Investigation of Site-Specific Differences in Glycan Microheterogeneity by N-Glycopeptide Mapping of VEGFR-IgG Fusion Protein
Abstract
1. Introduction
2. Results and Discussion
2.1. Cleaved Glycan Analysis by MALDI MS
2.2. Protein Sequencing by LC-MS/MS
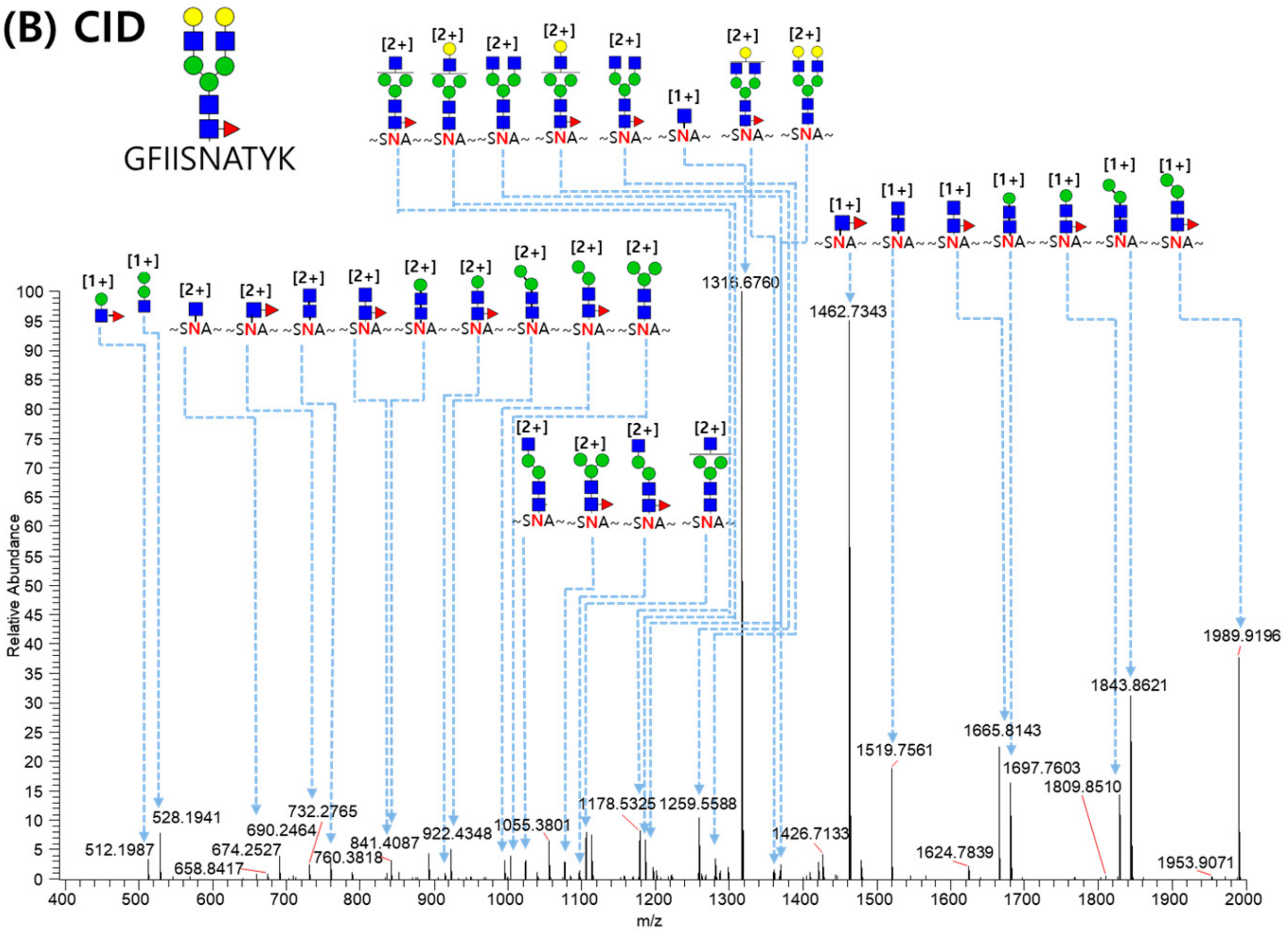
2.3. LC-MS/MS Glycopeptide Mapping of Fusion Protein
3. Materials and Methods
3.1. Materials
3.2. Stable Cell Line Generation of the VEGFR-IgG Fusion Protein
3.3. Stable Expression and Purification of VEGFR-IgG Fusion Protein
3.4. Cleaved Glycans Preparation
3.5. MALDI MS
3.6. Tryptic Digestion of Fusion Protein
3.7. LC-MS/MS Analysis
3.8. N-Glycopeptide Identification
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Aapro, M. Biosimilars in oncology: Current and future perspectives. GaBI J. 2013, 2, 91–93. [Google Scholar] [CrossRef]
- Lee, S.; Ballow, M. Monoclonal antibodies and fusion proteins and their complications: Targeting B cells in autoimmune diseases. J. Allergy Clin. Immun. 2010, 125, 814–820. [Google Scholar] [CrossRef] [PubMed]
- Liu, L. Pharmacokinetics of monoclonal antibodies and Fc-fusion proteins. Protein Cell 2018, 9, 15–32. [Google Scholar] [CrossRef] [PubMed]
