Phenolic Profile of Grape Canes: Novel Compounds Identified by LC-ESI-LTQ-Orbitrap-MS
Abstract
1. Introduction
2. Results and Discussion
2.1. General
2.2. Phenolic Acids
2.2.1. Hydroxybenzoic Acids and Derivatives
2.2.2. Hydroxycinnamic Acids Derivatives
2.3. Flavanols (Proanthocyanidins)
2.3.1. Procyanidins and Monomers
2.3.2. Prodelphinidins and Gallate Derivatives
2.4. Flavonols and Derivatives
2.5. Flavanonols and Derivatives
2.6. Flavanones and Derivatives
2.7. Stilbenes and Derivatives
2.7.1. Stilbene Monomers
2.7.2. Stilbene Dimers
2.7.3. Glycosylated Stilbenes
2.7.4. Stilbene Oligomers
3. Materials and Methods
3.1. Chemicals
3.2. Grape Cane Samples
3.3. Polyphenol Extraction from Grape Canes
3.4. LC-LTQ-Orbitrap-MS Analyses
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| FTMS | fourier transformation mass spectrometry |
| PDA | photodiode array detector |
| AGC | automatic gain control |
| PTFE | polytetrafluoroethylene |
| HCD | high-energy C-trap dissociation |
| FWHM | full width at half maximum |
| a.u. | arbitrary units |
| ESI | electrospray ionization |
| HPLC | high performance liquid chromatography |
| MS | mass spectrometry |
| LC | liquid chromatography |
| 13C-NMR | Carbon-13 nuclear magnetic resonance |
| EC | catechin |
| ECG | catechin gallate |
| EGC | gallocatechin |
| EGCG | gallocatechin gallate |
| QTrap | quadrupole ion trap |
| DP | degree of polymerization |
| QM | quinone methide |
| HRF | heterolytic ring fission |
| RDA | retro-Diels-Alder |
| IMF | Ion molecular formula |
| LC–LTQ-Orbitrap | liquid chromatography coupled with electrospray ionization hybrid linear trap quadrupole-Orbitrap mass spectrometry |
References
- Tanase, C.; Mocan, A.; Coșarcă, S.; Gavan, A.; Nicolescu, A.; Gheldiu, A.M.; Vodnar, D.C.; Muntean, D.L.; Crișan, O. Biological and chemical insights of beech (Fagus sylvatica L.) bark: A source of bioactive compounds with functional properties. Antioxidants 2019, 8, 417. [Google Scholar] [CrossRef] [PubMed]
- García, D.E.; Fuentealba, C.A.; Salazar, J.P.; Pérez, M.A.; Escobar, D.; Pizzi, A. Mild hydroxypropylation of polyflavonoids obtained under pilot-plant scale. Ind. Crops Prod. 2016, 87, 350–362. [Google Scholar] [CrossRef]
- Bocalandro, C.; Sanhueza, V.; Gómez-Caravaca, A.M.; González-Álvarez, J.; Fernández, K.; Roeckel, M.; Rodríguez-Estrada, M.T. Comparison of the composition of Pinus radiata bark extracts obtained at bench- and pilot-scales. Ind. Crops Prod. 2012, 38, 21–26. [Google Scholar] [CrossRef]
- García, D.E.; Delgado, N.; Aranda, F.L.; Toledo, M.A.; Cabrera-Barjas, G.; Sintjago, E.M.; Escobar-Avello, D.; Paczkowski, S. Synthesis of maleilated polyflavonoids and lignin as functional bio-based building-blocks. Ind. Crops Prod. 2018, 123, 154–163. [Google Scholar] [CrossRef]
- García, D.E.; Gavino, J.; Escobar, D.; Cancino, R.A. Maleinated polyflavonoids and lignin as functional additives for three kinds of thermoplastics. Iran. Polym. J. 2017, 26, 295–304. [Google Scholar] [CrossRef]
- Santos, J.; Delgado, N.; Fuentes, J.; Fuentealba, C.; Vega-Lara, J.; García, D.E. Exterior grade plywood adhesives based on pine bark polyphenols and hexamine. Ind. Crops Prod. 2018, 122, 340–348. [Google Scholar] [CrossRef]
- Coșarcă, S.L.; Moacă, E.A.; Tanase, C.; Muntean, D.L.; Pavel, I.Z.; Dehelean, C.A. Spruce and beech bark aqueous extracts: Source of polyphenols, tannins and antioxidants correlated to in vitro antitumor potential on two different cell lines. Wood Sci. Technol. 2019, 53, 313–333. [Google Scholar] [CrossRef]
- Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef]
- Sáez, V.; Pastene, E.; Vergara, C.; Mardones, C.; Hermosín-gutiérrez, I.; Gómez-alonso, S.; Gómez, M.V.; Theoduloz, C.; Riquelme, S.; Baer, D. Von Oligostilbenoids in Vitis vinifera L. Pinot Noir grape cane extract: Isolation, characterization, in vitro antioxidant capacity and anti-proliferative effect on cancer cells. Food Chem. 2018, 265, 101–110. [Google Scholar] [CrossRef]
- Montero, L.; Sáez, V.; von Baer, D.; Cifuentes, A.; Herrero, M. Profiling of Vitis vinifera L. canes (poly)phenolic compounds using comprehensive two-dimensional liquid chromatography. J. Chromatogr. A 2018, 1536, 205–215. [Google Scholar] [CrossRef]
