossRef]
- Xi, P.; Xu, Z.; Liu, X.; Cheng, F.; Zeng, Z. Synthesis, characterization and DNA-binding studies of 1-cyclohexyl-3-tosylurea and its Ni (II), and Cd (II) complexes. Spectrochimi. Acta A 2008, 71, 523–528. [Google Scholar] [CrossRef]
- Zhang, S.; Yang, H.; Zhao, L.; Gan, R.; Tang, P.; Sun, Q.; Xiong, X.; Li, H. Capecitabine as a minor groove binder of DNA: Molecular docking, molecular dynamics, and multi-spectroscopic studies. J. Biomol. Struct. Dyn. 2019, 37, 1451–1463. [Google Scholar] [CrossRef]
- Kuntz, I., Jr.; Gasparro, F.; Johnston, M., Jr.; Taylor, R. Molecular interactions and the Benesi-Hildebrand equation. J. Am. Chem. Soc. 1968, 90, 4778–4781. [Google Scholar] [CrossRef]
- Scott, R.L. Some comments on the Benesi-Hildebrand equation. Recl. Trav. Chim. Pays-Bas 1956, 75, 787–789. [Google Scholar] [CrossRef]
- Anjomshoa, M.; Fatemi, S.J.; Torkzadeh-Mahani, M.; Hadadzadeh, H. DNA- and BSA- binding studies and anticancer activity against human breast cancer cells (MCF-7) of the zinc (II) complex coordinated by 5,6-diphenyl-3-(2-pyridyl)-1,2,4-triazine. Spectrochimi. Acta A 2014, 127, 511–520. [Google Scholar] [CrossRef]
- Anjomshoa, M.; Hadadzadeh, H.; Fatemi, S.J.; Torkzadeh-Mahani, M. A mononuclear Ni (II) complex with 5,6-diphenyl-3-(2-pyridyl)-1,2,4-triazine: DNA- and BSA- binding and anticancer activity against human breast carcinoma cells. Spectrochimi. Acta A 2015, 136, 205–215. [Google Scholar] [CrossRef]
- Maheswari, P.U.; Roy, S.; den Dulk, H.; Barends, S.; van Wezel, G.; Kozlevcar, B.; Gamez, P.; Reedijk, J. The square-planar cytotoxic [CuII (pyrimol) Cl] complex acts as an efficient DNA cleaver without reductant. J. Am. Chem. Soc. 2006, 128, 710–711. [Google Scholar] [CrossRef]
- Tabassum, S.; Zaki, M.; Afzal, M.; Arjmand, F. Synthesis and characterization of Cu (II)-based anticancer chemotherapeutic agent targeting topoisomerase Iα: In vitro DNA binding, pBR322 cleavage, molecular docking studies and cytotoxicity against human cancer cell lines. Eur. J. Med. Chem. 2014, 74, 509–523. [Google Scholar] [CrossRef]
- Zhang, G.; Fu, P.; Wang, L.; Hu, M. Molecular spectroscopic studies of farrerol interaction with calf thymus DNA. J. Agric. Food Chem. 2011, 59, 8944–8952. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, X.; Yan, Y.; Xiang, W. Comparative studies on the interaction of genistein, 8-chlorogenistein, and 3′, 8-dichlorogenistein with bovine serum albumin. J. Agric. Food Chem. 2011, 59, 7506–7513. [Google Scholar] [CrossRef]
- Berova, N.; Nakanishi, K.; Woody, R.W.B. Circular Dichroism: Principles and Applications; John Wiley & Sons: Hoboken, NJ, USA, 2000. [Google Scholar]