- Liu, L. Antibody Glycosylation and Its Impact on the Pharmacokinetics and Pharmacodynamics of Monoclonal Antibodies and Fc-Fusion Proteins. J. Pharm. Sci. 2015, 104, 1866–1884. [Google Scholar] [CrossRef]
- Arnold, J.N.; Wormald, M.R.; Sim, R.B.; Rudd, P.M.; Dwek, R.A. The Impact of Glycosylation on the Biological Function and Structure of Human Immunoglobulins. Ann. Rev. Immunol. 2007, 25, 21–50. [Google Scholar] [CrossRef]
- Higel, F.; Seidl, A.; Sörgel, F.; Friess, W. N-glycosylation heterogeneity and the influence on structure, function and pharmacokinetics of monoclonal antibodies and Fc fusion proteins. Eur. J. Pharm. Biopharm. 2016, 100, 94–100. [Google Scholar] [CrossRef]
- Dolashka, P. Tandem mass spectrometry and glycoproteins. In Tandem Mass Spectrometry-Applications and Principles; Prasain, J., Ed.; Intechopen: London, UK, 2012; Chapter 5; pp. 105–126. [Google Scholar]
- Velkova, L.; Dolashki, A.; Dolashka, P. Carbohydrate structure of molluscan hemocyanins from snails Helix lucorum and Rapana venosa, determined by mass spectrometry. J. BioSci. Biotechnol. 2015, 75–85, SPECIAL EDITION. [Google Scholar]
- An, H.J.; Froehlich, J.W.; Lebrilla, C.B. Determination of glycosylation sites and site-specific heterogeneity in glycoproteins. Curr. Opin. Chem. Biol. 2009, 13, 421–426. [Google Scholar] [CrossRef]
