A Repurposing Approach for Uncovering the Anti-Tubercular Activity of FDA-Approved Drugs with Potential Multi-Targeting Profiles
Abstract
1. Introduction
2. Results
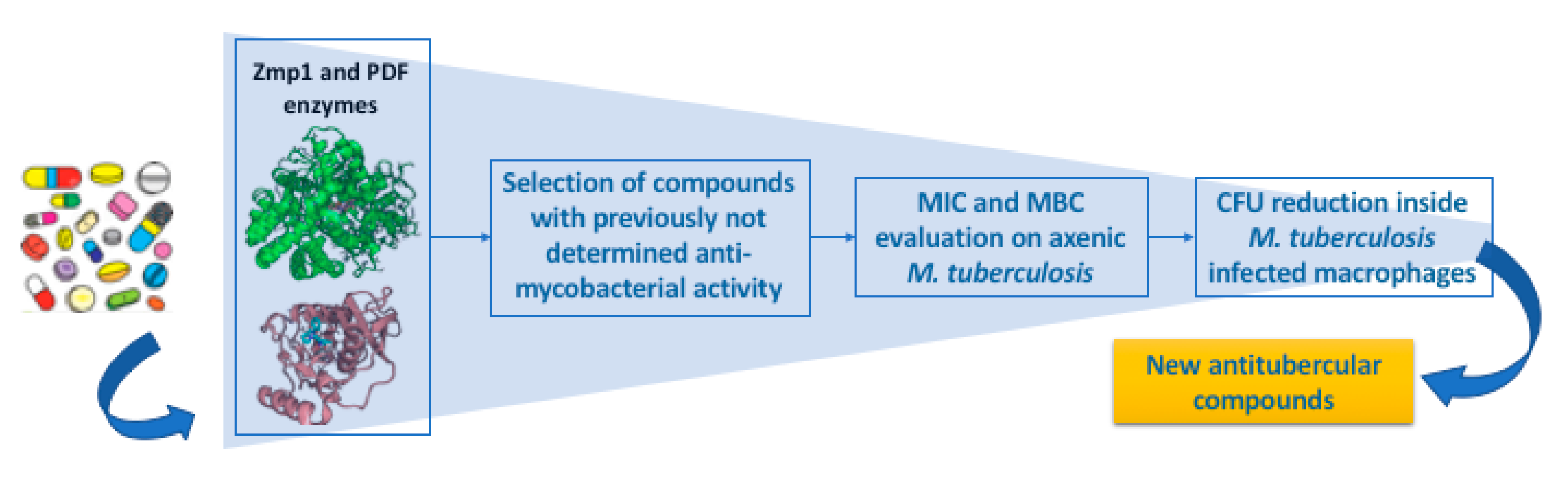
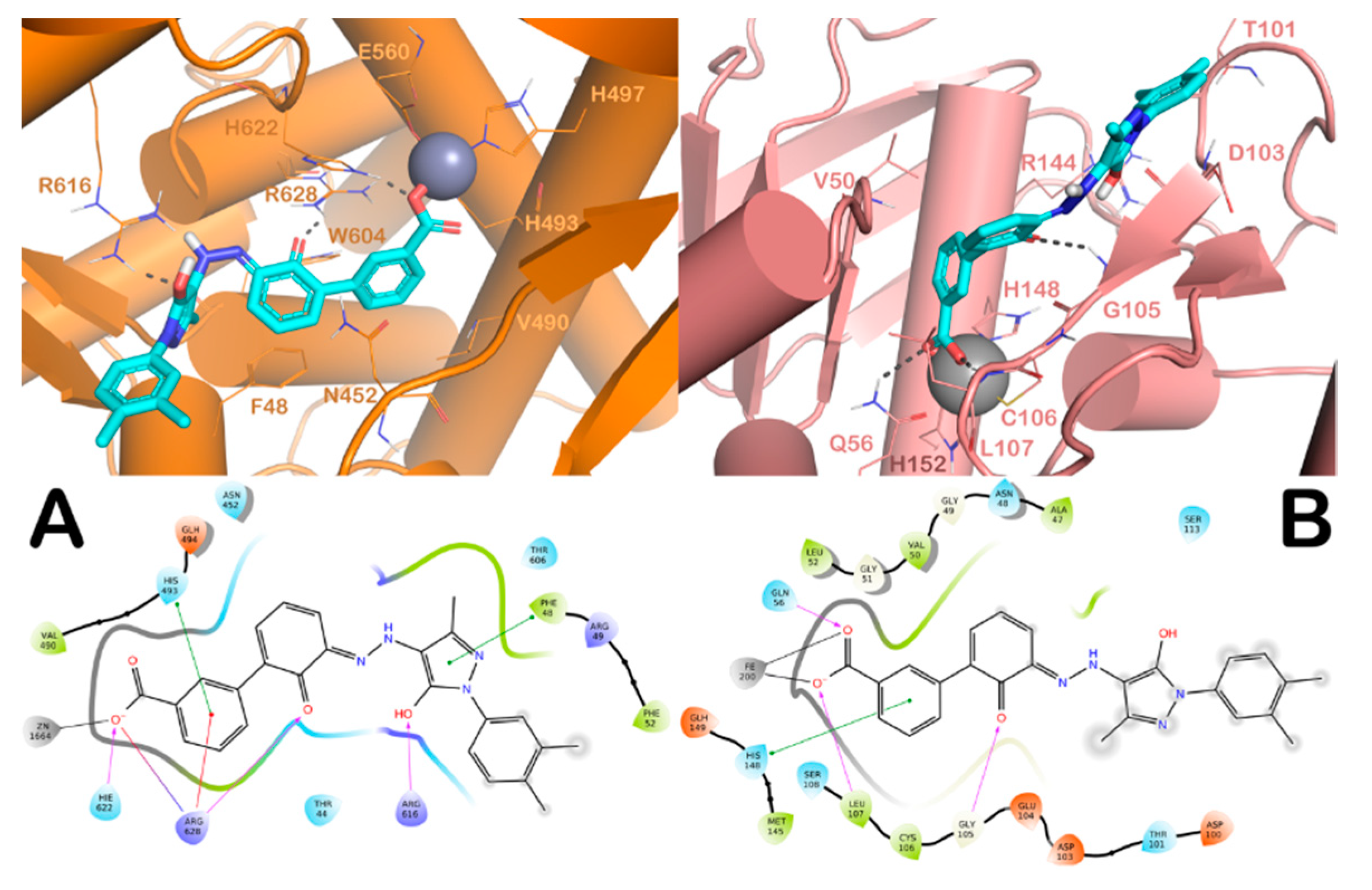
2.1. In Silico Screening and Antimycobacterial Activity of the Selected Compounds under Axenic Conditions
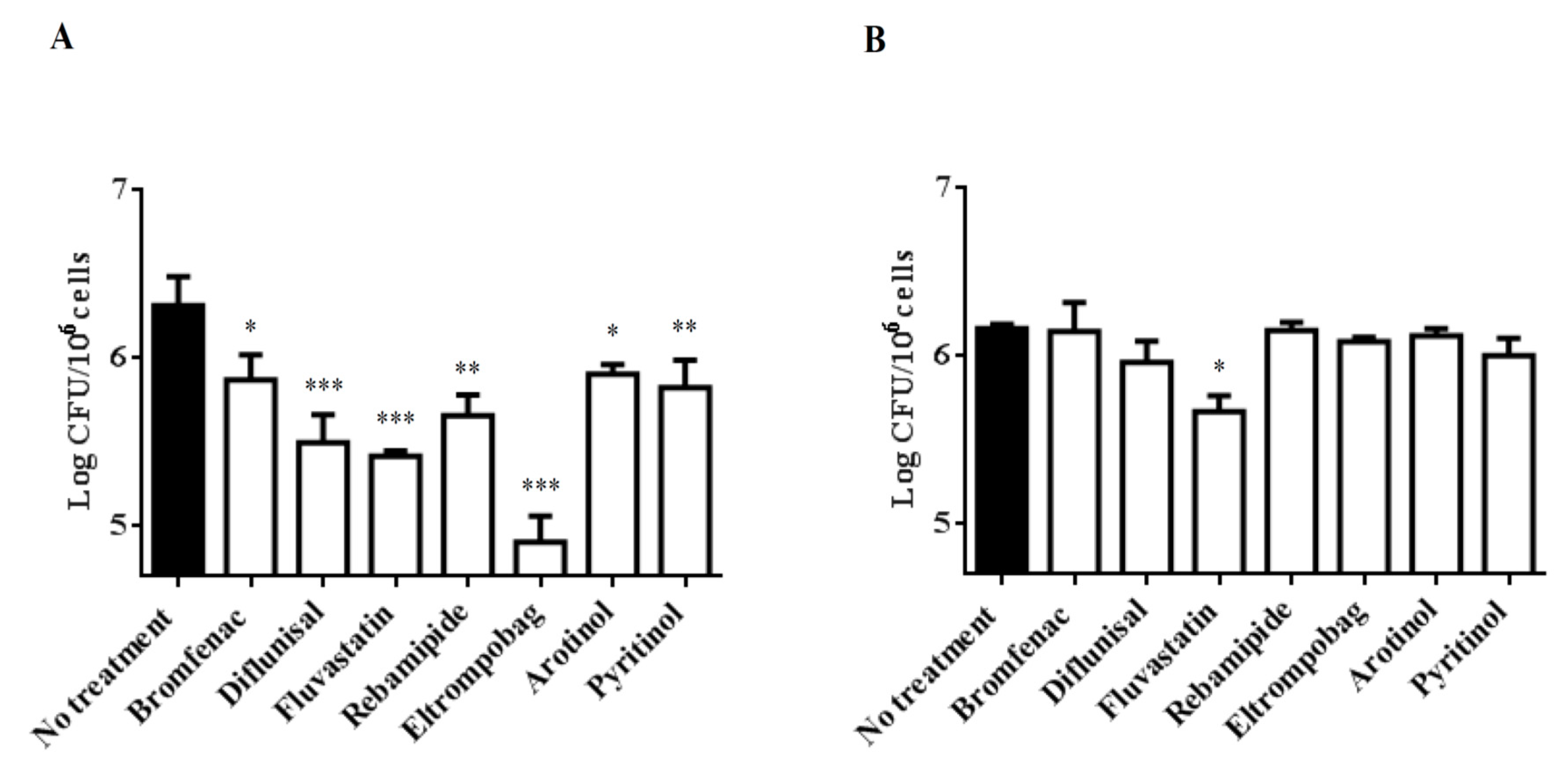
2.2. Treatment of Mtb-Infected PBMCs with Selected Drugs
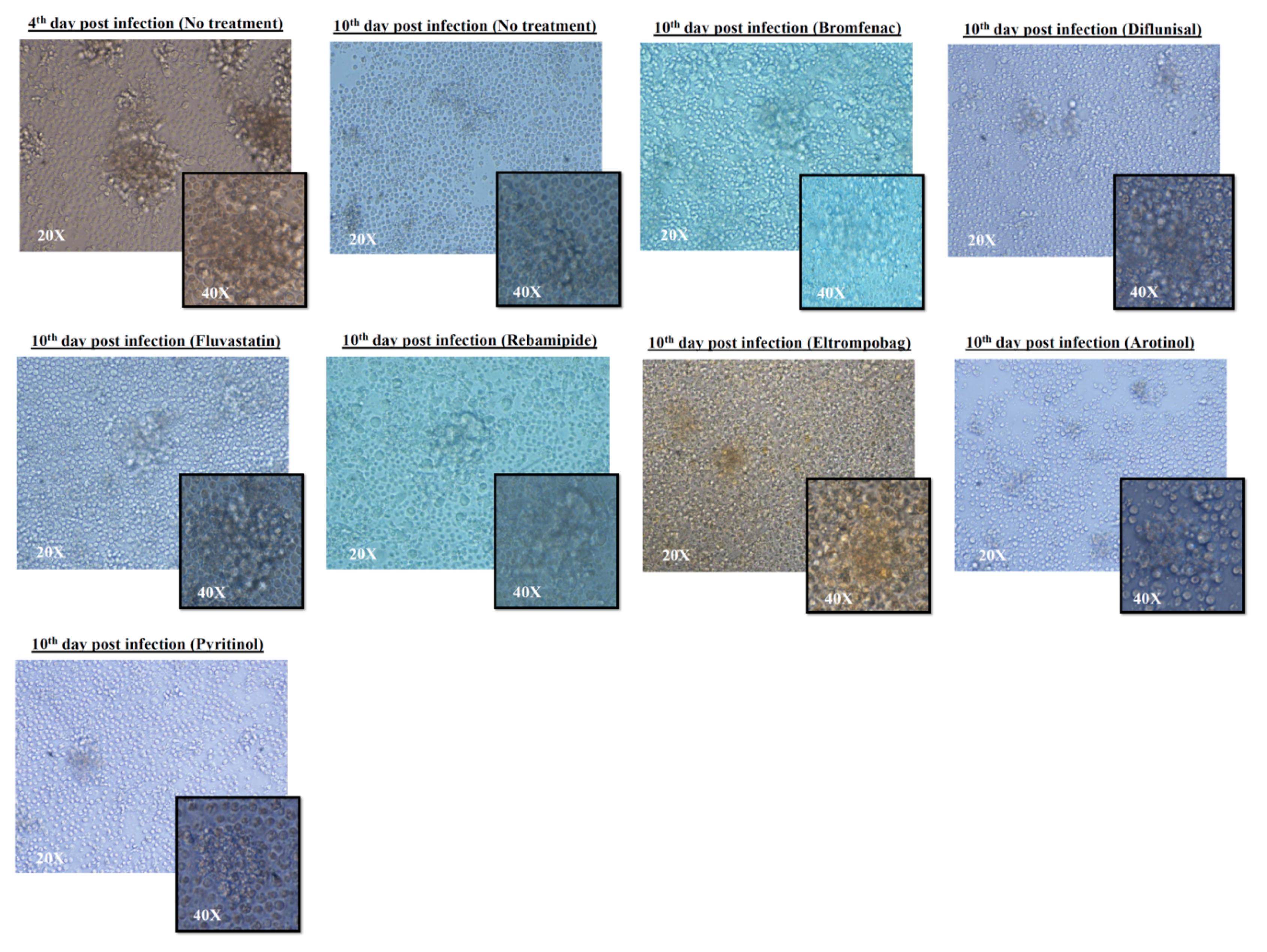
2.3. Granuloma-Like Structure (GLS) Formation before and after Treatment of Infected PBMCs
3. Discussion
4. Materials and Methods
4.1. In Silico Screening
4.1.1. Protein Preparation
4.1.2. Database Preparation
4.1.3. High-Throughput Docking (HTD) Details
4.2. Measurement of the Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC) of All Drugs
4.3. PBMCs Isolation from a Human Blood
4.4. Infection of the PBMCs Extracted Cells
4.5. Image Acquisition for Granuloma-Like Structure (GLS)
4.6. Statistics
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- World Health Organization. Global Tuberculosis Report 2018; WHO: Geneva, Switzerland, 2019. [Google Scholar]
- Dobbs, T.E.; Webb, R.M. Chemotherapy of Tuberculosis. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef] [PubMed]
- Dheda, K.; Gumbo, T.; Maartens, G.; Dooley, K.E.; Murray, M.; Furin, J.; Nardell, E.A.; Warren, R.M. Lancet Respiratory Medicine drug-resistant tuberculosis Commission, g., The Lancet Respiratory Medicine Commission: 2019 update: Epidemiology, pathogenesis, transmission, diagnosis, and management of multidrug-resistant and incurable tuberculosis. Lancet Respir. Med. 2019, 7, 820–826. [Google Scholar] [CrossRef]
- Dheda, K.; Gumbo, T.; Maartens, G.; Dooley, K.E.; McNerney, R.; Murray, M.; Furin, J.; Nardell, E.A.; London, L.; Lessem, E.; et al. The epidemiology, pathogenesis, transmission, diagnosis, and management of multidrug-resistant, extensively drug-resistant, and incurable tuberculosis. Lancet Respir. Med. 2017, 5, 291–360. [Google Scholar] [CrossRef]