- Gorena, T.; Saez, V.; Mardones, C.; Vergara, C.; Winterhalter, P.; Von Baer, D. Influence of post-pruning storage on stilbenoid levels in Vitis vinifera L. canes. Food Chem. 2014, 155, 256–263. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Garza, S.L.; Laveriano-Santos, E.P.; Marhuenda-Muñoz, M.; Storniolo, C.E.; Tresserra-Rimbau, A.; Vallverdú-Queralt, A.; Lamuela-Raventós, R.M. Health effects of resveratrol: Results from human intervention trials. Nutrients 2018, 10, 1892. [Google Scholar] [CrossRef] [PubMed]
- Bagchi, D.; Swaroop, A.; Preuss, H.G.; Bagchi, M. Free radical scavenging, antioxidant and cancer chemoprevention by grape seed proanthocyanidin: An overview. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2014, 768, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Cosme, F.; Pinto, T.; Vilela, A. Phenolic compounds and antioxidant activity in grape juices: A chemical and sensory view. Beverages 2018, 4, 22. [Google Scholar] [CrossRef]
- Zhang, A.; Wan, L.; Wu, C.; Fang, Y.; Han, G.; Li, H.; Zhang, Z.; Wang, H. Simultaneous determination of 14 phenolic compounds in grape canes by HPLC-DAD-UV Using Wavelength. Molecules 2013, 18, 14241–14257. [Google Scholar] [CrossRef]
- Farhadi, K.; Esmaeilzadeh, F.; Hatami, M.; Forough, M.; Molaie, R. Determination of phenolic compounds content and antioxidant activity in skin, pulp, seed, cane and leaf of five native grape cultivars in West Azerbaijan province, Iran. Food Chem. 2016, 199, 847–855. [Google Scholar] [CrossRef]
- Sáez, V.; Gayoso, C.; Riquelme, S.; Pérez, J.; Vergara, C.; Mardones, C.; von Baer, D. C18 core-shell column with in-series absorbance and fluorescence detection for simultaneous monitoring of changes in stilbenoid and proanthocyanidin concentrations during grape cane storage. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2018, 1074, 70–78. [Google Scholar] [CrossRef]
- Vergara, C.; Von Baer, D.; Mardones, C.; Wilkens, A.; Wernekinck, K.; Damm, A.; MacKe, S.; Gorena, T.; Winterhalter, P. Stilbene levels in grape cane of different cultivars in southern Chile: Determination by HPLC-DAD-MS/MS method. J. Agric. Food Chem. 2012, 60, 929–933. [Google Scholar] [CrossRef]
- Rusjan, D.; Mikulic-Petkovsek, M. Phenolic responses in 1-year-old canes of Vitis vinifera cv. Chardonnay induced by grapevine yellows (Bois noir). Aust. J. Grape Wine Res. 2015, 21, 123–134. [Google Scholar] [CrossRef]
- Ewald, P.; Delker, U.; Winterhalter, P. Quanti fi cation of stilbenoids in grapevine canes and grape cluster stems with a focus on long-term storage e ff ects on stilbenoid concentration in grapevine canes. Food Res. Int. 2017, 100, 326–331. [Google Scholar] [CrossRef]
- Tisserant, L.P.; Hubert, J.; Lequart, M.; Borie, N.; Maurin, N.; Pilard, S.; Jeandet, P.; Aziz, A.; Renault, J.H.; Nuzillard, J.M.; et al. 13C NMR and LC-MS Profiling of stilbenes from elicited grapevine hairy root cultures. J. Nat. Prod. 2016, 79, 2846–2855. [Google Scholar] [CrossRef] [PubMed]
- Cebrián, C.; Sánchez-Gómez, R.; Salinas, M.R.; Alonso, G.L.; Zalacain, A. Effect of post-pruning vine-shoots storage on the evolution of high-value compounds. Ind. Crops Prod. 2017, 109, 730–736. [Google Scholar] [CrossRef]
- Vallverdu-Queralt, A.; Verbaere, A.; Meudec, E.; Cheynier, V.; Sommerer, N. Straightforward method to quantify GSH, GSSG, GRP, and hydroxycinnamic acids in wines by UPLC-MRM-MS. J. Agric. Food Chem. 2015, 63, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Vallverdú-Queralt, A.; Regueiro, J.; Rinaldi-Alvarenga, J.F.; Martínez-Huélamo, M.; Leal, L.N.; Lamuela-Raventós, R. Characterization of the phenolic and antioxidant profiles of selected culinary herbs and spices: Caraway, turmeric, dill, marjoram and nutmeg. Food Sci. Technol. 2015, 35, 189–195. [Google Scholar] [CrossRef]
- Vallverdú-Queralt, A.; Boix, N.; Piqué, E.; Gómez-Catalan, J.; Medina-Remon, A.; Sasot, G.; Mercader-Martí, M.; Llobet, J.M.; Lamuela-Raventos, R.M. Identification of phenolic compounds in red wine extract samples and zebrafish embryos by HPLC-ESI-LTQ-Orbitrap-MS. Food Chem. 2015, 181, 146–151. [Google Scholar] [CrossRef] [PubMed]