- Nordén, B.; Kurucsev, T. Analysing DNA complexes by circular and linear dichroism. J. Mol. Recognit. 1994, 7, 141–155. [Google Scholar] [CrossRef]
- Garbett, N.C.; Ragazzon, P.A.; Chaires, J.B. Circular dichroism to determine binding mode and affinity of ligand–DNA interactions. Nat. Protoc. 2007, 2, 3166. [Google Scholar] [CrossRef]
- Kong, D.; Wang, J.; Zhu, L.; Jin, Y.; Li, X.; Shen, H.; Mi, H. Oxidative DNA cleavage by Schiff base tetraazamacrocyclic oxamido nickel (II) complexes. J. Inorg. Biochem. 2008, 102, 824–832. [Google Scholar] [CrossRef]
- Rahban, M.; Divsalar, A.; Saboury, A.A.; Golestani, A. Nanotoxicity and spectroscopy studies of silver nanoparticle: Calf thymus DNA and K562 as targets. J. Phys. Chem. C 2010, 114, 5798–5803. [Google Scholar] [CrossRef]
- Kashanian, S.; Askari, S.; Ahmadi, F.; Omidfar, K.; Ghobadi, S.; Tarighat, F.A. In vitro study of DNA interaction with clodinafop-propargyl herbicide. DNA Cell Biol. 2008, 27, 581–586. [Google Scholar] [CrossRef]
- Dehkordi, M.N.; Bordbar, A.; Lincoln, P.; Mirkhani, V. Spectroscopic study on the interaction of ct-DNA with manganese Salen complex containing triphenyl phosphonium groups. Spectrochimi. Acta A 2012, 90, 50–54. [Google Scholar] [CrossRef]
- Lyng, R.; Härd, T.; Norden, B. Induced CD of DNA intercalators: Electric dipole allowed transitions. Biopolymers 1987, 26, 1327–1345. [Google Scholar] [CrossRef]
- Modukuru, N.; Snow, K.; Perrin Jr, B.S.; Bhambhani, A.; Duff, M.; Kumar, C.V. Tuning the DNA binding modes of an anthracene derivative with salt. J. Photochem. Photobiol. A Chem. 2006, 177, 43–54. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [Green Version]
- Holt, P.A.; Chaires, J.B.; Trent, J.O. Molecular docking of intercalators and groove-binders to nucleic acids using Autodock and Surflex. J. Chem. Inf. Model. 2008, 48, 1602–1615. [Google Scholar] [CrossRef] [Green Version]
- Reichmann, M.; Rice, S.; Thomas, C.; Doty, P. A further examination of the molecular weight and size of desoxypentose nucleic acid. J. Am. Chem. Soc. 1954, 76, 3047–3053. [Google Scholar] [CrossRef]
- Agilent. CrysAlis PRO, Version 171.40.53; Agilent Technologies: Yarnton, UK, 2012. [Google Scholar]
- Bruker. APEX2, Version 1.0-27; Bruker AXS Inc.: Madison, WI, USA, 2005. [Google Scholar]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crys. 2015, C71, 3–8. [Google Scholar]
- Sheldrick, G.M. A short history of SHELX. Acta Cryst. 2008, A64, 112–122. [Google Scholar] [CrossRef] [Green Version]
- Farrugia, L.J. WinGX Suite for Small-Molecule Single-Crystal Crystallography. J. Appl. Crystallogr. 1999, 32, 837–838. [Google Scholar] [CrossRef]