- Siddiqi, S.; Takhar, P.; Baldeviano, C.; Glover, W.; Zhang, Y. Isoniazid induces its own resistance in nonreplicating Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2007, 51, 2100–2104. [Google Scholar] [CrossRef]
- Raghunandanan, S.; Jose, L.; Kumar, R.A. Dormant Mycobacterium tuberculosis converts isoniazid to the active drug in a Wayne’s model of dormancy. J. Antibiot. (Tokyo) 2018, 71, 939–949. [Google Scholar] [CrossRef]
- Smith, T.; Wolff, K.A.; Nguyen, L. Molecular biology of drug resistance in Mycobacterium tuberculosis. Curr. Top. Microbiol. Immunol. 2013, 374, 53–80. [Google Scholar]
- Torfs, E.; Piller, T.; Cos, P.; Cappoen, D. Opportunities for Overcoming Mycobacterium tuberculosis Drug Resistance: Emerging Mycobacterial Targets and Host-Directed Therapy. Int. J. Mol. Sci. 2019, 20, 2868. [Google Scholar] [CrossRef]
- Oldfield, E.; Feng, X. Resistance-resistant antibiotics. Trends Pharmacol. Sci. 2014, 35, 664–674. [Google Scholar] [CrossRef]
- De Oliveira Viana, J.; Ishiki, H.M.; Scotti, M.T.; Scotti, L. Multi-Target Antitubercular Drugs. Curr. Top. Med. Chem. 2018, 18, 750–758. [Google Scholar] [CrossRef]
- Li, K.; Schurig-Briccio, L.A.; Feng, X.; Upadhyay, A.; Pujari, V.; Lechartier, B.; Fontes, F.L.; Yang, H.; Rao, G.; Zhu, W.; et al. Multitarget drug discovery for tuberculosis and other infectious diseases. J. Med. Chem. 2014, 57, 3126–3139. [Google Scholar] [CrossRef]
- Fong, W.; To, K.K.W. Drug repurposing to overcome resistance to various therapies for colorectal cancer. Cell. Mol. Life Sci. 2019, 76, 3383–3406. [Google Scholar] [CrossRef] [PubMed]
- Rommer, P.S.; Sellner, J. Repurposing multiple sclerosis drugs: A review of studies in neurological and psychiatric conditions. Drug Discov. Today 2019, 24, 1398–1404. [Google Scholar] [CrossRef] [PubMed]
- Cha, Y.; Erez, T.; Reynolds, I.J.; Kumar, D.; Ross, J.; Koytiger, G.; Kusko, R.; Zeskind, B.; Risso, S.; Kagan, E.; et al. Drug repurposing from the perspective of pharmaceutical companies. Br. J. Pharmacol. 2018, 175, 168–180. [Google Scholar] [CrossRef]