- Regueiro, J.; Sánchez-González, C.; Vallverdú-Queralt, A.; Simal-Gándara, J.; Lamuela-Raventós, R.; Izquierdo-Pulido, M. Comprehensive identification of walnut polyphenols by liquid chromatography coupled to linear ion trap-Orbitrap mass spectrometry. Food Chem. 2014, 152, 340–348. [Google Scholar] [CrossRef] [PubMed]
- Sasot, G.; Martínez-Huélamo, M.; Vallverdú-Queralt, A.; Mercader-Martí, M.; Estruch, R.; Lamuela-Raventós, R.M. Identification of phenolic metabolites in human urine after the intake of a functional food made from grape extract by a high resolution LTQ-Orbitrap-MS approach. Food Res. Int. 2017, 100, 435–444. [Google Scholar] [CrossRef]
- Fayeulle, N.; Vallverdu-Queralt, A.; Meudec, E.; Hue, C.; Boulanger, R.; Cheynier, V.; Sommerer, N. Characterization of new flavan-3-ol derivatives in fermented cocoa beans. Food Chem. 2018, 259, 207–212. [Google Scholar] [CrossRef]
- Vallverdú-Queralt, A.; Meudec, E.; Eder, M.; Lamuela-Raventos, R.M.; Sommerer, N.; Cheynier, V. The hidden face of wine polyphenol polymerization highlighted by high-resolution mass spectrometry. ChemistryOpen 2017, 6, 336–339. [Google Scholar] [CrossRef]
- Vallverdú-queralt, A.; Meudec, E.; Eder, M.; Lamuela-raventos, R.M.; Sommerer, N.; Cheynier, V. Targeted filtering reduces the complexity of UHPLC-Orbitrap-HRMS data to decipher polyphenol polymerization. Food Chem. 2017, 227, 255–263. [Google Scholar] [CrossRef]
- Di Lecce, G.; Arranz, S.; Jáuregui, O.; Tresserra-Rimbau, A.; Quifer-Rada, P.; Lamuela-Raventós, R.M. Phenolic profiling of the skin, pulp and seeds of Albariño grapes using hybrid quadrupole time-of-flight and triple-quadrupole mass spectrometry. Food Chem. 2014, 145, 874–882. [Google Scholar] [CrossRef] [PubMed]
- Godevac, D.; Teševic, V.; Veličković, M.; Vujisica, L.; Vajs, V.; Milosavljević, S. Polyphenolic compounds in seeds from some grape cultivars grown in Serbia. J. Serbian Chem. Soc. 2010, 75, 1641–1652. [Google Scholar] [CrossRef]
- Pugajeva, I.; Perkons, I.; Górnaś, P. Identification and determination of stilbenes by Q-TOF in grape skins, seeds, juice and stems. J. Food Compos. Anal. 2018, 74, 44–52. [Google Scholar] [CrossRef]
- Teixeira, N.; Mateus, N.; de Freitas, V.; Oliveira, J. Wine industry by-product: Full polyphenolic characterization of grape stalks. Food Chem. 2018, 268, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Rockenbach, I.I.; Jungfer, E.; Ritter, C.; Santiago-Schübel, B.; Thiele, B.; Fett, R.; Galensa, R. Characterization of flavan-3-ols in seeds of grape pomace by CE, HPLC-DAD-MSn and LC-ESI-FTICR-MS. Food Res. Int. 2012, 48, 848–855. [Google Scholar] [CrossRef]
- Perestrelo, R.; Lu, Y.; Santos, S.A.O.; Silvestre, A.J.D.; Neto, C.P.; Câmara, J.S.; Rocha, S.M. Phenolic profile of Sercial and Tinta Negra Vitis vinifera L. grape skins by HPLC-DAD-ESI-MSn: Novel phenolic compounds in Vitis vinifera L. grape. Food Chem. 2012, 135, 94–104. [Google Scholar] [CrossRef]
- Tinikul, R.; Chenprakhon, P.; Maenpuen, S.; Chaiyen, P. Biotransformation of plant-derived phenolic acids. Biotechnol. J. 2018, 13, 1–12. [Google Scholar] [CrossRef]
- Saibabu, V.; Fatima, Z.; Khan, L.A.; Hameed, S. Therapeutic potential of dietary phenolic acids. Adv. Pharmacol. Sci. 2015, 2015, 1–10. [Google Scholar] [CrossRef]
- Moreira, M.M.; Barroso, M.F.; Porto, J.V.; Ramalhosa, M.J.; Švarc-Gajić, J.; Estevinho, L.; Morais, S.; Delerue-Matos, C. Potential of Portuguese vine shoot wastes as natural resources of bioactive compounds. Sci. Total Environ. 2018, 634, 831–842. [Google Scholar] [CrossRef]
- Hooi Poay, T.; Sui Kiong, L.; Cheng Hock, C. Characterisation of galloylated cyanogenic glucosides and hydrolysable tannins from leaves of Phyllagathis rotundifolia by LC-ESI-MS/MS. Phytochem. Anal. 2011, 22, 516–525. [Google Scholar] [CrossRef]
- Sun, J.; Liang, F.; Bin, Y.; Li, P.; Duan, C. Screening non-colored phenolics in red wines using liquid chromatography/ultraviolet and mass spectrometry/mass spectrometry libraries. Molecules 2007, 12, 679–693. [Google Scholar] [CrossRef] [PubMed]
- Gasperotti, M.; Masuero, D.; Vrhovsek, U.; Guella, G.; Mattivi, F. Profiling and accurate quantification of Rubus ellagitannins and ellagic acid conjugates using direct uplc-q-tof hdms and hplc-dad analysis. J. Agric. Food Chem. 2010, 58, 4602–4616. [Google Scholar] [CrossRef] [PubMed]