- Ruhaak, L.R.; Zauner, G.; Huhn, C.; Bruggink, C.; Deelder, A.M.; Wuhrer, M. Glycan labeling strategies and their use in identification and quantification. Anal. Bioanal. Chem. 2010, 397, 3457–3481. [Google Scholar] [CrossRef]
- Bongers, J.; Devincentis, J.; Fu, J.; Huang, P.; Kirkley, D.H.; Leister, K.; Liu, P.; Ludwig, R.; Rumney, K.; Tao, L.; et al. Characterization of glycosylation sites for a recombinant IgG1 monoclonal antibody and a CTLA4-Ig fusion protein by liquid chromatography–mass spectrometry peptide mapping. J. Chromatogr. A 2011, 1218, 8140–8149. [Google Scholar] [CrossRef]
- Reusch, D.; Haberger, M.; Falck, D.; Peter, B.; Maier, B.; Gassner, J.; Hook, M.; Wagner, K.; Bonnington, L.; Bulau, P.; et al. Comparison of methods for the analysis of therapeutic immunoglobulin G Fc-glycosylation profiles—Part 2: Mass spectrometric methods. mAbs 2015, 7, 732–742. [Google Scholar] [CrossRef] [PubMed]
- Lynaugh, H.; Li, H.; Gong, B. Rapid Fc glycosylation analysis of Fc fusions with IdeS and liquid chromatography mass spectrometry. mAbs 2013, 5, 641–645. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Guo, Q.; Guo, H.; Liu, T.; Zheng, Y.; Gu, P.; Chen, X.; Wang, H.; Hou, S.; Guo, Y. Versatile characterization of glycosylation modification in CTLA4-Ig fusion proteins by liquid chromatography-mass spectrometry. mAbs 2014, 6, 1474–1485. [Google Scholar] [CrossRef] [PubMed]
- Park, G.W.; Kim, J.Y.; Hwang, H.; Lee, J.Y.; Ahn, Y.H.; Lee, H.K.; Ji, E.S.; Kim, K.H.; Jeong, H.K.; Yun, K.N.; et al. Integrated GlycoProteome Analyzer (I-GPA) for Automated Identification and Quantitation of Site-Specific N-Glycosylation. Sci. Rep. 2016, 6, 21175. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Lee, H.K.; Park, G.W.; Hwang, H.; Jeong, H.K.; Yun, K.N.; Ji, E.S.; Kim, K.H.; Kim, J.S.; Kim, J.W.; et al. Characterization of Site-Specific N-Glycopeptide Isoforms of α-1-Acid Glycoprotein from an Interlaboratory Study Using LC–MS/MS. J. Proteome Res. 2016, 15, 4146–4164. [Google Scholar] [CrossRef] [PubMed]
- Shibuya, M. Vascular endothelial growth factor and its receptor system: Physiological functions in angiogenesis and pathological roles in various diseases. J. Biochem. 2013, 153, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Fischer, C.; Mazzone, M.; Jonckx, B.; Carmeliet, P. FLT1 and its ligands VEGFB and PlGF: Drug targets for anti-angiogenic therapy? Nat. Rev. Cancer 2008, 8, 942–956. [Google Scholar] [CrossRef]