- Zheng, W.; Sun, W.; Simeonov, A. Drug repurposing screens and synergistic drug-combinations for infectious diseases. Br. J. Pharmacol. 2018, 175, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Paolino, M.; Brindisi, M.; Vallone, A.; Butini, S.; Campiani, G.; Nannicini, C.; Giuliani, G.; Anzini, M.; Lamponi, S.; Giorgi, G.; et al. Development of Potent Inhibitors of the Mycobacterium tuberculosis Virulence Factor Zmp1 and Evaluation of Their Effect on Mycobacterial Survival inside Macrophages. ChemMedChem 2018, 13, 422–430. [Google Scholar] [CrossRef] [PubMed]
- Teo, J.W.; Thayalan, P.; Beer, D.; Yap, A.S.; Nanjundappa, M.; Ngew, X.; Duraiswamy, J.; Liung, S.; Dartois, V.; Schreiber, M.; et al. Peptide deformylase inhibitors as potent antimycobacterial agents. Antimicrob. Agents Chemother. 2006, 50, 3665–3673. [Google Scholar] [CrossRef] [PubMed]
- Pichota, A.; Duraiswamy, J.; Yin, Z.; Keller, T.H.; Alam, J.; Liung, S.; Lee, G.; Ding, M.; Wang, G.; Chan, W.L.; et al. Peptide deformylase inhibitors of Mycobacterium tuberculosis: Synthesis, structural investigations, and biological results. Bioorg. Med. Chem. Lett. 2008, 18, 6568–6572. [Google Scholar] [CrossRef]
- Ferraris, D.M.; Sbardella, D.; Petrera, A.; Marini, S.; Amstutz, B.; Coletta, M.; Sander, P.; Rizzi, M. Crystal structure of Mycobacterium tuberculosis zinc-dependent metalloprotease-1 (Zmp1), a metalloprotease involved in pathogenicity. J. Biol. Chem. 2011, 286, 32475–32482. [Google Scholar] [CrossRef]
- Lazarevic, V.; Martinon, F. Linking inflammasome activation and phagosome maturation. Cell. Host Microbe 2008, 3, 199–200. [Google Scholar] [CrossRef]
- Master, S.S.; Rampini, S.K.; Davis, A.S.; Keller, C.; Ehlers, S.; Springer, B.; Timmins, G.S.; Sander, P.; Deretic, V. Mycobacterium tuberculosis prevents inflammasome activation. Cell Host Microbe 2008, 3, 224–232. [Google Scholar] [CrossRef]