- Heleno, S.A.; Martins, A.; Queiroz, M.J.R.P.; Ferreira, I.C.F.R. Bioactivity of phenolic acids: Metabolites versus parent compounds: A review. Food Chem. 2015, 173, 501–513. [Google Scholar] [CrossRef] [PubMed]
- Taofiq, O.; González-Paramás, A.M.; Barreiro, M.F.; Ferreira, I.C.F.R.; McPhee, D.J. Hydroxycinnamic acids and their derivatives: Cosmeceutical significance, challenges and future perspectives, a review. Molecules 2017, 22, 281. [Google Scholar] [CrossRef]
- Khoza, B.S.; Gbashi, S.; Steenkamp, P.A.; Njobeh, P.B.; Madala, N.E. Identification of hydroxylcinnamoyl tartaric acid esters in Bidens pilosa by UPLC-tandem mass spectrometry. S. Afr. J. Bot. 2016, 103, 95–100. [Google Scholar] [CrossRef]
- Santos-Buelga, C.; Scalbert, A. Proanthocyanidins and tannin-like compounds-nature, occurrence, dietary intake and effects on nutrition and health. J. Sci. Food Agric. 2000, 80, 1094–1117. [Google Scholar] [CrossRef]
- Bonn, G.K. Structural elucidation of catechin and epicatechin in sorrel leaf extracts using liquid-chromatography coupled to diode array-, fluorescence-, and mass Original Paper. J. Sep. Sci. 2004, 27, 524–528. [Google Scholar]
- Bravo, M.N.; Silva, S.; Coelho, A.V.; Boas, L.V.; Bronze, M.R. Analysis of phenolic compounds in Muscatel wines produced in Portugal. Anal. Chim. Acta 2006, 563, 84–92. [Google Scholar] [CrossRef]
- De Villiers, A.; Venter, P.; Pasch, H. Recent advances and trends in the liquid-chromatography-mass spectrometry analysis of flavonoids. J. Chromatogr. A 2015, 1430, 16–78. [Google Scholar] [CrossRef]
- Yan, T.; Hu, G.; Wang, A.; Hong, Y. Formerly natural product letters characterisation of proanthocyanidins from Schisandra chinensis seed coats by UPLC-QTOF/MS. Nat. Prod. Res. 2014, 28, 1834–1842. [Google Scholar] [CrossRef]
- Prodanov, M.; Vacas, V.; Hernández, T.; Estrella, I.; Amador, B.; Winterhalter, P. Chemical characterisation of Malvar grape seeds (Vitis vinifera L.) by ultrafiltration and RP-HPLC-PAD-MS. J. Food Compos. Anal. 2013, 31, 284–292. [Google Scholar] [CrossRef]
- Delcambre, A.; André, Y.; Saucier, C. Sequencing of red wine proanthocyanidins by UHPLC-ESI-Q-ToF. J. Appl. Bioanal. 2015, 1, 46–54. [Google Scholar] [CrossRef]
- Dou, J.; Lee, V.S.Y.; Tzen, J.T.C.; Lee, M.R. Identification and comparison of phenolic compounds in the preparation of oolong tea manufactured by semifermentation and drying processes. J. Agric. Food Chem. 2007, 55, 7462–7468. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zou, T.; Gao, J.; Gu, L. Depolymerization of cranberry procyanidins using (+)-catechin, (−)-epicatechin, and (−)-epigallocatechin gallate as chain breakers. Food Chem. 2013, 141, 488–494. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, R.; Jayasinghe, L.; Kuhnert, N. Identification and characterization of proanthocyanidins of 16 members of the Rhododendron genus (Ericaceae) by tandem LC-MS. J. Mass Spectrom. 2012, 47, 502–515. [Google Scholar] [CrossRef]
- Li, H.; Deinzer, M.L. Tandem Mass Spectrometry for Sequencing Proanthocyanidins. Anal. Chem. 2007, 79, 1739–1748. [Google Scholar] [CrossRef]
- Li, F.; Wang, Y.; Li, D.; Chen, Y.; Qiao, X.; Fardous, R.; Lewandowski, A.; Liu, J.; Chan, T.H.; Dou, Q.P. Perspectives on the recent developments with green tea polyphenols in drug discovery. Expert Opin. Drug Discov. 2018, 13, 643–660. [Google Scholar]
- Higdon, J.V.; Frei, B. Tea Catechins and polyphenols: Health effects, metabolism, and antioxidant functions. Crit. Rev. Food Sci. Nutr. 2003, 43, 89–143. [Google Scholar] [CrossRef]
- Sur, S.; Panda, C.K. Molecular aspects of cancer chemopreventive and therapeutic efficacies of tea and tea polyphenols. Nutrition 2017, 43, 8–15. [Google Scholar] [CrossRef]
- Singh, N.A.; Mandal, A.K.A.; Khan, Z.A. Potential neuroprotective properties of epigallocatechin-3-gallate (EGCG). Nutr. J. 2016, 15, 1–17. [Google Scholar] [CrossRef]