- Iyer, S.; Darley, P.I.; Acharya, K.R. Structural Insights into the Binding of Vascular Endothelial Growth Factor-B by VEGFR-1 D2: Recognition and Specificity. J. Biol. Chem. 2010, 285, 23779–23789. [Google Scholar] [CrossRef]
- Lee, S.H.; Jeong, D.; Han, Y.S.; Baek, M.J. Pivotal role of vascular endothelial growth factor pathway in tumor angiogenesis. Ann. Surg. Treat. Res. 2015, 89, 1–8. [Google Scholar] [CrossRef]
- Bork, K.; Horstkorte, R.; Weidemann, W. Increasing the sialylation of therapeutic glycoproteins: The potential of the sialic acid biosynthetic pathway. J. Pharm. Sci. 2009, 98, 3499–3508. [Google Scholar] [CrossRef]
- Leppänen, V.M.; Prota, A.E.; Jeltsch, M.; Anisimov, A.; Kalkkinen, N.; Strandin, T.; Lankinen, H.; Goldman, A.; Ballmer-Hofer, K.; Alitalo, K. Structural determinants of growth factor binding and specificity by VEGF receptor 2. Proc. Natl. Acad. Sci. USA 2010, 107, 2425–2430. [Google Scholar] [CrossRef] [PubMed]
- Chandler, K.B.; Leon, D.R.; Meyer, R.D.; Rahimi, N.; Costello, C.E. Site-Specific N -Glycosylation of Endothelial Cell Receptor Tyrosine Kinase VEGFR-2. J. Proteome Res. 2017, 16, 677–688. [Google Scholar] [CrossRef] [PubMed]
