Prediction of Severity of Drug-Drug Interactions Caused by Enzyme Inhibition and Activation
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. OpeRational ClassificAtion (ORCA) System
4.2. Training Set Creation
4.3. PASS
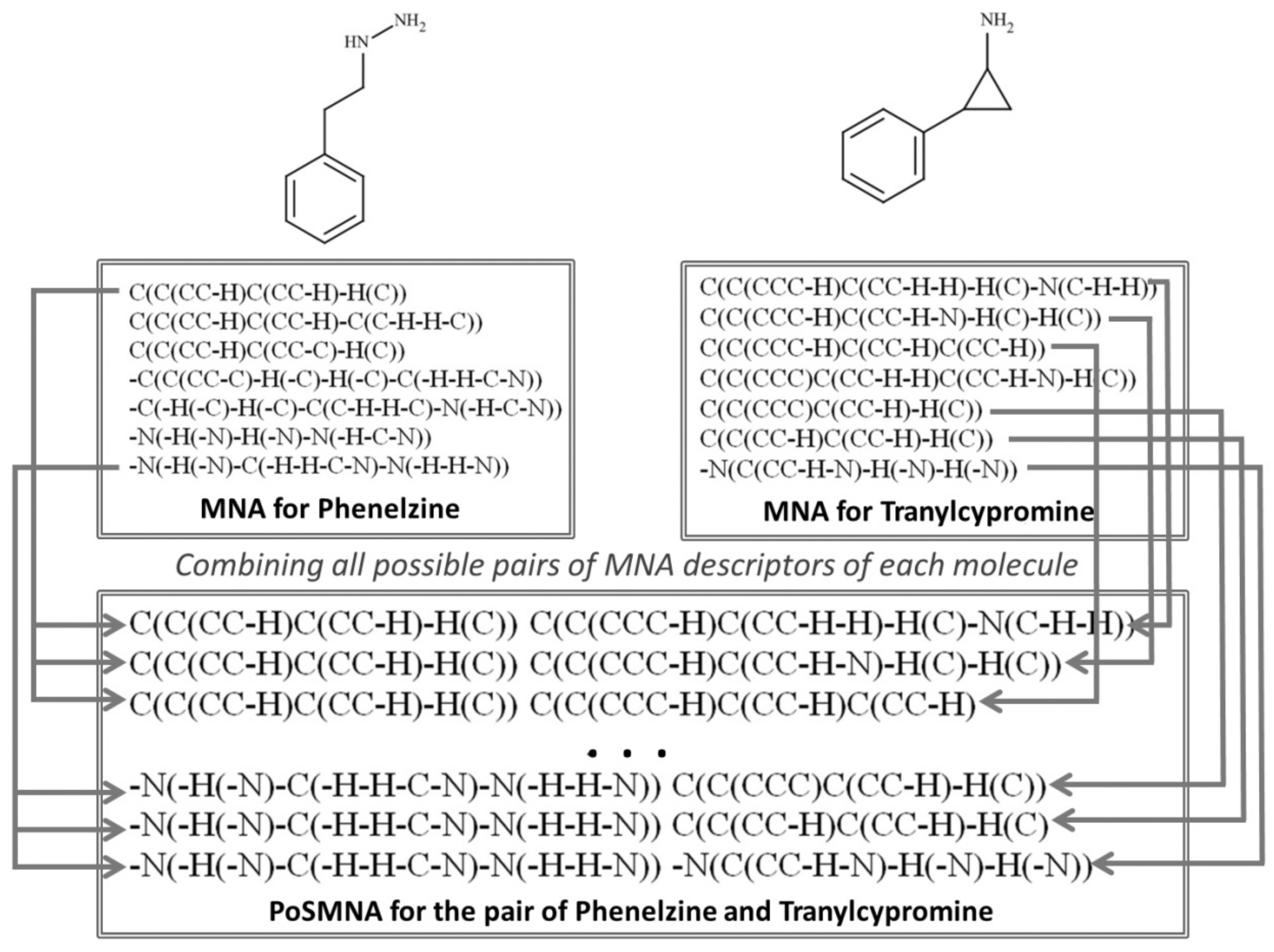
4.4. Pairs of Substances Multilevel Neighbourhoods of Atoms Descriptors
Author Contributions
Funding
Conflicts of Interest
References
- Lyubimov, A.V. Encyclopedia of Drug Metabolism and Interactions; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2012. [Google Scholar]
- Tornio, A.; Filppula, A.M.; Niemi, M.; Backman, J.T. Clinical Studies on Drug–Drug Interactions Involving Metabolism and Transport: Methodology, Pitfalls, and Interpretation. Clin. Pharmacol. Ther. 2019, 105, 1345–1361. [Google Scholar] [CrossRef] [PubMed]
- Dmitriev, A.V.; Lagunin, A.A.; Karasev, D.A.; Rudik, A.V.; Pogodin, P.V.; Filimonov, D.A.; Poroikov, V.V. Prediction of Drug-Drug Interactions Related to Inhibition or Induction of Drug-Metabolizing Enzymes. Curr. Top. Med. Chem. 2019, 19, 319–336. [Google Scholar] [CrossRef] [PubMed]
- Alfaro, C.L. Emerging role of drug interaction studies in drug development: The good, the bad, and the unknown. Psychopharmacol. Bull. 2001, 35, 80–93. [Google Scholar] [PubMed]
- Hochleitner, J.; Akram, M.; Ueberall, M.; Davis, R.A.; Waltenberger, B.; Stuppner, H.; Sturm, S.; Ueberall, F.; Gostner, J.M.; Schuster, D. A combinatorial approach for the discovery of cytochrome P450 2D6 inhibitors from nature. Sci. Rep. 2017, 7, 8071. [Google Scholar] [CrossRef] [PubMed]
- Kaserer, T.; Höferl, M.; Müller, K.; Elmer, S.; Ganzera, M.; Jäger, W.; Schuster, D. In Silico Predictions of Drug—Drug Interactions Caused by CYP1A2, 2C9 and 3A4 Inhibition—A Comparative Study of Virtual Screening Performance. Mol. Inform. 2015, 34, 431–457. [Google Scholar] [CrossRef] [PubMed]
- Torimoto-Katori, N.; Huang, R.; Kato, H.; Ohashi, R.; Xia, M. In Silico Prediction of hPXR Activators Using Structure-Based Pharmacophore Modeling. J. Pharm. Sci. 2017, 106, 1752–1759. [Google Scholar] [CrossRef]
- Duke, J.D.; Han, X.; Wang, Z.; Subhadarshini, A.; Karnik, S.D.; Li, X.; Hall, S.D.; Jin, Y.; Callaghan, J.T.; Overhage, M.J.; et al. Literature based drug interaction prediction with clinical assessment using electronic medical records: Novel myopathy associated drug interactions. PLoS Comput. Biol. 2012, 8, e1002614. [Google Scholar] [CrossRef]