- Pantelić, M.M.; Dabić Zagorac, D.; Davidović, S.M.; Todić, S.R.; Bešlić, Z.S.; Gašić, U.M.; Tešić, Ž.L.; Natić, M.M. Identification and quantification of phenolic compounds in berry skin, pulp, and seeds in 13 grapevine varieties grown in Serbia. Food Chem. 2016, 211, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Takemoto, M.; Takemoto, H. Synthesis of theaflavins and their functions. Molecules 2018, 23, 918. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Kelm, M.A.; Hammerstone, J.F.; Beecher, G.; Holden, J.; Haytowitz, D.; Prior, R.L. Screening of foods containing proanthocyanidins and their structural characterization using LC-MS/MS and thiolytic degradation. J. Agric. Food Chem. 2003, 51, 7513–7521. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, N.; Azevedo, J.; Mateus, N.; De Freitas, V. Proanthocyanidin screening by LC-ESI-MS of Portuguese red wines made with teinturier grapes. Food Chem. 2016, 190, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, W.; Eberhardt, A.; Galensa, R. Investigation of proanthocyanidins by HPLC with electrospray ionization mass spectrometry. Eur. Food Res. Technol. 2000, 211, 56–64. [Google Scholar] [CrossRef]
- Semwal, D.K.; Semwal, R.B.; Combrinck, S.; Viljoen, A. Myricetin: A dietary molecule with diverse biological activities. Nutrients 2016, 8, 90. [Google Scholar] [CrossRef] [PubMed]
- Medina Remon, A.; Estruch, R.; Tresserra-Rimbau, A.; Vallverdu-Queralt, A.; Lamuela-Raventos, R.M. The effect of polyphenol consumption on blood pressure. Mini Rev. Med. Chem. 2013, 13, 1137–1149. [Google Scholar] [CrossRef] [PubMed]
- Aaby, K.; Ekeberg, D.; Skrede, G. Characterization of phenolic compounds in strawberry (Fragaria x ananassa) fruits by different HPLC detectors and contribution of individual compounds to total antioxidant capacity. J. Agric. Food Chem. 2007, 55, 4395–4406. [Google Scholar] [CrossRef]
- Rehman, K.; Chohan, T.A.; Waheed, I.; Gilani, Z.; Akash, M.S.H. Taxifolin prevents postprandial hyperglycemia by regulating the activity of α-amylase: Evidence from an in vivo and in silico studies. J. Cell. Biochem. 2019, 120, 425–438. [Google Scholar] [CrossRef]
- Tresserra-Rimbau, A.; Guasch-Ferre, M.; Salas-Salvado, J.; Toledo, E.; Corella, D.; Castaner, O.; Guo, X.; Gomez-Gracia, E.; Lapetra, J.; Aros, F.; et al. Intake of total polyphenols and some classes of polyphenols is inversely associated with diabetes in elderly people at high cardiovascular disease risk. J. Nutr. 2016, 146, 767–777. [Google Scholar]
- Yang, P.; Xu, F.; Li, H.; Wang, Y.; Li, F.; Shang, M.; Liu, G.; Wang, X.; Cai, S. Detection of 191 taxifolin metabolites and their distribution in rats using HPLC-ESI-IT-TOF-MS. Molecules 2016, 21, 1209. [Google Scholar]
- Zhao, M.; Xu, J.; Qian, D.; Guo, J.; Jiang, S.; Shang, E. Identi fi cation of astilbin metabolites produced by human intestinal bacteria using UPLC-Q-TOF/MS. Biomed. Chromatogr. 2014, 28, 1024–1029. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.E.; Yang, H.; Son, G.W.; Park, H.R.; Park, C.; Jin, Y.; Park, Y.S. Eriodictyol protects endothelial cells against oxidative stress-induced cell death through modulating ERK/Nrf2/ARE-dependent heme oxygenase-1 expression. Int. J. Mol. Sci. 2015, 16, 14526–14539. [Google Scholar] [CrossRef] [PubMed]
- Fabre, N.; Rustan, I.; De Hoffmann, E.; Quetin-Leclercq, J. Determination of flavone, flavonol, and flavanone aglycones by negative ion liquid chromatography electrospray ion trap mass spectrometry. J. Am. Soc. Mass Spectrom. 2001, 12, 707–715. [Google Scholar] [CrossRef]
- Pawlus, A.D.; Sahli, R.; Bisson, J.; Rivière, C.; Delaunay, J.C.; Richard, T.; Gomès, E.; Bordenave, L.; Waffo-Téguo, P.; Mérillon, J.M. Stilbenoid profiles of canes from Vitis and Muscadinia species. J. Agric. Food Chem. 2013, 61, 501–511. [Google Scholar] [CrossRef] [PubMed]
- Catalgol, B.; Batirel, S.; Taga, Y.; Ozer, N.K. Resveratrol: French paradox revisited. Front. Pharmacol. 2012, 3, 1–18. [Google Scholar] [CrossRef]
- Buiarelli, F.; Coccioli, F.; Jasionowska, R.; Merolle, M.; Terracciano, A. Analysis of some stilbenes in Italian wines by liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2007, 21, 2955–2964. [Google Scholar] [CrossRef]
- Moss, R.; Mao, Q.; Taylor, D.; Saucier, C. Investigation of monomeric and oligomeric wine stilbenoids in red wines by ultra-high-performance liquid chromatography/electrospray ionization quadrupole time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2013, 27, 1815–1827. [Google Scholar] [CrossRef]