- Vidarsson, G.; Dekkers, G.; Rispens, T. IgG Subclasses and Allotypes: From Structure to Effector Functions. Front. Immunol. 2014, 5, 520. [Google Scholar] [CrossRef] [PubMed]
- Zauner, G.; Selman, M.H.; Bondt, A.; Rombouts, Y.; Blank, D.; Deelder, A.M.; Wuhrer, M. Glycoproteomic Analysis of Antibodies. Mol. Cell. Proteomics 2013, 12, 856–865. [Google Scholar] [CrossRef] [PubMed]
- Gudelj, I.; Lauc, G.; Pezer, M. Immunoglobulin G glycosylation in aging and diseases. Cell. Immunol. 2018, 333, 65–79. [Google Scholar] [CrossRef]
- Kobata, A. The N-Linked sugar chains of human immunoglobulin G: Their unique pattern, and their functional roles. BBA Gen. Subj. 2008, 1780, 472–478. [Google Scholar] [CrossRef]
- Scanlan, C.N.; Burton, D.R.; Dwek, R.A. Making autoantibodies safe. Proc. Natl. Acad. Sci. USA 2008, 105, 4081–4082. [Google Scholar] [CrossRef]
- Higel, F.; Sandl, T.; Kao, C.Y.; Pechinger, N.; Sörgel, F.; Friess, W.; Wolschin, F.; Seidl, A. N-glycans of complex glycosylated biopharmaceuticals and their impact on protein clearance. Eur. J. Pharm. Biopharm. 2019, 139, 123–131. [Google Scholar] [CrossRef]