- Takeda, T.; Hao, M.; Cheng, T.; Bryant, S.H.; Wang, Y. Predicting drug-drug interactions through drug structural similarities and interaction networks incorporating pharmacokinetics and pharmacodynamics knowledge. J. Cheminform. 2017, 9, 16. [Google Scholar] [CrossRef]
- Vilar, S.; Uriarte, E.; Santana, L.; Lorberbaum, T.; Hripcsak, G.; Friedman, C.; Tatonetti, N.P. Similarity-based modeling in large-scale prediction of drug-drug interactions. Nat. Protoc. 2014, 9, 2147–2163. [Google Scholar] [CrossRef]
- Cheng, F.; Zhao, Z. Machine learning-based prediction of drug-drug interactions by integrating drug phenotypic, therapeutic, chemical, and genomic properties. J. Am. Med. Inform. Assoc. 2014, 21, e278–e286. [Google Scholar] [CrossRef]
- Zhang, P.; Wang, F.; Hu, J.; Sorrentino, R. Label Propagation Prediction of Drug-Drug Interactions Based on Clinical Side Effects. Sci. Rep. 2015, 5, 12339. [Google Scholar] [CrossRef] [PubMed]
- Ferdousi, R.; Safdari, R.; Omidi, Y. Computational prediction of drug-drug interactions based on drugs functional similarities. J. Biomed. Inform. 2017, 70, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Poroikov, V.V.; Filimonov, D.A.; Borodina, Y.V.; Lagunin, A.A.; Kos, A. Robustness of Biological Activity Spectra Predicting by Computer Program PASS for Noncongeneric Sets of Chemical Compounds. J. Chem. Inf. Comput. Sci. 2000, 40, 1349–1355. [Google Scholar] [CrossRef]
- Rudik, A.V.; Dmitriev, A.V.; Lagunin, A.A.; Filimonov, D.A.; Poroikov, V.V. Prediction of Reacting Atoms for the Major Biotransformation Reactions of Organic Xenobiotics. J. Cheminform. 2016, 8, 68. [Google Scholar] [CrossRef]
- Rudik, A.; Dmitriev, A.; Lagunin, A.; Filimonov, D.; Poroikov, V. SOMP: Web Server for in Silico Prediction of Sites of Metabolism for Drug-like Compounds. Bioinformatics 2015, 31, 2046–2048. [Google Scholar] [CrossRef]
- Zakharov, A.V.; Varlamova, E.V.; Lagunin, A.A.; Dmitriev, A.V.; Muratov, E.N.; Fourches, D.; Kuz’min, V.E.; Poroikov, V.V.; Tropsha, A.; Nicklaus, M.C. QSAR Modeling and Prediction of Drug-Drug Interactions. Mol. Pharm. 2016, 13, 545–556. [Google Scholar] [CrossRef]
- Dmitriev, A.V.; Filimonov, D.A.; Rudik, A.V.; Pogodin, P.V.; Karasev, D.A.; Lagunin, A.A.; Poroikov, V.V. Drug-drug interaction prediction using PASS. SAR QSAR Environ. Res. 2019, 30, 655–664. [Google Scholar] [CrossRef]