- Mattivi, F.; Vrhovsek, U.; Malacarne, G.; Masuero, D.; Zulini, L.; Stefanini, M.; Mose, C.; Velasco, R.; Guella, G. Profiling of resveratrol oligomers, important stress metabolites, accumulating in the leaves of hybrid Vitis vinifera (Merzling × Teroldego) genotypes infected with Plasmopara viticola. J. Agric. Food Chem. 2011, 59, 5364–5375. [Google Scholar] [CrossRef]
- Kong, Q.J.; Ren, X.Y.; Hu, N.; Sun, C.R.; Pan, Y.J. Identification of isomers of resveratrol dimer and their analogues from wine grapes by HPLC/MSn and HPLC/DAD-UV. Food Chem. 2011, 127, 727–734. [Google Scholar] [CrossRef]
- Cichewicz, R.H.; Kouzi, S.A.; Hamann, M.T. Dimerization of resveratrol by the grapevine pathogen Botrytis cinerea. J. Nat. Prod. 2000, 63, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Vallverdú-Queralt, A.; Jáuregui, O.; Medina-Remón, A.; Andrés-Lacueva, C.; Lamuela-Raventós, R.M. Improved characterization of tomato polyphenols using liquid chromatography electrospray ionization linear ion trap quadrupole Orbitrap mass spectrometry and liquid chromatography/electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2010, 24, 2986–2992. [Google Scholar] [CrossRef] [PubMed]
- Puessa, T.; Floren, J.; Kuldkepp, P.; Raal, A. Survey of grapevine Vitis vinifera stem polyphenolsby liquid chromatography-diode array detection-tandem mass spectrometry. J. Agric. Food Chem. 2006, 54, 7488–7494. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples are not available from the authors. |
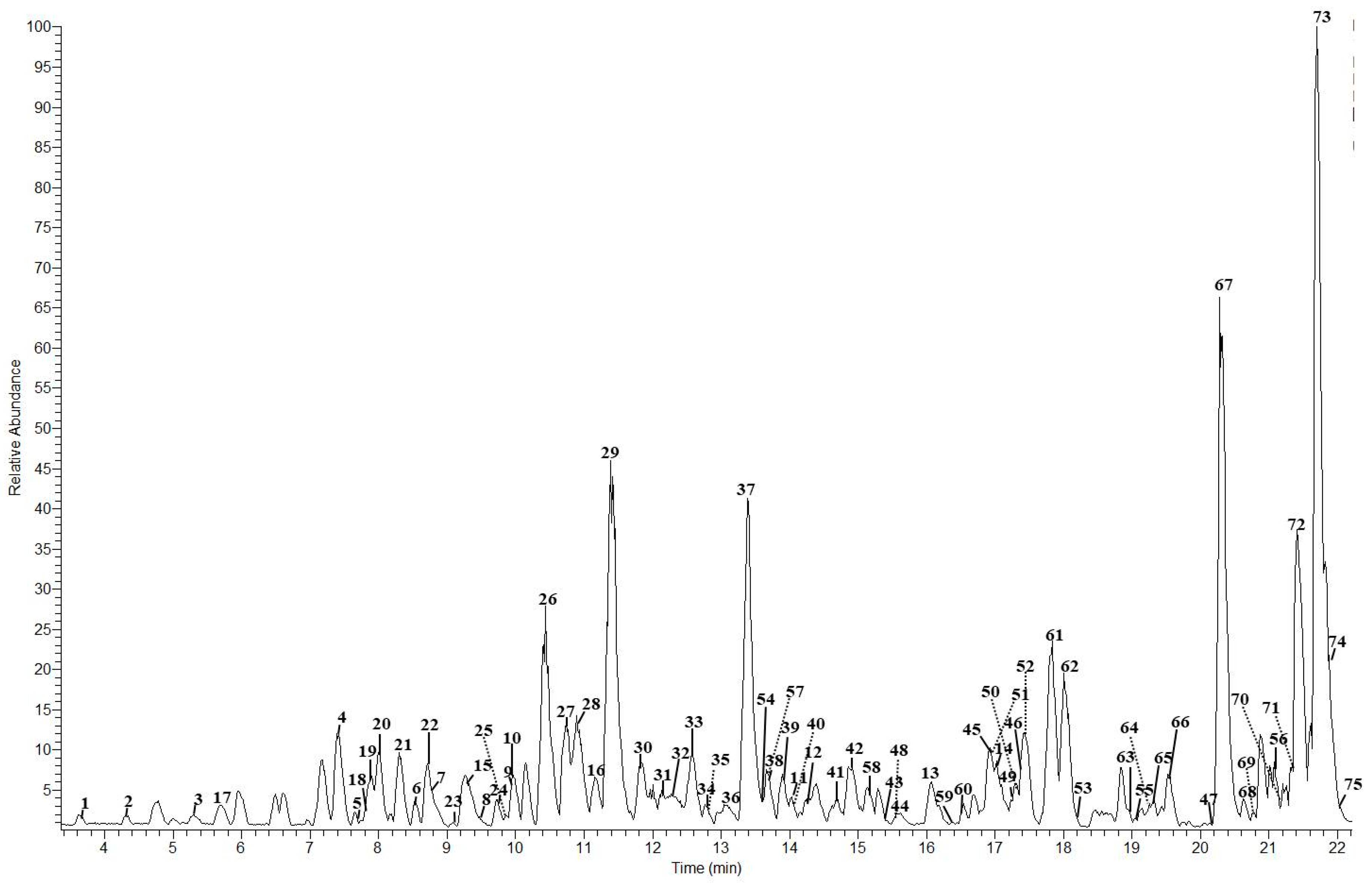
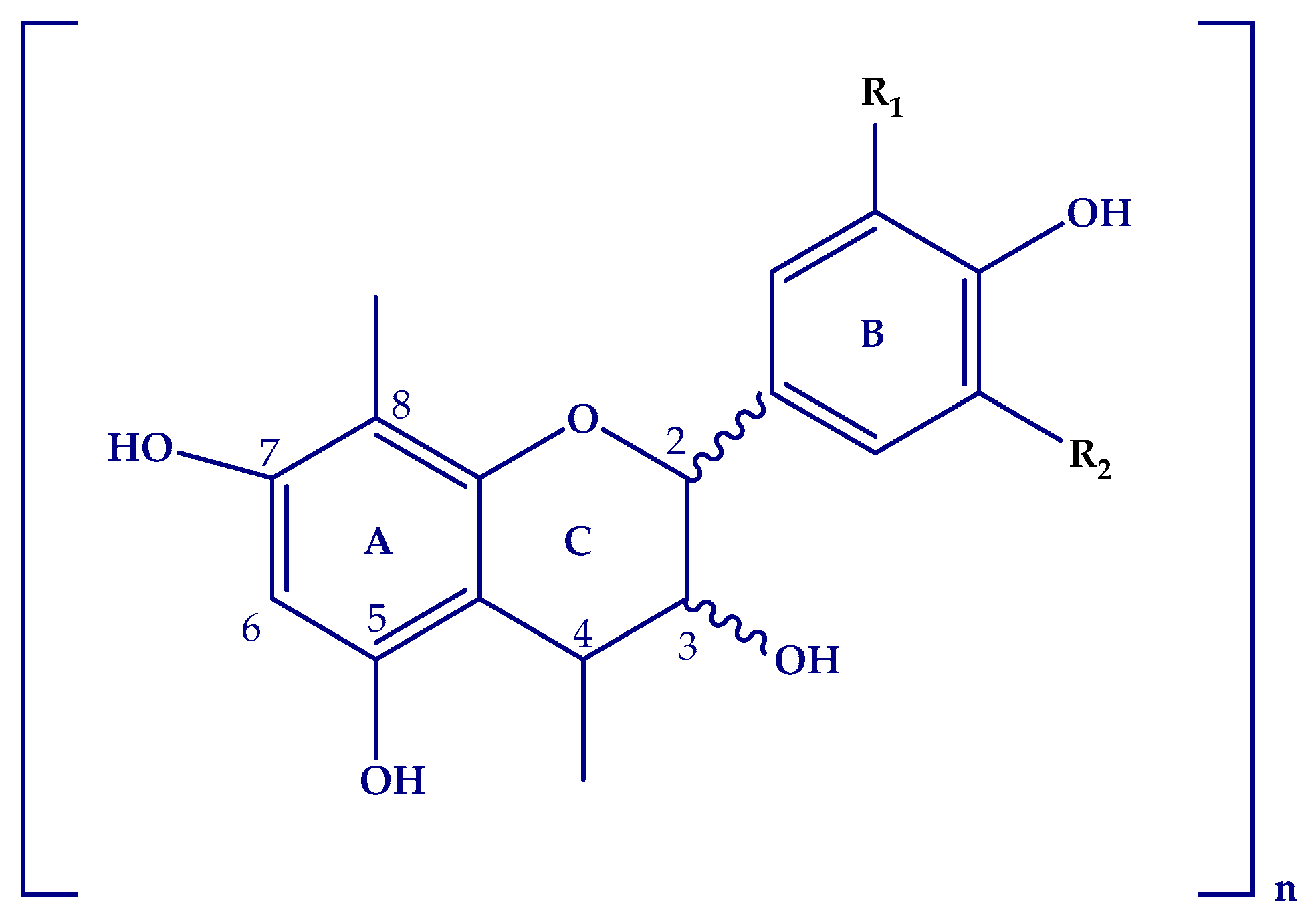
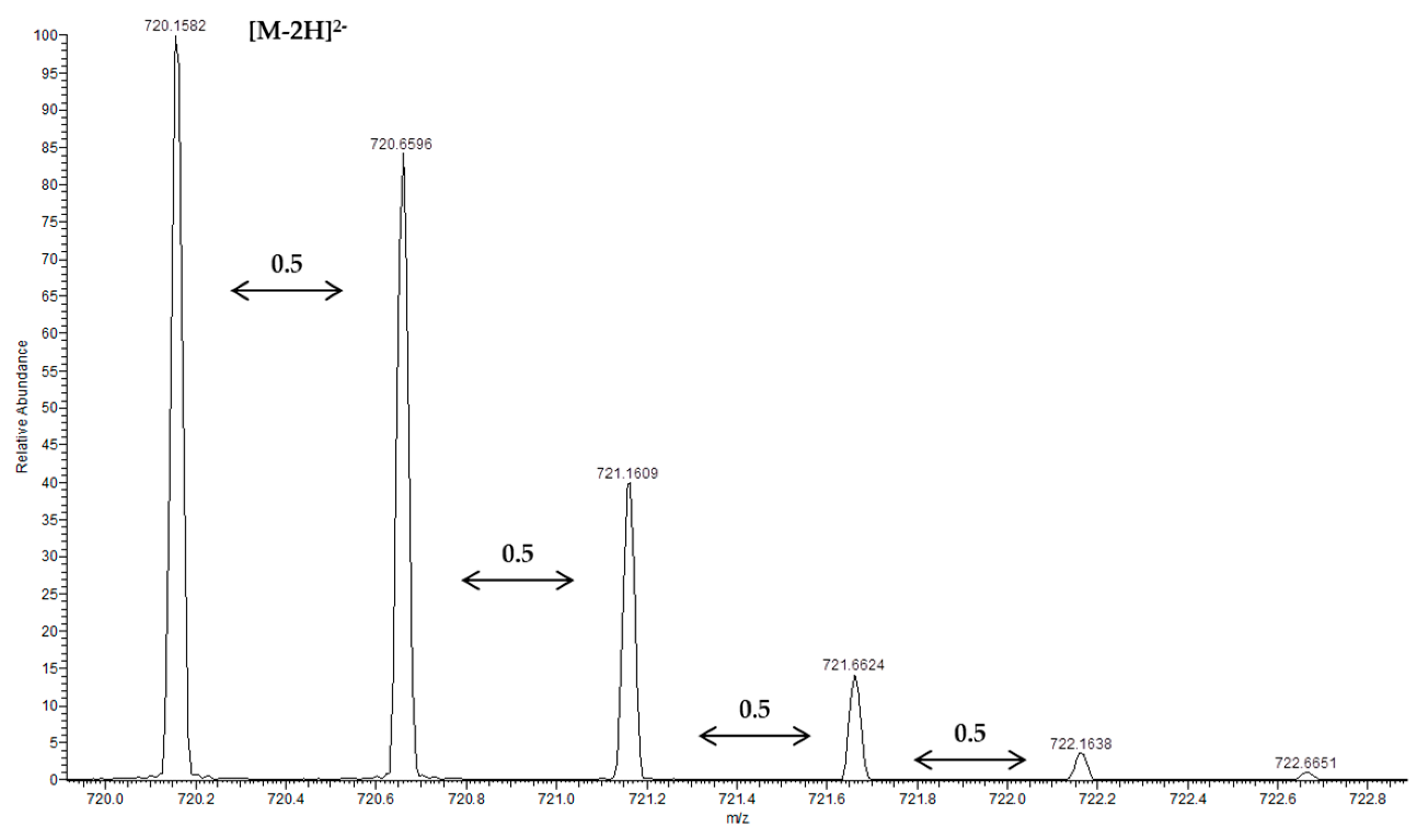
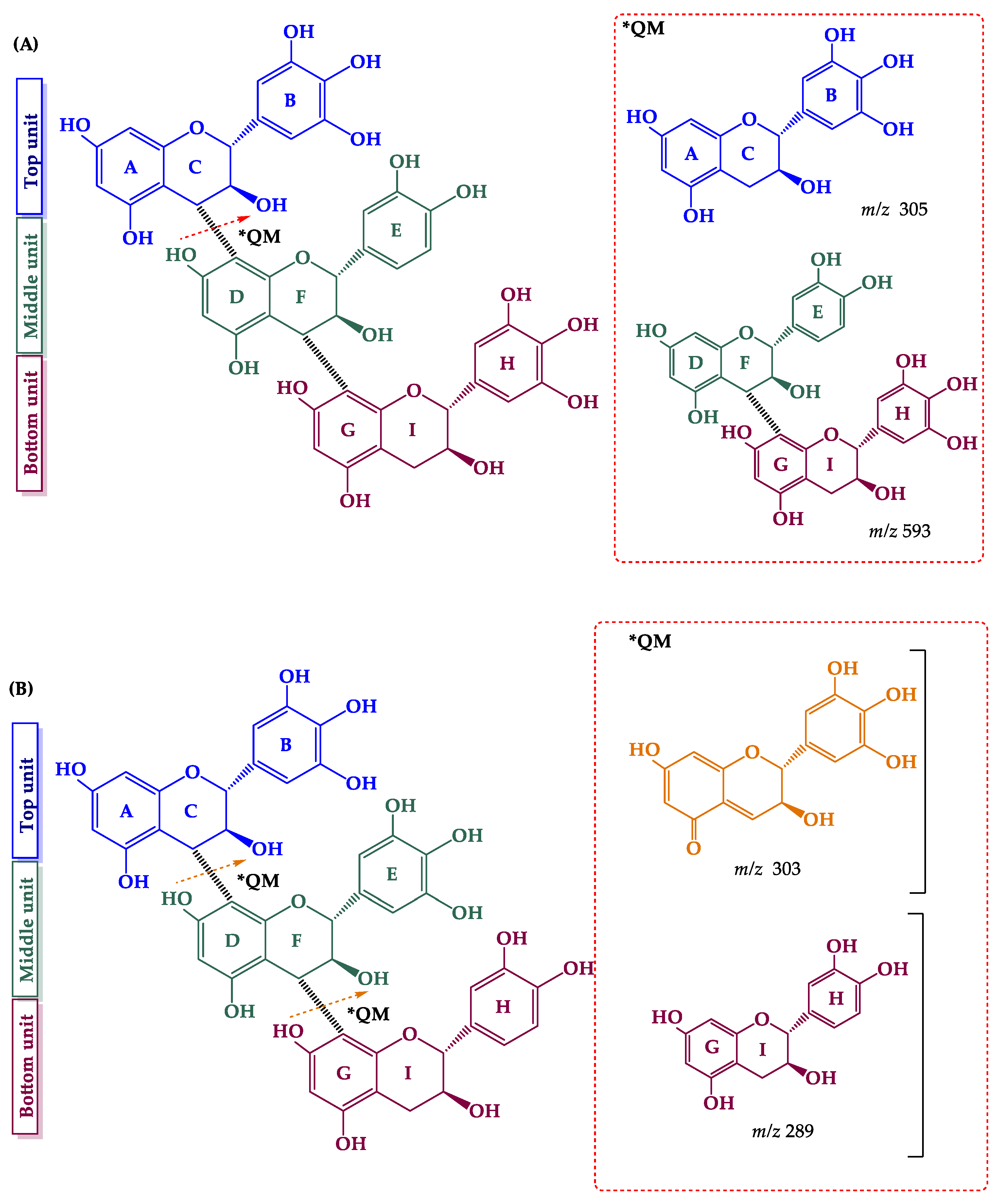
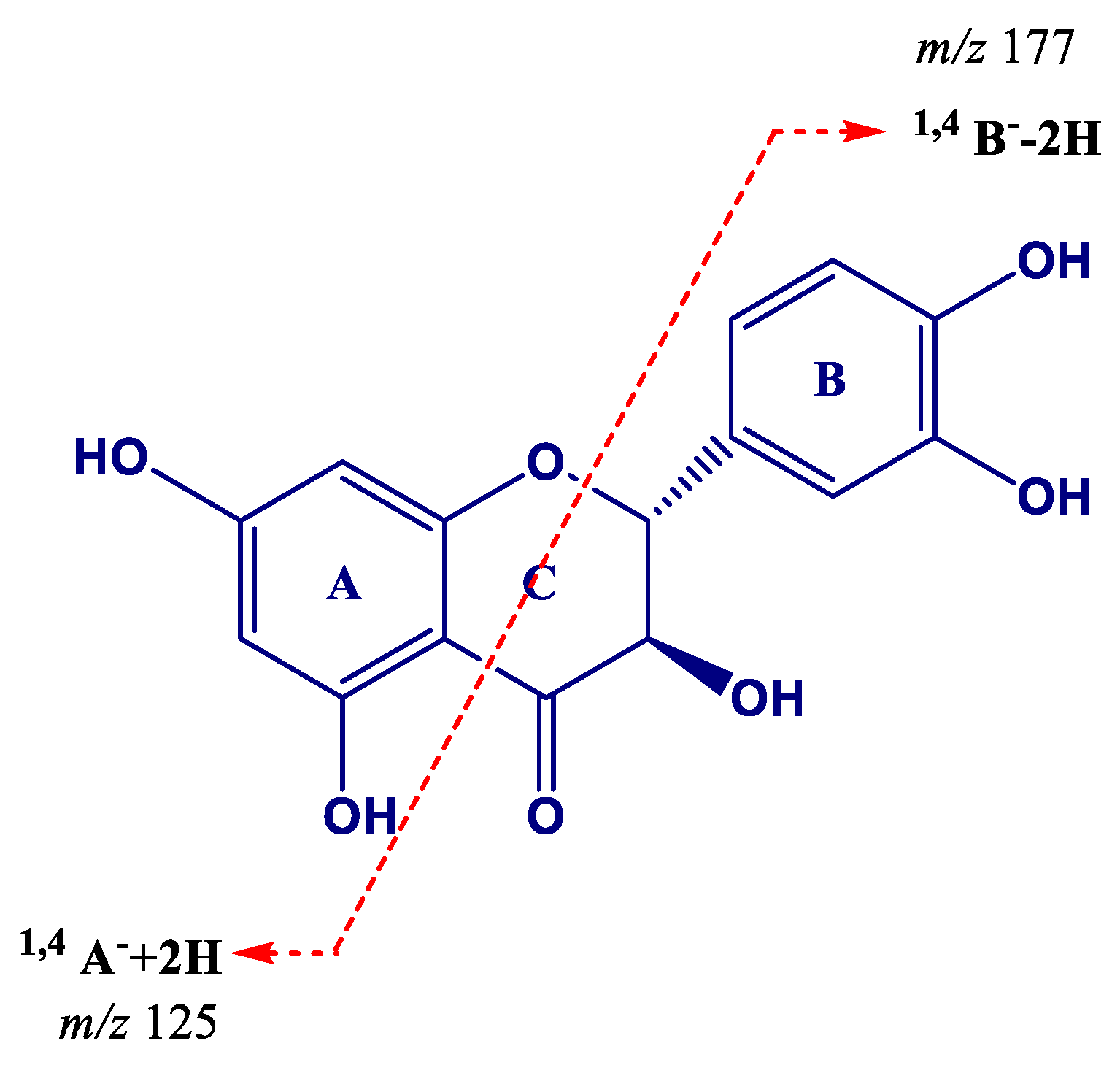
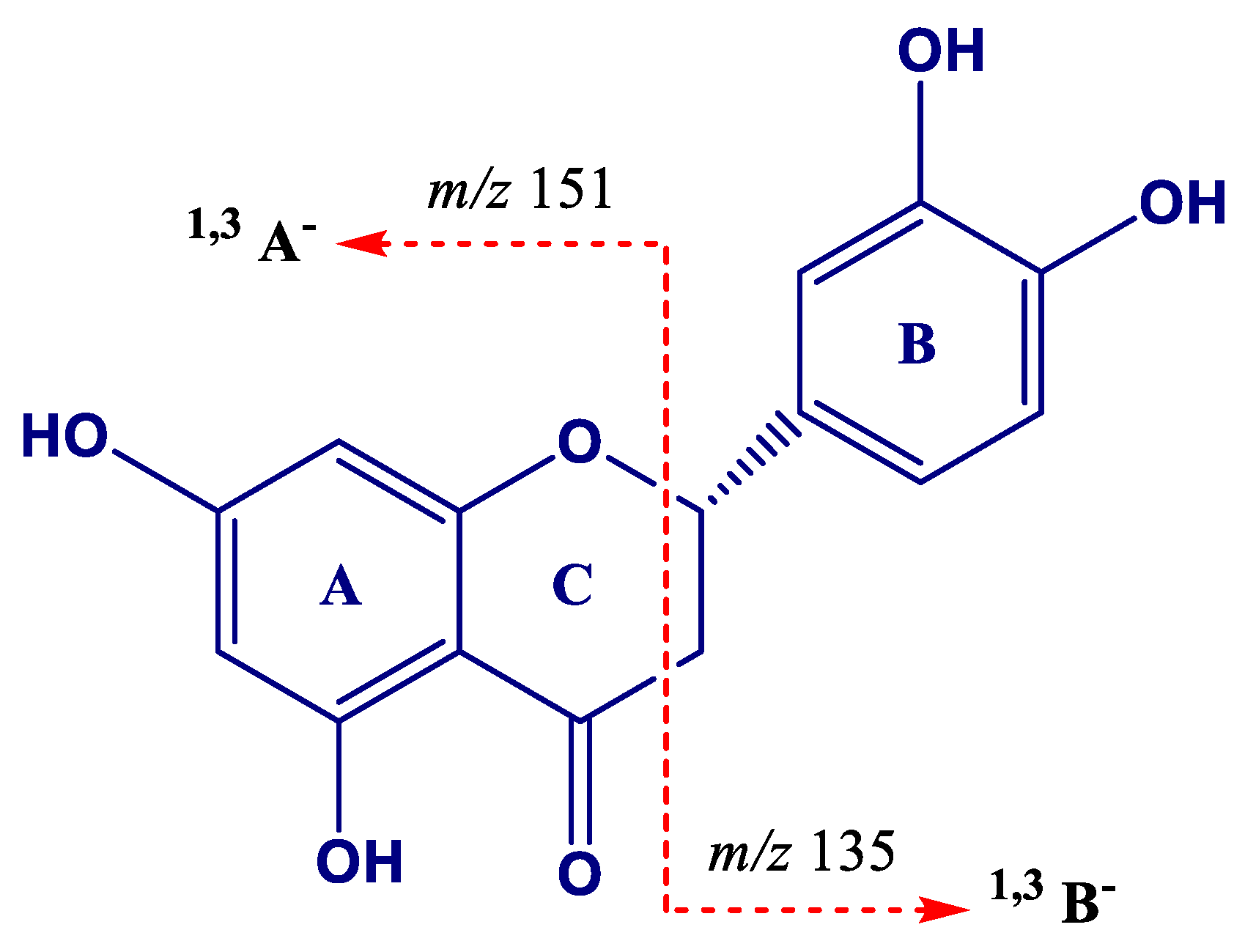
| Peak | Compounds | R.T. (min) | Accurate Mass [M − H]− | MS/MS Ions (% Intensity) | Δm (ppm) | Ion Molecular Formula (IMF) |
|---|---|---|---|---|---|---|
| Hydroxybenzoic Acids and Derivatives | ||||||
| 1 | Monogalloyl-glucose (1) | 3.60 | 331.0668 | 271.0446(60), 211.0237(20), 169.0133(100) | −0.936 | C13H15O10 |
| 2 | Gallic acid * | 4.30 | 169.0141 | 125.0239(100) | −0.985 | C7H5O5 |
| 3 | Monogalloyl-glucose (2) | 5.38 | 331.0664 | 271.0446(60), 211.0237(20), 169.0133(100) | −1.510 | C13H15O10 |
| 4 | Protocatechuic acid-O-hexoside (1) | 7.39 | 315.0719 | 153.0185(100) | −1.001 | C13H15O9 |
| 5 | Protocatechuic acid | 7.66 | 153.0192 | 109.0290(100) | −1.058 | C7H5O4 |
| 6 | Protocatechuic acid-O-hexoside (2) | 8.52 | 315.0718 | 153.0186(100) | −0.969 | C13H15O9 |
| 7 | Syringic acid hexoside | 8.75 | 359.0981 | 197.0446(100) | −0.724 | C15H19O10 |
| 8 | 2-Hydroxybenzoic acid | 9.47 | 137.0243 | 93.0341(100) | −1.075 | C7H5O3 |
| 9 | Hydroxybenzoyl hexoside | 9.96 | 299.0770 | 137.0236(100) | −1.039 | C13H15O8 |
| 10 | 4-Hydroxybenzoic acid * | 10.02 | 137.0242 | 93.0340(100) | −1.221 | C7H5O3 |
| 11 | Ellagic acid hexoside | 13.99 | 463.0518 | 300.9974(100) | 0.079 | C20H15O13 |
| 12 | Gallic acid ethyl ester | 14.28 | 197.0454 | 169.0135(100) | −1.252 | C9H9O5 |
| 13 | Ellagic acid pentoside | 16.03 | 433.0416 | 300.9975(100), 299.9898(40) | −0.766 | C19H13O12 |
| 14 | Ellagic acid * | 16.94 | 300.9980 | 257.0079(100), 229.0132(60), 185.0235(30) | −1.131 | C14H5O8 |
| Hydroxycinnamic Acids Derivatives | ||||||
| 15 | Caftaric acid | 9.27 | 311.0406 | 179.0343(45), 149.0965(100) | −0.852 | C13H11O9 |
| 16 | Coutaric acid | 11.18 | 295.0456 | 163.0392(100) | −1.052 | C13H11O8 |
| Flavanols (Proanthocyanidins) | ||||||
| 17 | (Epi)gallocatechin (EGC)→(epi)gallocatechin (1) | 5.69 | 609.1244 | 483.0917(20), 441.0812(85), 423.0707(100), 305.0656(45) | −0.966 | C30H25O14 |
| 18 | (Epi)gallocatechin→(epi)gallocatechin (2) | 7.81 | 609.1240 | 483.0913(25), 441.0808(100), 423.0703(90), 305.0653(40) | −1.574 | C30H25O14 |
| 19 | Procyanidin trimer (1) | 7.90 | 865.1982 | 739.1638(15), 713.1479(20), 695.1379(100), 577.1329(25), 451.1016(15), 425.0867(15), 407.0761(20), 289.0709(10) | −1.118 | C45H37O18 |
| 20 | (Epi)gallocatechin→(epi)catechin (1) | 8.01 | 593.1305 | 467.0970(30), 425.0868(100), 407.0763(80), 303.0501(10), 289.0710(70) | 0.718 | C30H25O13 |
| 21 | (Epi)gallocatechin (1) | 8.30 | 305.0665 | 261.0761(45), 221.0448(75), 219.0658(65), 179.0344(100) | 0.303 | C15H13O7 |