- Rajendiran, V.; Murali, M.; Suresh, E.; Palaniandavar, M.; Periasamy, V.S.; Akbarsha, M.A. Non-covalent DNA binding and cytotoxicity of certain mixed-ligand ruthenium (II) complexes of 2,2′-dipyridylamine and diimines. Dalton Trans. 2008, 2157–2170. [Google Scholar] [CrossRef]
- DeLano, W.L. PyMOL Molecular Viewer; DeLano Scientific LLC: Palo Alto, CA, USA, 2008. [Google Scholar]
- Seeliger, D.; de Groot, B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput-Aided. Mol. Des. 2010, 24, 417–422. [Google Scholar] [CrossRef] [Green Version]
Sample Availability: Samples of the compounds 1–10 are available from the authors. |

| Table. Cont. | 2 | 3 | 4 | 5 | 9 | 10 |
|---|---|---|---|---|---|---|
| Empirical formula | C22H17Cl2N3Zn | C22H17Cl2N3OZn | C27H19Cl2N3Zn | C32H33Cl2N3O2S2Zn | C22H19Cl2N3O4Zn | C22H14Cl2N3O2SZn |
| Formula weight | 459.66 | 475.66 | 521.72 | 692.02 | 525.69 | 520.69 |
| Temperature | 298(2) K | 298(2) K | 298(2) K | 298(2) K | 298(2) K | 298(2) K |
| Crystal system | Monoclinic | Monoclinic | Orthorhombic | Triclinic | Monoclinic | Monoclinic |
| space group | P21/c | P21/c | Pbcn | P-1 | I2/a | P21/n |
| a (Å) | 12.6962(6) | 8.5597(17) | 12.025(2) | 10.3442(12) | 11.065(2) | 10.3908(5) |
| b (Å) | 9.5863(3) | 14.455(3) | 17.748(4) | 12.2318(13) | 17.610(3) | 18.0058(9) |
| c (Å) | 17.4340(9) | 16.558(3) | 11.024(2) | 13.2856(14) | 11.531(6) | 12.7990(7) |
| α (°) | 90 | 90 | 90 | 97.470(5) | 90 | 90 |
| β (°) | 111.229(5) | 99.10(3) | 90 | 95.354(6) | 107.31(3) | 111.638(2) |
| γ (°) | 90 | 90 | 90 | 97.459(5) | 90 | 90 |
| Volume (Å3) | 1977.90(15) | 2023.0(7) | 2352.7(8) | 1642.2(3) | 2145.1(12) | 2225.9(2) |
| Z | 4 | 4 | 4 | 2 | 4 | 4 |
| Calculated desity(Mg/m3) | 1.544 | 1.562 | 1.473 | 1.399 | 1.628 | 1.554 |
| Absorption coefficient(mm−1) | 1.525 | 1.497 | 1.292 | 1.071 | 1.430 | 1.462 |
| F(000) | 936 | 968 | 1064 | 716 | 1072 | 1052 |
| Crystal size(mm−1) | 0.49 × 0.48 × 0.29 | 0.48 × 0.46 × 0.34 | 0.48 × 0.36 × 0.27 | 0.43 × 0.28 × 0.25 | 0.48 × 0.46 × 0.34 | 0.39 × 0.22 × 0.21 |
| θmax, θmin (°) | 27.50, 2.74 | 27.49, 2.86 | 29.61, 2.95 | 27.92, 2.13 | 29.59, 3.22 | 29.60, 2.05 |
| Index range h | −16 → 16 | −11 → 9 | −16 → 15 | −13 → 13 | −8 → 14 | −14 → 13 |
| K | −10 → 12 | −17 → 18 | −24 → 22 | −15 → 16 | −22 → 20 | −24 → 20 |
| L | −15 → 22 | −20 → 21 | −14 → 14 | −17 → 17 | −15 → 14 | −17 → 17 |
| Reflections collected/unique | 16,461/4492 [R(int) = 0.0181] | 13,327/4516 [R(int) = 0.0265] | 23,550/3051 [R(int) = 0.0273] | 24,876/7674 [R(int) = 0.0222] | 5545/2558 [R(int) = 0.0188] | 23,202/6085 [R(int) = 0.0472] |