- Masamrekh, R.A.; Kuzikov, A.V.; Haurychenka, Y.I.; Shcherbakov, K.A.; Veselovsky, A.V.; Filimonov, D.A.; Dmitriev, A.V.; Zavialova, M.G.; Gilep, A.A.; Shkel, T.V.; et al. In vitro interactions of abiraterone, erythromycin and CYP3A4: Implications for drug-drug interactions. Fundam. Clin. Pharmacol. 2019. [Google Scholar] [CrossRef]
- Quinn, K.J.; Shah, N.H. A dataset quantifying polypharmacy in the United States. Sci. Data 2017, 4, 170167. [Google Scholar] [CrossRef]
- Fulton, M.M.; Allen, E.R. Polypharmacy in the elderly: A literature review. J. Am. Acad. Nurse Pract. 2005, 17, 123–132. [Google Scholar] [CrossRef]
- Hansten, P.D.; Horn, J.R.; Hazlet, T.K. ORCA: OpeRational ClassificAtion of Drug Interactions. J. Am. Pharm. Assoc. (Wash.) 2001, 41, 161–165. [Google Scholar] [CrossRef]
- Hansten, P.D.; Horn, J.R. Drug Interaction Analysis and Management 2013; Wolters Kluwer Health: St. Louis, MO, USA, 2013. [Google Scholar]
Sample Availability: Not apply. |
| DDIs Class | N | DDIs IAP | DDIs IAP 20-Fold |
|---|---|---|---|
| Class 1 | 59 | 0.876 | 0.876 |
| Class 2 | 236 | 0.788 | 0.777 |
| Class 3 | 1139 | 0.683 | 0.679 |
| Class 4 | 523 | 0.671 | 0.668 |
| Class 5 | 133 | 0.752 | 0.754 |
| Average | 0.754 | 0.751 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dmitriev, A.; Filimonov, D.; Lagunin, A.; Karasev, D.; Pogodin, P.; Rudik, A.; Poroikov, V. Prediction of Severity of Drug-Drug Interactions Caused by Enzyme Inhibition and Activation. Molecules 2019, 24, 3955. https://doi.org/10.3390/molecules24213955
Dmitriev A, Filimonov D, Lagunin A, Karasev D, Pogodin P, Rudik A, Poroikov V. Prediction of Severity of Drug-Drug Interactions Caused by Enzyme Inhibition and Activation. Molecules. 2019; 24(21):3955. https://doi.org/10.3390/molecules24213955
Chicago/Turabian StyleDmitriev, Alexander, Dmitry Filimonov, Alexey Lagunin, Dmitry Karasev, Pavel Pogodin, Anastasiya Rudik, and Vladimir Poroikov. 2019. "Prediction of Severity of Drug-Drug Interactions Caused by Enzyme Inhibition and Activation" Molecules 24, no. 21: 3955. https://doi.org/10.3390/molecules24213955
APA StyleDmitriev, A., Filimonov, D., Lagunin, A., Karasev, D., Pogodin, P., Rudik, A., & Poroikov, V. (2019). Prediction of Severity of Drug-Drug Interactions Caused by Enzyme Inhibition and Activation. Molecules, 24(21), 3955. https://doi.org/10.3390/molecules24213955