| 22 | (Epi)catechin→(epi)gallocatechin | 8.71 | 593.1301 | 467.0973(55), 441.0817(40), 423.0715(100), 305.0660(60), 287.0554(10) | 0.010 | C30H25O13 |
| 23 | (Epi)gallocatechin→(epi)catechin→(epi)gallocatechin | 9.09 | 897.1869 | 771.1526(25), 729.1426(15), 711.1321(100), 593.1277(30), 305.0651(5) | −1.445 | C45H37O20 |
| 24 | (Epi)gallocatechin→(epi)catechin (2) | 9.73 | 593.1296 | 467.0970(25), 425.0868(100), 407.0761(50), 303.0510(5), 289.0709(40) | −0.715 | C30H25O13 |
| 25 | (Epi)gallocatechin→(epi)gallocatechin→(epi)catechin | 9.86 | 897.1868 | 771.1538(15), 729.1435(25), 711.1331(100), 593.1280(20), 303.0499(10), 289.0706(10) | −1.779 | C45H37O20 |
| 26 | Procyanidin dimer (1) | 10.44 | 577.1342 | 451.1026 (95), 425.0872 (80), 407.0766 (95), 289.0711(100) | −1.610 | C30H25O12 |
| 27 | (Epi)gallocatechin (2) | 10.73 | 305.0662 | 261.0762(40), 221.0449(75), 219.0659(60), 179.0345(100) | 1.122 | C15H13O7 |
| 28 | Procyanidin dimer (2) | 10.89 | 577.1347 | 451.1027(75), 425.0873(100), 407.0768(90), 289.0713(70) | −0.657 | C30H25O12 |
| 29 | Catechin * | 11.39 | 289.0715 | 245.0810(100), 205.0498(40), 179.1342(20) | −0.800 | C15H13O6 |
| 30 | Procyanidin trimer (2) | 11.95 | 865.1978 | 739.1652(40), 713.1495 (30), 695.1393(100), 577.1340(50), 451.1024(30), 425.0870(25), 407.0764(55), 289.0710 (20) | −0.841 | C45H37O18 |
| 31 | Procyanidin tetramer | 12.13 | 576.1271 [M − 2H]2− | 1027.2257(35), 865.1948(30), 863.1793(65), 739.1640(30), 451.1016(45), 407.0756(25), 289.0705(100), 287.0548(40) | −0.424 | C60H50O24 |
| 32 | Procyanidin dimer (3) | 12.28 | 577.1348 | 451.1020(50), 425.0867(100), 407.0761(90), 289.0708(45) | −0.883 | C30H25O12 |
| 33 | Procyanidin dimer (4) | 12.57 | 577.1356 | 451.1026 (65), 425.0872(100), 407.0766(95), 289.0711(60) | −0.815 | C30H25O12 |
| 34 | (Epi)gallocatechin→(epi)catechin (3) | 12.77 | 593.1290 | 467.0968(30), 425.0865(100), 407.0758(70), 303.0500(10), 289.0707(40) | −1.713 | C30H25O13 |
| 35 | (Epi)catechin→(epi)gallocatechin gallate (EGCG) | 12.81 | 745.1400 | 593.1257(80), 575.1136(55), 457.0757(25), 441.0809(5), 423.0703(100), 305.0655(15) | −2.003 | C37H29O17 |
| 36 | Procyanidin pentamer (1) | 13.04 | 720.1580 [M − 2H]2− | 1315.2897(25), 1153.2595 (25), 1151.2442(55), 1027.2273(25), 865.1955(60), 863.1794(100), 739.1645(25), 635.6298(80), 577.1333(90), 575.1178(80), 451.5990(30), 407.0758(45), 289.0707(70), 287.0550 (40) | 0.504 | C75H62O30 |
| 37 | Epicatechin * | 13.36 | 289.0715 | 245.0810(100), 205.0498(40), 179.1342(10) | −0.766 | C15H13O6 |
| 38 | (Epi)gallocatechin gallate (EGCG) | 13.74 | 457.0770 | 331.0445(70), 305.0653(35), 169.0135(100) | −1.454 | C22H17O11 |
| 39 | (Epi)catechin gallate (ECG)→(epi)catechin (1) | 13.86 | 729.1459 | 603.1140(5), 577.1345(100), 439.066(5), 425.0876(25), 407.0769(50), 289.0713(10) | −0.230 | C37H29O16 |
| 40 | Procyanidin dimer (5) | 14.01 | 577.1346 | 451.1025(55), 425.0872(90), 407.0766(100), 289.0711(50) | −0.883 | C30H25O12 |
| 41 | Procyanidin trimer (3) | 14.39 | 865.1979 | 739.1643(40), 713.1486(35), 695.1385(100), 577.1334(55), 451.1020(30), 425.0866(25), 407.0760(50), 289.0708(15) | −0.760 | C45H37O18 |
| 42 | (Epi)catechin→(epi)catechin gallate | 14.84 | 729.1449 | 603.1133(30), 577.1312(35), 559.0917(30), 451.1024(45), 441.0818(30), 407.0764(100), 289.0710(30) | −1.629 | C37H29O16 |
| 43 | Procyanidin trimer (4) | 15.43 | 865.1959 | 739.1643(50), 713.1489(20), 695.1384(100), 577.1334(50), 451.1020(25), 425.0866(25), 407.0759(55), 289.0707(10) | −3.094 | C45H37O18 |
| 44 | Procyanidin pentamer (2) | 15.59 | 720.1578 [M − 2H]2− | 1315.2906(25), 1153.2585(20), 1151.2437(45), 1027.2261(20), 865.1949(50), 863.1793(75), 739.1639(20), 635.6275(100), 577.1330(60), 575.1175(60), 451.1016(20), 407.0756(25), 289.0706(45), 287.0548(25) | −0.079 | C75H62O30 |
| 45 | Epicatechin gallate * | 16.89 | 441.0825 | 289.0710(100), 169.0137(30) | −0.476 | C27H17O10 |
| 46 | (Epi)catechin gallate→(epi)catechin (2) | 17.54 | 729.1441 | 603.1125(5), 577.1332(100), 439.0657(5), 425.0866(25), 407.0760(30), 289.0705(5) | 2.184 | C37H29O16 |
| 47 | Theaflavin | 20.88 | 563.1191 | 545.1064(100), 519.1272(45), 425.0857(40), 407.0751(65), 397.0908(30), 379.0805(60) | −1.295 | C29H23O12 |
| Flavonols | ||||||