- Sharma, A.; Khuller, G.K.; Sharma, S. Peptide Deformylase—A promising therapeutic target for tuberculosis and antibacterial drug discovery. Expert Opin. Ther. Targets 2009, 13, 753–765. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Khuller, G.K.; Kanwar, A.J.; Sharma, S. Therapeutic potential of peptide deformylase inhibitors against experimental tuberculosis. J. Infect. 2010, 60, 498–501. [Google Scholar] [CrossRef] [PubMed]
- Barbier, M.; Wirth, T. The Evolutionary History, Demography, and Spread of the Mycobacterium tuberculosis Complex. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef]
- Romagnoli, A.; Petruccioli, E.; Palucci, I.; Camassa, S.; Carata, E.; Petrone, L.; Mariano, S.; Sali, M.; Dini, L.; Girardi, E.; et al. Clinical isolates of the modern Mycobacterium tuberculosis lineage 4 evade host defense in human macrophages through eluding IL-1β-induced autophagy. Cell Death Dis. 2018, 9, 624. [Google Scholar] [CrossRef]
- Bhavanam, S.; Rayat, G.R.; Keelan, M.; Kunimoto, D.; Drews, S.J. Characterization of immune responses of human PBMCs infected with Mycobacterium tuberculosis H37Ra: Impact of donor declared BCG vaccination history on immune responses and M. tuberculosis growth. PLoS ONE 2018, 13, e0203822. [Google Scholar] [CrossRef]
- Je, S.; Quan, H.; Na, Y.; Cho, S.N.; Kim, B.J.; Seok, S.H. An in vitro model of granuloma-like cell aggregates substantiates early host immune responses against Mycobacterium massiliense infection. Biol. Open 2016, 5, 1118–1127. [Google Scholar] [CrossRef]
- Cox, H.S.; Kubica, T.; Doshetov, D.; Kebede, Y.; Rusch-Gerdess, S.; Niemann, S. The Beijing genotype and drug resistant tuberculosis in the Aral Sea region of Central Asia. Respir. Res. 2005, 6, 134. [Google Scholar] [CrossRef]
- Bard, J.A.M.; Goodall, E.A.; Greene, E.R.; Jonsson, E.; Dong, K.C.; Martin, A. Structure and Function of the 26S Proteasome. Annu. Rev. Biochem. 2018, 87, 697–724. [Google Scholar] [CrossRef]
- Palucci, I.; Delogu, G. Host Directed Therapies for Tuberculosis: Futures Strategies for an Ancient Disease. Chemotherapy 2018, 63, 172–180. [Google Scholar] [CrossRef]