| Data/restraints/parameters | 4492/0/253 | 4516/0/262 | 3051/0/153 | 7674/0/389 | 2558/0/148 | 6085/0/280 |
| Goodness-of-fit on F2 | 1.032 | 1.019 | 1.038 | 1.027 | 1.046 | 1.008 |
| Final R indices [I > 2σ(I)] | R1 = 0.0254, wR2 = 0.0717 | R1 = 0.0324, wR2 = 0.0805 | R1 = 0.0426, wR2 = 0.1298 | R1 = 0.0407, wR2 = 0.1151 | R1 = 0.0430, wR2 = 0.1268 | R1 = 0.0582, wR2 = 0.1600 |
| R indices (all data) | R1 = 0.0311, wR2 = 0.0756 | R1 = 0.0422, wR2 = 0.0861 | R1 = 0.0528, wR2 = 0.1414 | R1 = 0.0494, wR2 = 0.1220 | R1 = 0.0490, wR2 = 0.1355 | R1 = 0.1109, wR2 = 0.1844 |
| Largest diff. peak and hole(e Å−3) | 0.306 and −0.375 | 0.324 and −0.473 | 1.169 and −0.440 | 1.486 and −0.651 | 0.372 and −0.893 | 0.828 and −0.442 |
| CCDC number | 1,568,389 | 1,568,387 | 1,568,390 | 1,568,391 | 1,568,392 | 1,568,388 |
| Compound | Solid State | DMF Solution | ||||
|---|---|---|---|---|---|---|
| Excitation (nm) | Emission Bands (nm) | Excitation (nm) | Emission Bands (nm) | |||
| 1 | 300a | 409a | 270 | 370 | 509 | |
| 2 | 300 | 412 | 350 | 413 | 504 | |
| 3 | 300 | 420 | 533 | 380 | 443 | 513 |
| 4 | 300 | 417 | 360 | 438 | ||
| 5 | 300 | 461 | 300 | 460 | ||
| 6 | 330b | 420b | 470 | 528 | ||
| 7 | 300b | 415b | 370 | 510 | ||
| 8 | 360b | 483b | 380 | 464 | ||
| 9 | 300 | 381 | 400 | 270 | 371 | 513 |
| 10 | 300 | 385 | 310 | 375 | 511 | |
| Compound | A549 | Bel-7402 | MCF-7 | Eca-109 | ||||
|---|---|---|---|---|---|---|---|---|
| IC50 (μM) | 95% Confidence Intervals (μM) | IC50 (μM) | 95% Confidence Intervals (μM) | IC50 (μM) | 95% Confidence Intervals (μM) | IC50 (μM) | 95% Confidence Intervals (μM) | |
| 1 | 0.440 | 0.384–0.504 | 1.309 | 1.098–1.560 | 1.486 | 0.855–2.582 | 1.251 | 1.024–1.528 |
| 2 | 0.933 | 0.821–1.061 | 1.842 | 1.741–1.949 | 2.769 | 1.584–4.840 | 2.017 | 1.507–2.701 |
| 3 | 0.756 | 0.672–0.852 | 1.470 | 1.202–1.797 | 1.358 | 0.911–2.025 | 1.821 | 1.475–2.247 |
| 4 | 1.042 | 0.858–1.265 | 1.883 | 1.691–2.096 | 0.589 | 0.336–1.035 | 3.320 | 2.996–3.680 |
| 5 | 0.586 | 0.476–0.722 | 1.435 | 1.150–1.790 | 1.187 | 0.655–2.152 | 1.722 | 1.360–2.181 |
| 6 | 0.435 | 0.370–0.513 | 0.660 | 0.447–0.975 | 1.956 | 1.148–3.332 | 1.198 | 0.908–1.580 |
| 7 | 0.633 | 0.568–0.705 | 1.636 | 1.459–1.835 | 0.374 | 0.229–0.609 | 1.045 | 0.862–1.266 |
| 8 | 1.228 | 0.938–1.608 | 1.557 | 1.396–1.736 | 2.428 | 1.870–3.153 | 1.215 | 0.852–1.732 |
| 9 | 1.270 | 1.165–1.385 | 1.804 | 1.521–2.139 | 3.548 | 3.016–4.173 | 1.280 | 0.985–1.665 |
| 10 | 0.333 | 0.279–0.396 | 0.730 | 0.604–0.883 | 1.764 | 0.999–3.116 | 1.193 | 0.841–1.692 |