| 48 | Myricetin-O-hexoside | 15.49 | 479.0821 | 317.0288(60), 316.0211(100) | −1.239 | C21H19O13 |
| 49 | Quercetin-O-glucoside * | 17.23 | 463.0876 | 301.0337(100), 299.0174(30) | −1.402 | C21H19O12 |
| 50 | Quercetin-3-O-glucuronide | 17.28 | 477.0674 | 301.0341(100) | −0.700 | C21H17O13 |
| Flavanonol | ||||||
| 51 | Taxifolin | 17.01 | 303.0505 | 285.0391(100), 177.0185(10), 125.0239(10) | −1.636 | C15H11O7 |
| 52 | Astilbin (1) | 17.39 | 449.1090 | 303.0497(100), 285.0391(90), 151.0030(30) | −0.211 | C21H21O11 |
| 53 | Astilbin (2) | 18.22 | 449.1086 | 303.0497(100), 285.0391(85), 151.0030(25) | −0.612 | C21H21O11 |
| Flavanones | ||||||
| 54 | Eriodictyol-O-hexoside (1) | 13.58 | 449.1090 | 287.0545(100) | −0.367 | C21H21O11 |
| 55 | Eriodictyol-O-hexoside (2) | 19.03 | 449.1087 | 287.0548(100) | −0.433 | C21H21O11 |
| 56 | Eriodictyol | 21.08 | 287.0556 | 151.0033(100), 135.0448(10) | −1.677 | C15H11O6 |
| Stilbenoids | ||||||
| 57 | Resveratrol C-hexoside | 13.66 | 389.1241 | 269.0812(100), 241.0864(10), 299.0915(5) | −0.208 | C20H21O8 |
| 58 | Restrisol (A or B) | 15.15 | 471.1441 | 377.1015(90), 349.1067(100), 255.0651(80) | −1.754 | C28H23O7 |
| 59 | Oxidized stilbenoid dimer (1) | 16.25 | 471.1438 | 349.1066(100) | −2.391 | C28H23O7 |
| 60 | Oxidized stilbenoid dimer (2) | 16.85 | 471.1443 | 349.1066(100) | −1.351 | C28H23O7 |
| 61 | Stilbenoid dimer (1)(heterodimer) | 17.78 | 469.1283 | 451.1181(100), 375.0866(30), 363.0869(35) | −1.484 | C28H21O7 |
| 62 | (E)-Piceatannol * | 18.01 | 243.0659 | 225.0551(100), 201.0551(65), 159.0447(20) | −1.202 | C14H11O4 |
| 63 | Oxidized stilbenoid dimer (3) | 18.93 | 471.1446 | 349.1067(100) | −2.709 | C28H23O7 |
| 64 | Stilbenoid dimer (2)(heterodimer) | 19.23 | 469.1287 | 375.0855(20), 363.0857(100) | −1.122 | C28H21O7 |
| 65 | Viniferin diglycoside | 19.33 | 777.2387 | 615.1848(95), 453.1327(100) | −1.670 | C40H41O16 |
| 66 | Pallidol | 19.51 | 453.1340 | 359.0915(100), 265.0497(10) | −0.820 | C28H21O5 |
| 67 | (E)-resveratrol * | 20.27 | 227.0707 | 185.0602(65), 143.0496(20) | −2.543 | C14H11O3 |
| 68 | Stilbene dimer (resveratrol+resveratrol) | 20.62 | 453.1339 | 359.0912(100) | -1.593 | C28H21O6 |
| 69 | Resveratrol dimer-O-hexoside | 20.85 | 615.1866 | 453.1325(100) | −0.951 | C34H31O11 |
| 70 | Stilbenoid tetramer (Hopeaphenol *) | 21.17 | 905.2582 | 811.2159(100), 717.1748(80) | −2.331 | C56H41O12 |
| 71 | Stilbenoid dimer (3)(Scirpusin A) | 21.35 | 469.1283 | 451.1182(25), 385.1066(50), 375.0860(100), 359.0912(30), 347.0919(15) | −2.188 | C28H21O7 |
| 72 | Stilbenoid tetramer (Isohopeaphenol *) | 21.41 | 905.2580 | 811.2158(100), 717.1747(80) | −2.596 | C56H41O12 |
| 73 | (E)-ε-viniferin * | 21.68 | 453.1335 | 359.0924(100), 347.0919(40) | −1.813 | C28H21O6 |
| 74 | (E)-ω-viniferin | 21.87 | 453.1335 | 435.1230(35), 411.1229(25), 359.0918(100), 347.0918(55) | −1.593 | C28H21O6 |
| 75 | Stilbenoid tetramer | 22.05 | 905.2576 | 887.2472(25), 811.2159(50), 799.2164(100), 359.0917(30) | −3.004 | C56H41O12 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Escobar-Avello, D.; Lozano-Castellón, J.; Mardones, C.; Pérez, A.J.; Saéz, V.; Riquelme, S.; von Baer, D.; Vallverdú-Queralt, A. Phenolic Profile of Grape Canes: Novel Compounds Identified by LC-ESI-LTQ-Orbitrap-MS. Molecules 2019, 24, 3763. https://doi.org/10.3390/molecules24203763
Escobar-Avello D, Lozano-Castellón J, Mardones C, Pérez AJ, Saéz V, Riquelme S, von Baer D, Vallverdú-Queralt A. Phenolic Profile of Grape Canes: Novel Compounds Identified by LC-ESI-LTQ-Orbitrap-MS. Molecules. 2019; 24(20):3763. https://doi.org/10.3390/molecules24203763
Chicago/Turabian StyleEscobar-Avello, Danilo, Julián Lozano-Castellón, Claudia Mardones, Andy J. Pérez, Vania Saéz, Sebastián Riquelme, Dietrich von Baer, and Anna Vallverdú-Queralt. 2019. "Phenolic Profile of Grape Canes: Novel Compounds Identified by LC-ESI-LTQ-Orbitrap-MS" Molecules 24, no. 20: 3763. https://doi.org/10.3390/molecules24203763
APA StyleEscobar-Avello, D., Lozano-Castellón, J., Mardones, C., Pérez, A. J., Saéz, V., Riquelme, S., von Baer, D., & Vallverdú-Queralt, A. (2019). Phenolic Profile of Grape Canes: Novel Compounds Identified by LC-ESI-LTQ-Orbitrap-MS. Molecules, 24(20), 3763. https://doi.org/10.3390/molecules24203763