- Parihar, S.P.; Guler, R.; Khutlang, R.; Lang, D.M.; Hurdayal, R.; Mhlanga, M.M.; Suzuki, H.; Marais, A.D.; Brombacher, F. Statin therapy reduces the mycobacterium tuberculosis burden in human macrophages and in mice by enhancing autophagy and phagosome maturation. J. Infect. Dis. 2014, 209, 754–763. [Google Scholar] [CrossRef]
- Schrödinger. Maestro; Version 10.3; Schrödinger, LLC: New York, NY, USA, 2015. [Google Scholar]
- Zaccagnini, L.; Brogi, S.; Brindisi, M.; Gemma, S.; Chemi, G.; Legname, G.; Campiani, G.; Butini, S. Identification of novel fluorescent probes preventing PrPSc replication in prion diseases. Eur. J. Med. Chem. 2017, 127, 859–873. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger. MacroModel; Version 10.9; Schrödinger, LLC: New York, NY, USA, 2015. [Google Scholar]
- Schrödinger. LigPrep; Version 3.3; Schrödinger, LLC: New York, NY, USA, 2015. [Google Scholar]
- Brindisi, M.; Brogi, S.; Giovani, S.; Gemma, S.; Lamponi, S.; De Luca, F.; Novellino, E.; Campiani, G.; Docquier, J.D.; Butini, S. Targeting clinically-relevant metallo-beta-lactamases: From high-throughput docking to broad-spectrum inhibitors. J. Enzyme Inhib. Med. Chem. 2016, 31 (Suppl. S1), 98–109. [Google Scholar] [CrossRef] [PubMed]
- Brindisi, M.; Brogi, S.; Relitti, N.; Vallone, A.; Butini, S.; Gemma, S.; Novellino, E.; Colotti, G.; Angiulli, G.; Di Chiaro, F.; et al. Structure-based discovery of the first non-covalent inhibitors of Leishmania major tryparedoxin peroxidase by high throughput docking. Sci. Rep. 2015, 5, 9705. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Maxwell, D.S.; TiradoRives, J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 1996, 118, 11225–11236. [Google Scholar] [CrossRef]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef]
- Schrödinger. Prime; Version 3.9; Schrödinger, LLC: New York, NY, USA, 2015. [Google Scholar]
- Brindisi, M.; Gemma, S.; Kunjir, S.; Di Cerbo, L.; Brogi, S.; Parapini, S.; D’Alessandro, S.; Taramelli, D.; Habluetzel, A.; Tapanelli, S.; et al. Synthetic spirocyclic endoperoxides: New antimalarial scaffolds. Medchemcomm 2015, 6, 357–362. [Google Scholar] [CrossRef][Green Version]