- Brezinsky, S.C.G.; Chiang, G.G.; Szilvasi, A.; Mohan, S.; Shapiro, R.I.; MacLean, A.; Sisk, W.; Thill, G.J. A simple method for enriching populations of transfected CHO cells for cells of higher specific productivity. J. Immunol. Methods 2003, 277, 141–155. [Google Scholar] [CrossRef]
- Kronewitter, S.R.; An, H.J.; De Leoz, M.L.; Lebrilla, C.B.; Miyamoto, S.; Leiserowitz, G.S. The development of retrosynthetic glycan libraries to profile and classify the human serum N-linked glycome. Proteomics 2009, 9, 2986–2994. [Google Scholar] [CrossRef]
- Ozohanics, O.; Turiák, L.; Puerta, A.; Vékey, K.; Drahos, L. High-performance liquid chromatography coupled to mass spectrometry methodology for analyzing site-specific N-glycosylation patterns. J. Chromatogr. 2012, 1259, 200–212. [Google Scholar] [CrossRef] [PubMed]
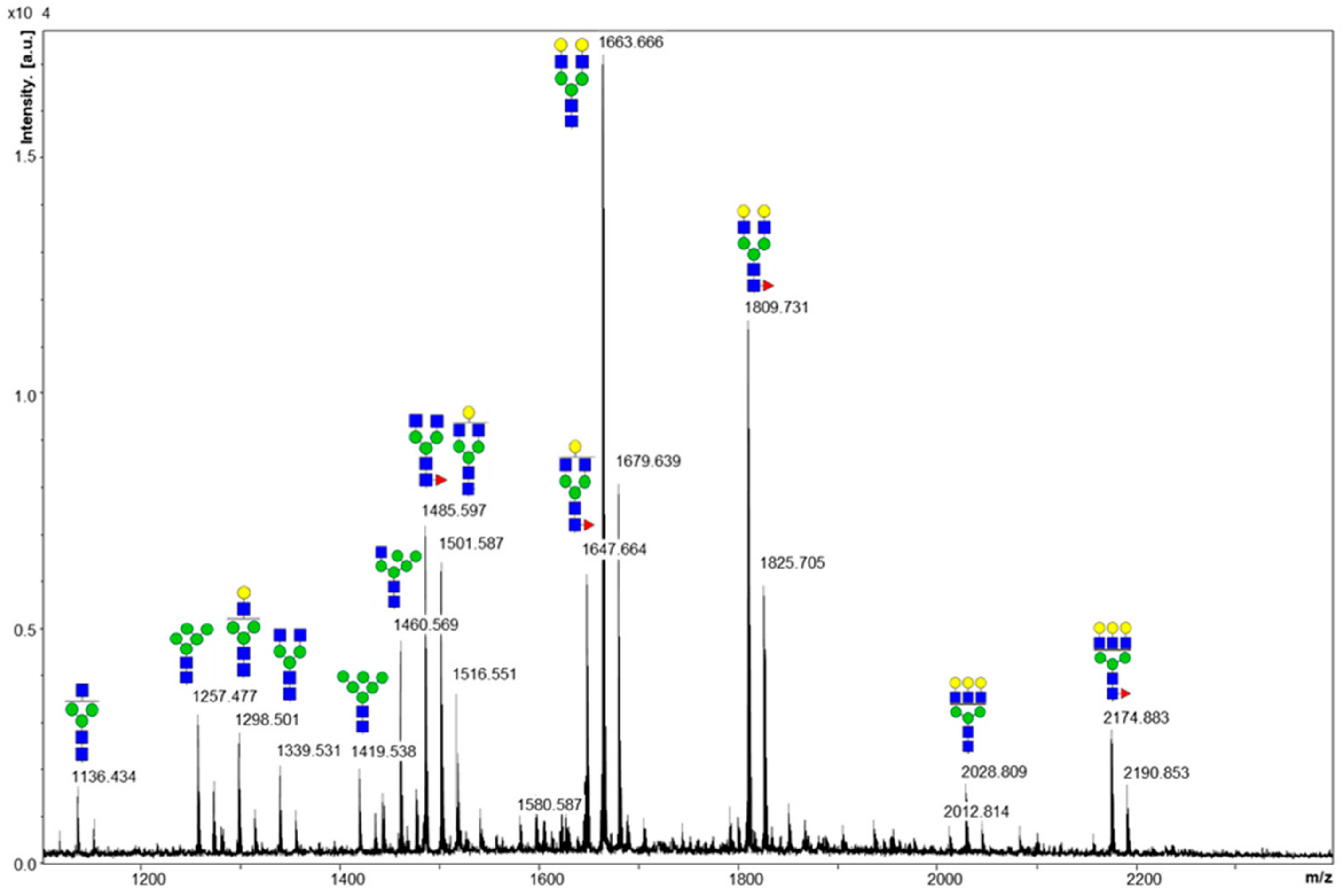
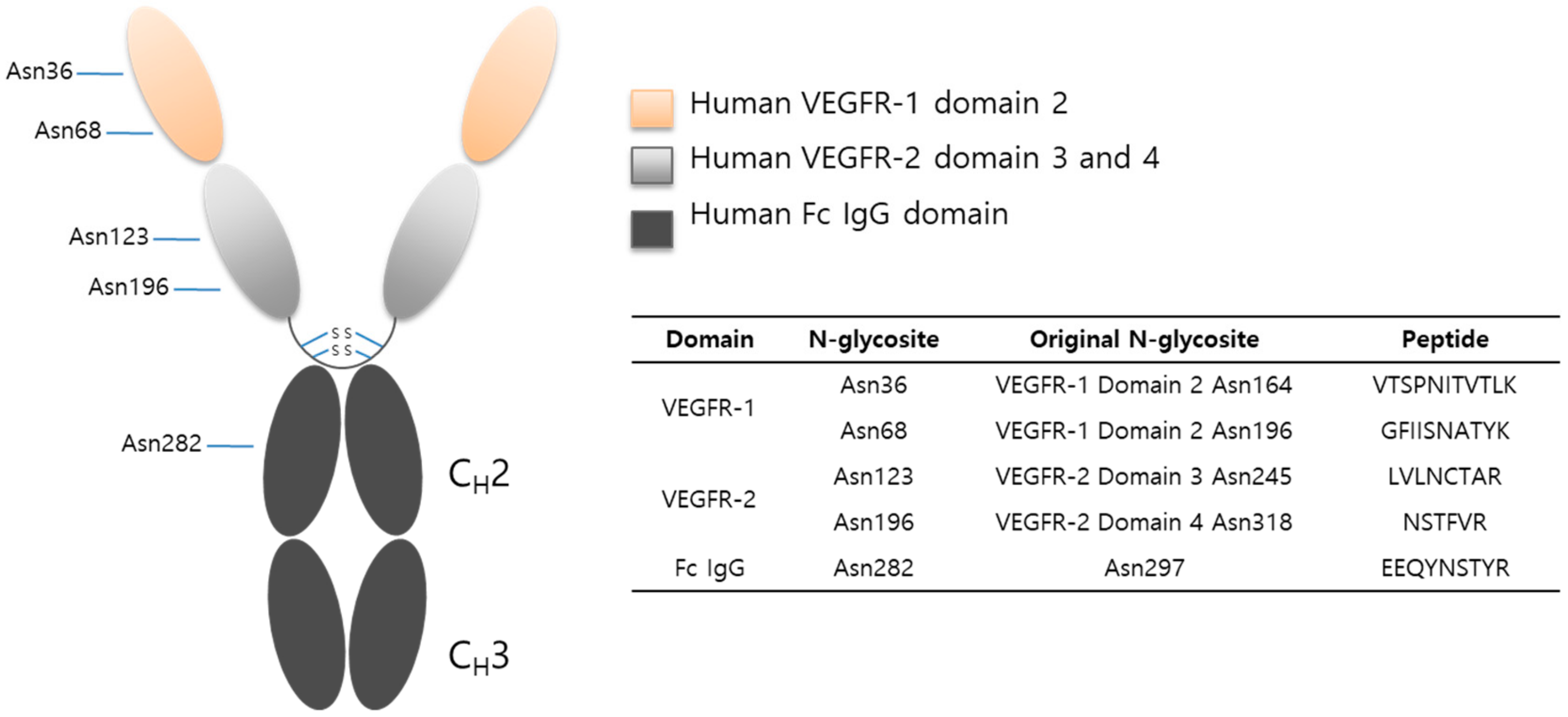

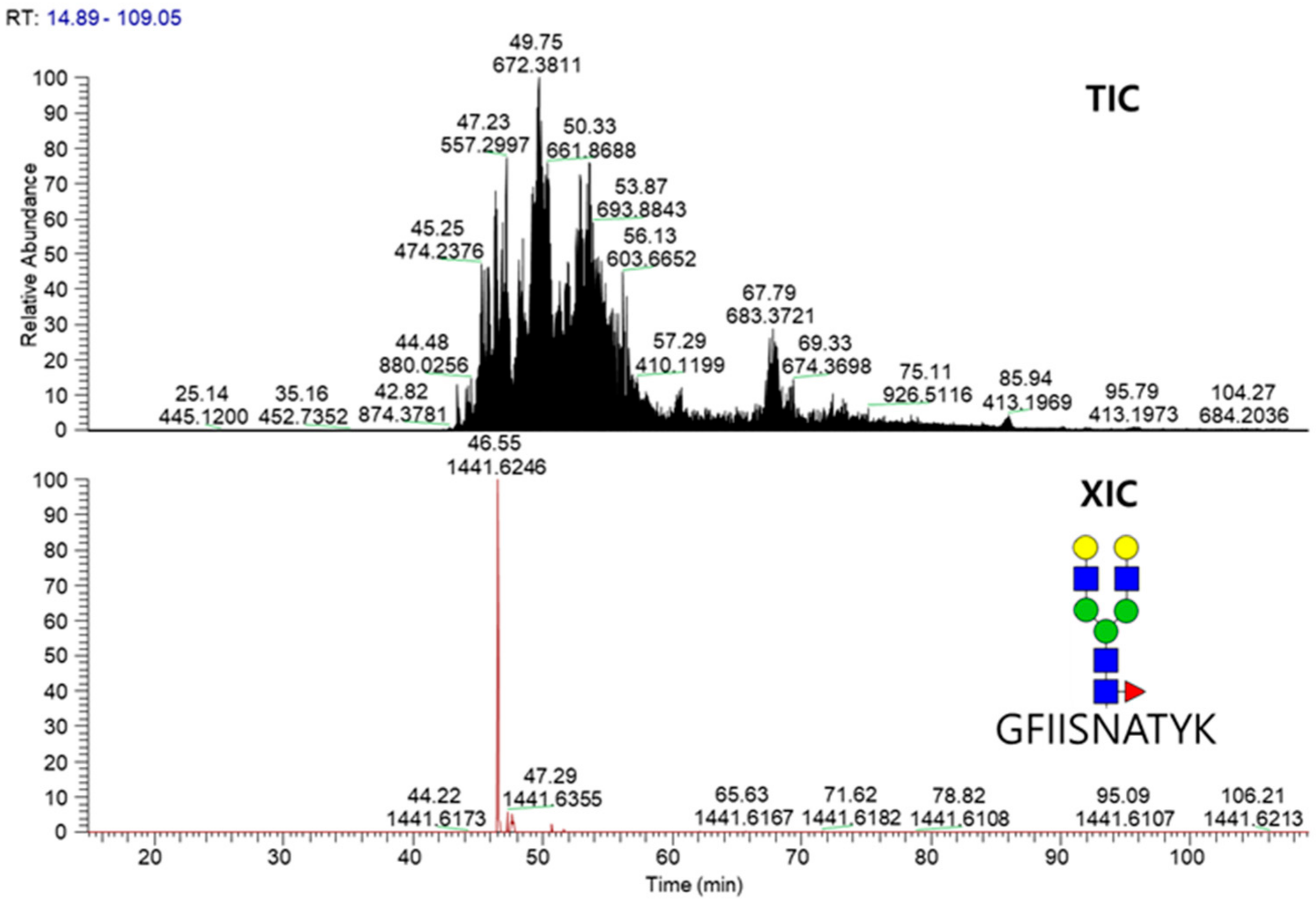
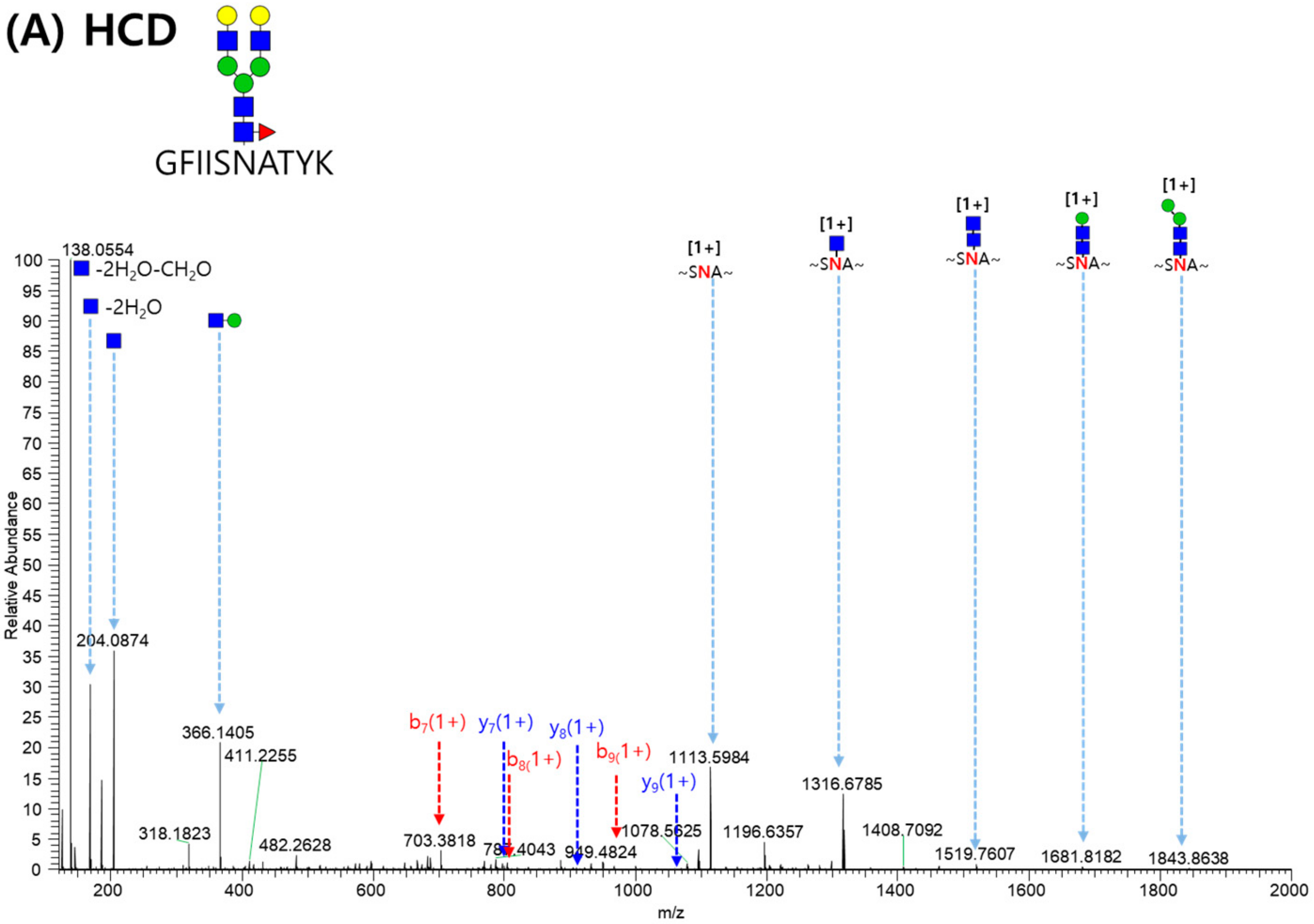
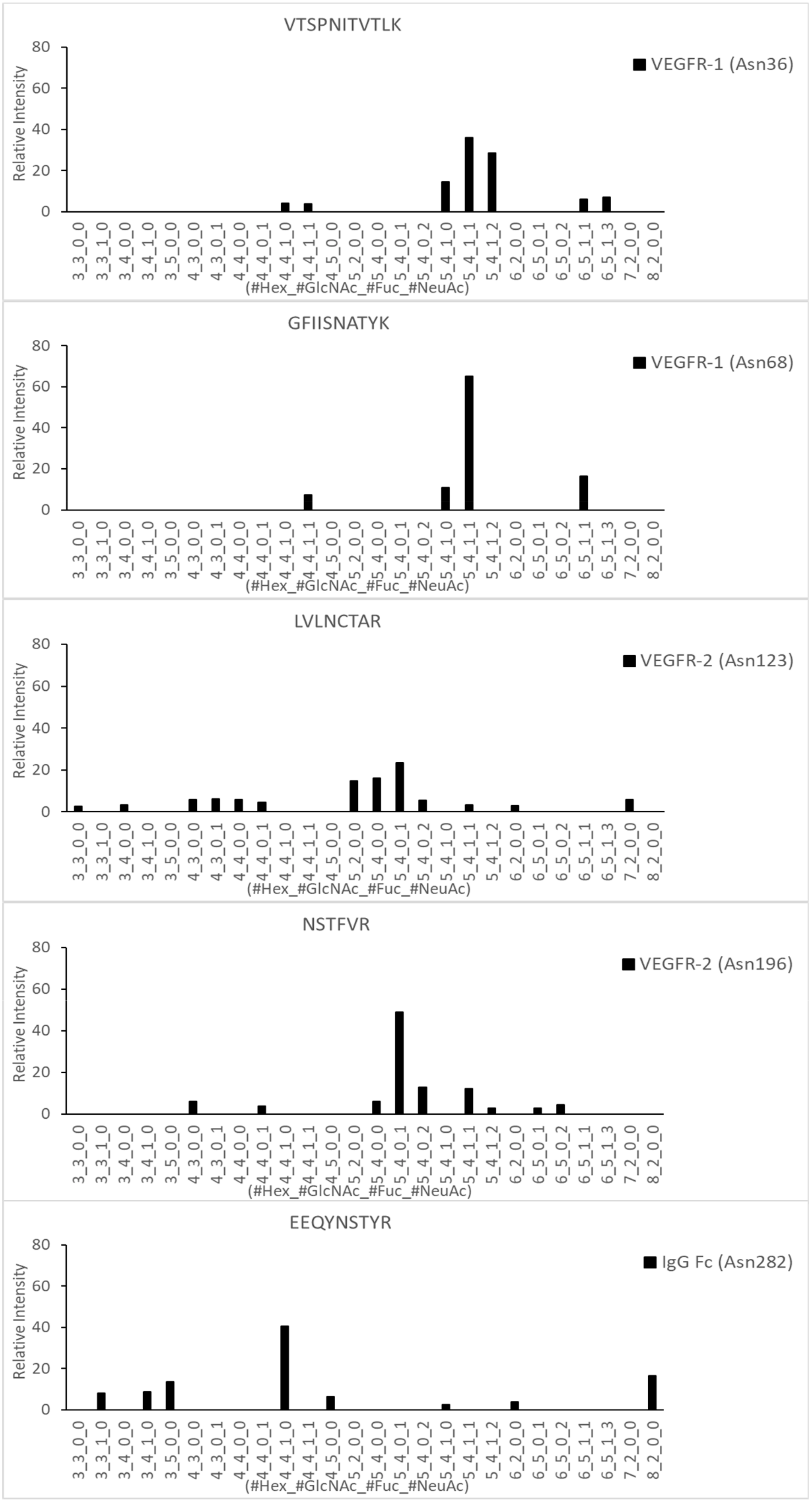
| Domain | Site | Original Site | Peptide | Fuc% | Sia% |
|---|---|---|---|---|---|
| VEGFR-1 | Asn57 | VEGFR-1 Domain 2 Asn164 | VTSPNITVTLK | 100.0 | 81.6 |
| Asn90 | VEGFR-1 Domain 2 Asn196 | GFIISNATYK | 100.0 | 89.0 | |