| Cisplatin | 5.082 | 4.639–5.568 | 3.088 | 2.856–3.340 | 11.49 | 9.435–14.00 | 11.99 | 10.79–13.32 |
| Compound | Kb (M−1) | Log Kb | Ra |
|---|---|---|---|
| 1 | 6.89 × 102 | 2.84 | 0.9949 |
| 2 | 4.78 × 104 | 4.68 | 0.9830 |
| 3 | 9.95 × 101 | 0.00 | 0.9949 |
| 4 | 3.96 × 104 | 4.60 | 0.9923 |
| 5 | 9.79 × 104 | 4.99 | 0.9898 |
| 6 | 1.36 × 104 | 4.13 | 0.9973 |
| 7 | 1.86 × 104 | 4.27 | 0.9830 |
| 8 | 8.55 × 104 | 4.93 | 0.9911 |
| 9 | 1.08 × 104 | 4.03 | 0.9830 |
| 10 | 3.49 × 103 | 3.54 | 0.9987 |
| Compound | Affinity (kcal/mol) | |||
|---|---|---|---|---|
| 1BNA | 4JD8 | 2DA8M | 2ROUM | |
| 1 | −9.09 | −8.43 | −8.30 | −8.13 |
| 2 | −9.63 | −8.88 | −8.42 | −8.16 |
| 3 | −9.20 | −8.21 | −7.49 | −8.00 |
| 4 | −11.43 | −8.77 | −8.94 | −8.71 |
| 5 | −11.83 | −9.14 | −8.90 | −8.83 |
| 6 | −9.56 | −8.40 | −8.21 | −7.92 |
| 7 | −9.32 | −8.64 | −8.50 | −8.28 |
| 8 | −9.29 | −8.85 | −8.12 | −8.60 |
| 9 | −8.70 | −7.71 | −7.74 | −7.38 |
| 10 | −10.83 | −8.65 | −8.33 | −8.35 |
| Compound | DNA | Bonds Formed | Bond Distance (Å) | Bond Energy (kcal/mol) |
|---|---|---|---|---|
| 3 | 1BNA | N–H…O (DG4) | 1.877 | −0.818 |
| 6 | 1BNA | O–H…O (DA18) | 2.047 | −5.96 |
| 4JD8 | N–H…O (DA5) | 2.065 | −5.551 | |
| 2DA8M | N–H…O (DT5) | 2.113 | −5.749 | |
| 2ROUM | N–H…O (DT2) | 1.946 | −3.502 | |
| 7 | 1BNA | O–H…O (DT19) | 2.164 | −5.727 |
| 4JD8 | O–H…O (DT2) | 2.229 | −0.263 | |
| 2DA8M | O–H…O (DT5) | 1.820 | −1.640 | |
| 2ROUM | O–H…O (DC7) | 1.975 | −5.769 | |
| 8 | 1BNA | O–H…O (DT19) | 1.828 | −4.119 |
| 2DA8M | N–H…O (DT3) | 1.862 | −4.078 | |
| 2ROUM | N–H…O (DC7) | 2.210 | −3.302 | |
| 9 | 1BNA | N–H…O (DG16) | 1.837 | −7.674 |
| 10 | 1BNA | N–H…O (DG16) | 1.765 | −7.869 |
| 2DA8M | O–H…O (DA4) | 2.115 | −4.156 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Liu, R.; Jiang, J.; Liang, X.; Huang, L.; Huang, G.; Chen, H.; Pan, L.; Ma, Z. Zinc(II) Terpyridine Complexes: Substituent Effect on Photoluminescence, Antiproliferative Activity, and DNA Interaction. Molecules 2019, 24, 4519. https://doi.org/10.3390/molecules24244519
Li J, Liu R, Jiang J, Liang X, Huang L, Huang G, Chen H, Pan L, Ma Z. Zinc(II) Terpyridine Complexes: Substituent Effect on Photoluminescence, Antiproliferative Activity, and DNA Interaction. Molecules. 2019; 24(24):4519. https://doi.org/10.3390/molecules24244519
Chicago/Turabian StyleLi, Jiahe, Rongping Liu, Jinzhang Jiang, Xing Liang, Ling Huang, Gang Huang, Hailan Chen, Lixia Pan, and Zhen Ma. 2019. "Zinc(II) Terpyridine Complexes: Substituent Effect on Photoluminescence, Antiproliferative Activity, and DNA Interaction" Molecules 24, no. 24: 4519. https://doi.org/10.3390/molecules24244519