- Brogi, S.; Fiorillo, A.; Chemi, G.; Butini, S.; Lalle, M.; Ilari, A.; Gemma, S.; Campiani, G. Structural characterization of Giardia duodenalis thioredoxin reductase (gTrxR) and computational analysis of its interaction with NBDHEX. Eur. J. Med. Chem. 2017, 135, 479–490. [Google Scholar] [CrossRef]
- D’Alessandro, S.; Alfano, G.; Di Cerbo, L.; Brogi, S.; Chemi, G.; Relitti, N.; Brindisi, M.; Lamponi, S.; Novellino, E.; Campiani, G.; et al. Bridged bicyclic 2,3-dioxabicyclo[3.3.1]nonanes as antiplasmodial agents: Synthesis, structure-activity relationships and studies on their biomimetic reaction with Fe(II). Bioorg. Chem. 2019, 89, 103020. [Google Scholar] [CrossRef]
- Delogu, G.; Pusceddu, C.; Bua, A.; Fadda, G.; Brennan, M.J.; Zanetti, S. Rv1818c-encoded PE_PGRS protein of Mycobacterium tuberculosis is surface exposed and influences bacterial cell structure. Mol. Microbiol. 2004, 52, 725–733. [Google Scholar] [CrossRef]
- De Maio, F.; Maulucci, G.; Minerva, M.; Anoosheh, S.; Palucci, I.; Iantomasi, R.; Palmieri, V.; Camassa, S.; Sali, M.; Sanguinetti, M.; et al. Impact of protein domains on PE_PGRS30 polar localization in Mycobacteria. PLoS ONE 2014, 9, e112482. [Google Scholar] [CrossRef]
- De Maio, F.; Battah, B.; Palmieri, V.; Petrone, L.; Corrente, F.; Salustri, A.; Palucci, I.; Bellesi, S.; Papi, M.; Rubino, S.; et al. PE_PGRS3 of Mycobacterium tuberculosis is specifically expressed at low phosphate concentration, and its arginine-rich C-terminal domain mediates adhesion and persistence in host tissues when expressed in Mycobacterium smegmatis. Cell Microbiol. 2018, 20, e12952. [Google Scholar] [CrossRef] [PubMed]