| VEGFR-2 | Asn149 | VEGFR-2 Domain 3 Asn245 | LVLNCTAR | 3.4 | 42.7 |
| Asn224 | VEGFR-2 Domain 4 Asn318 | NSTFVR | 15.2 | 87.8 | |
| IgG Fc | Asn316 | IgG Fc Domain Asn297 | EEQYNSTYR | 59.8 | 0.0 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hahm, Y.H.; Lee, J.Y.; Ahn, Y.H. Investigation of Site-Specific Differences in Glycan Microheterogeneity by N-Glycopeptide Mapping of VEGFR-IgG Fusion Protein. Molecules 2019, 24, 3924. https://doi.org/10.3390/molecules24213924
Hahm YH, Lee JY, Ahn YH. Investigation of Site-Specific Differences in Glycan Microheterogeneity by N-Glycopeptide Mapping of VEGFR-IgG Fusion Protein. Molecules. 2019; 24(21):3924. https://doi.org/10.3390/molecules24213924
Chicago/Turabian StyleHahm, Young Hye, Ju Yeon Lee, and Yeong Hee Ahn. 2019. "Investigation of Site-Specific Differences in Glycan Microheterogeneity by N-Glycopeptide Mapping of VEGFR-IgG Fusion Protein" Molecules 24, no. 21: 3924. https://doi.org/10.3390/molecules24213924
APA StyleHahm, Y. H., Lee, J. Y., & Ahn, Y. H. (2019). Investigation of Site-Specific Differences in Glycan Microheterogeneity by N-Glycopeptide Mapping of VEGFR-IgG Fusion Protein. Molecules, 24(21), 3924. https://doi.org/10.3390/molecules24213924