- Straniero, V.; Pallavicini, M.; Chiodini, G.; Zanotto, C.; Volonte, L.; Radaelli, A.; Bolchi, C.; Fumagalli, L.; Sanguinetti, M.; Menchinelli, G.; et al. 3-(Benzodioxan-2-ylmethoxy)-2,6-difluorobenzamides bearing hydrophobic substituents at the 7-position of the benzodioxane nucleus potently inhibit methicillin-resistant Sa and Mtb cell division. Eur. J. Med. Chem. 2016, 120, 227–243. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, N.; Pawar, S.; Sirakova, T.D.; Deb, C.; Warren, W.L.; Kolattukudy, P.E. Human granuloma in vitro model, for TB dormancy and resuscitation. PLoS ONE 2013, 8, e53657. [Google Scholar] [CrossRef] [PubMed]
- Palucci, I.; Battah, B.; Salustri, A.; De Maio, F.; Petrone, L.; Ciccosanti, F.; Sali, M.; Bondet, V.; Duffy, D.; Fimia, G.M.; et al. IP-10 contributes to the inhibition of mycobacterial growth in an ex vivo whole blood assay. Int. J. Med. Microbiol. 2019, 309, 299–306. [Google Scholar] [CrossRef]
- Birkness, K.A.; Guarner, J.; Sable, S.B.; Tripp, R.A.; Kellar, K.L.; Bartlett, J.; Quinn, F.D. An in vitro model of the leukocyte interactions associated with granuloma formation in Mycobacterium tuberculosis infection. Immunol. Cell Biol. 2007, 85, 160–168. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds reported in Table 1 are not available from the authors. |
| Drug | MIC μM (H37Rv) | MBC μM (H37Rv) | MIC μM (H3) | MBC μM (H3) | MIC μM (Beijing) | MBC μM (Beijing) |
|---|---|---|---|---|---|---|
| Bromfenac | 100 | >100 | 100 | >100 | 50 | 100 |
| Naratriptan | >100 | n.d. | >100 | n.d. | >100 | n.d. |
| Sofalcone | 100 | >100 | 50 | 100 | 50 | 100 |
| Fosinopril | 50 | 100 | >100 | n.d. | 100 | >100 |
| Carfilzomib | 1.56 | 3.125 | * | n.d. | * | n.d. |
| Pemetrexed | 25 | 50 | >100 | n.d. | >100 | n.d. |
| Sitagliptin | >100 | n.d. | >100 | n.d. | >100 | n.d. |
| Tamsulosin | >100 | n.d. | >100 | n.d. | >100 | n.d. |
| Nelarabine | 50 | 100 | 50 | 100 | >100 | n.d. |
| Pazopanib | >100 | n.d. | >100 | n.d. | >100 | n.d. |
| Fluvastatin | 25 | 50 | 50 | 100 | 25 | 50 |
| Pyritinol | 50 | 100 | >100 | n.d. | 50 | 100 |
| Ledipasvir | >100 | n.d. | 12.5 | 50 | >100 | n.d. |
| Cellcept | 12.5 | 25 | 50 | 100 | 50 | 100 |
| Biotin | >100 | n.d. | >100 | n.d. | >100 | n.d. |
| Picosulfuric acid | 25 | 50 | >100 | n.d. | >100 | n.d. |
| Peramivir | 100 | >100 | >100 | n.d. | >100 | n.d. |
| Rebamipide | 100 | >100 | >100 | n.d. | 100 | >100 |
| Pitavastatin | 50 | 100 | >100 | n.d. | 100 | >100 |
| Eltrombopag | 6.25 | 12.5 | 50 | 100 | 12.5 | 25 |
| Nintedanib | 25 | 50 | 25 | 50 | 12.5 | 25 |
| Mitiglinide | 25 | 50 | >100 | n.d. | 50 | 100 |
| Bicalutamide | >100 | n.d. | >100 | n.d. | >100 | n.d. |
| Arotinolol | 3.125 | 6.25 | 50 | 100 | 25 | 50 |
| Diflunisal | 50 | 100 | 50 | 100 | 25 | 50 |
| Amifostine | 50 | 100 | 100 | >100 | >100 | n.d. |
| Fosamax | 12.5 | 25 | >100 | n.d. | >100 | n.d. |
| Cidofovir | >100 | n.d. | >100 | n.d. | >100 | n.d. |
| Ranelic acid | >100 | n.d. | >100 | n.d. | >100 | n.d. |
| 1.25–12.5 μM | |
| 25–50 μM | |
| 100 μM | |
| >100 μM | |
| n.d. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Battah, B.; Chemi, G.; Butini, S.; Campiani, G.; Brogi, S.; Delogu, G.; Gemma, S. A Repurposing Approach for Uncovering the Anti-Tubercular Activity of FDA-Approved Drugs with Potential Multi-Targeting Profiles. Molecules 2019, 24, 4373. https://doi.org/10.3390/molecules24234373
Battah B, Chemi G, Butini S, Campiani G, Brogi S, Delogu G, Gemma S. A Repurposing Approach for Uncovering the Anti-Tubercular Activity of FDA-Approved Drugs with Potential Multi-Targeting Profiles. Molecules. 2019; 24(23):4373. https://doi.org/10.3390/molecules24234373
Chicago/Turabian StyleBattah, Basem, Giulia Chemi, Stefania Butini, Giuseppe Campiani, Simone Brogi, Giovanni Delogu, and Sandra Gemma. 2019. "A Repurposing Approach for Uncovering the Anti-Tubercular Activity of FDA-Approved Drugs with Potential Multi-Targeting Profiles" Molecules 24, no. 23: 4373. https://doi.org/10.3390/molecules24234373
APA StyleBattah, B., Chemi, G., Butini, S., Campiani, G., Brogi, S., Delogu, G., & Gemma, S. (2019). A Repurposing Approach for Uncovering the Anti-Tubercular Activity of FDA-Approved Drugs with Potential Multi-Targeting Profiles. Molecules, 24(23), 4373. https://doi.org/10.3390/molecules24234373









