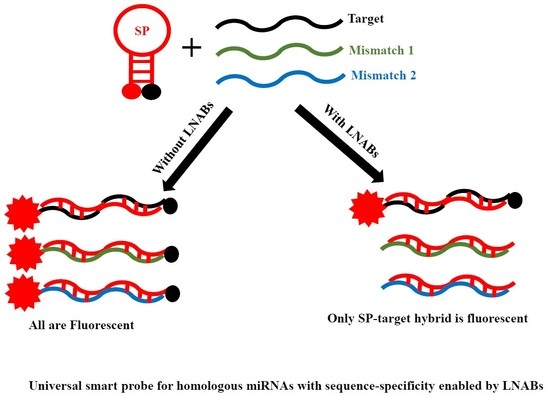
Detection of Several Homologous MicroRNAs by a Single Smart Probe System Consisting of Linear Nucleic Acid Blockers
Abstract
1. Introduction
2. Results and Discussion
2.1. The SP and Target Sequences
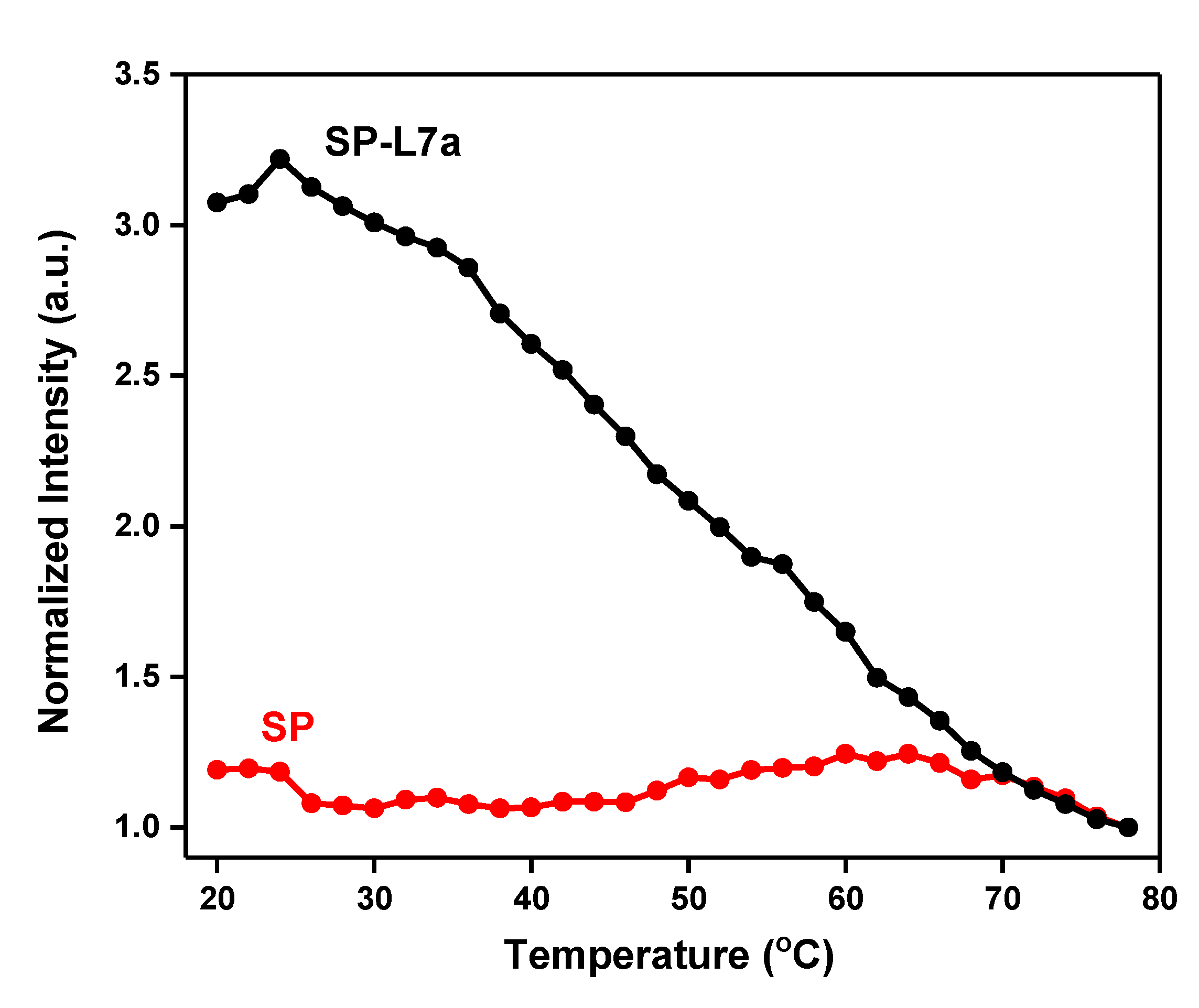
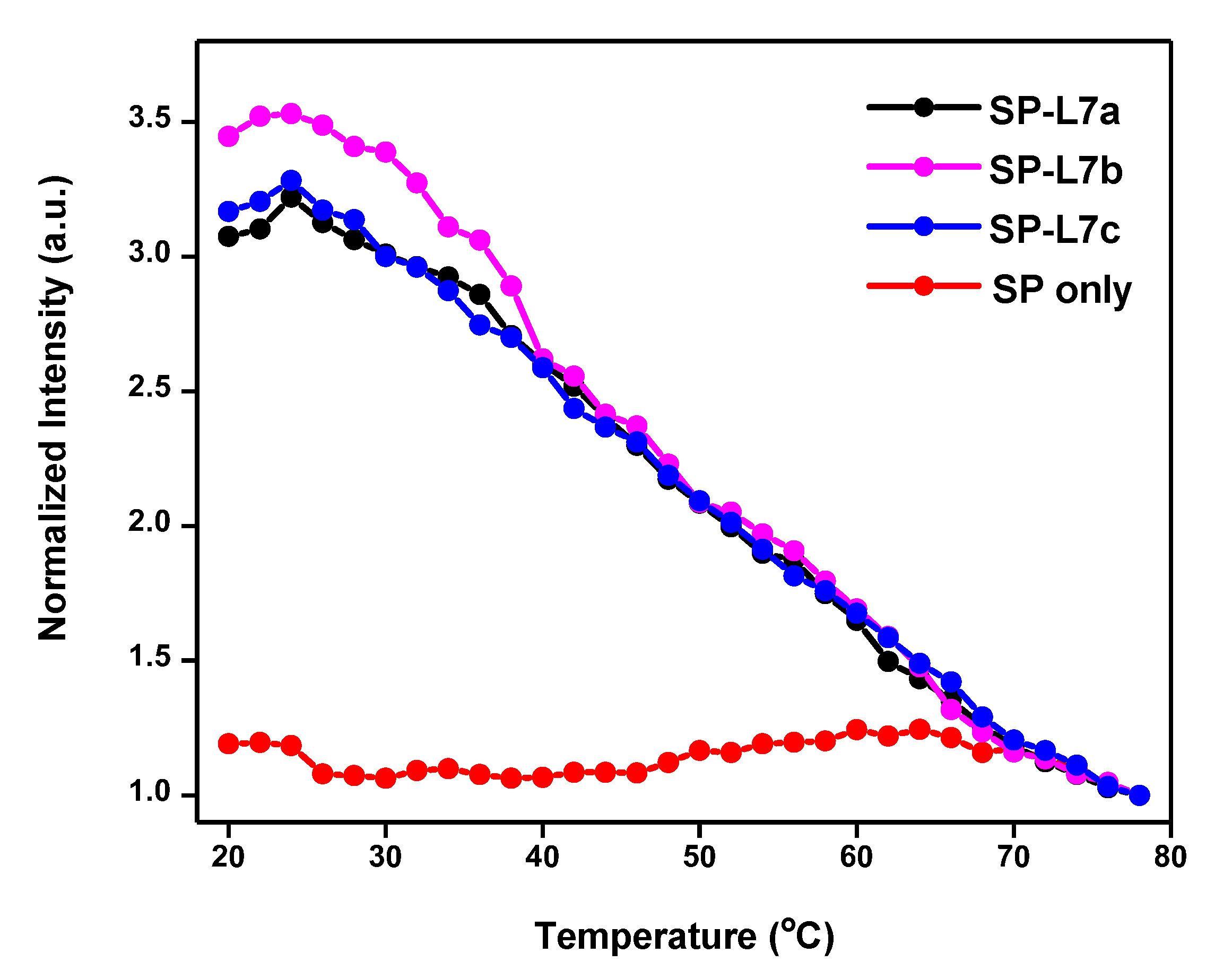
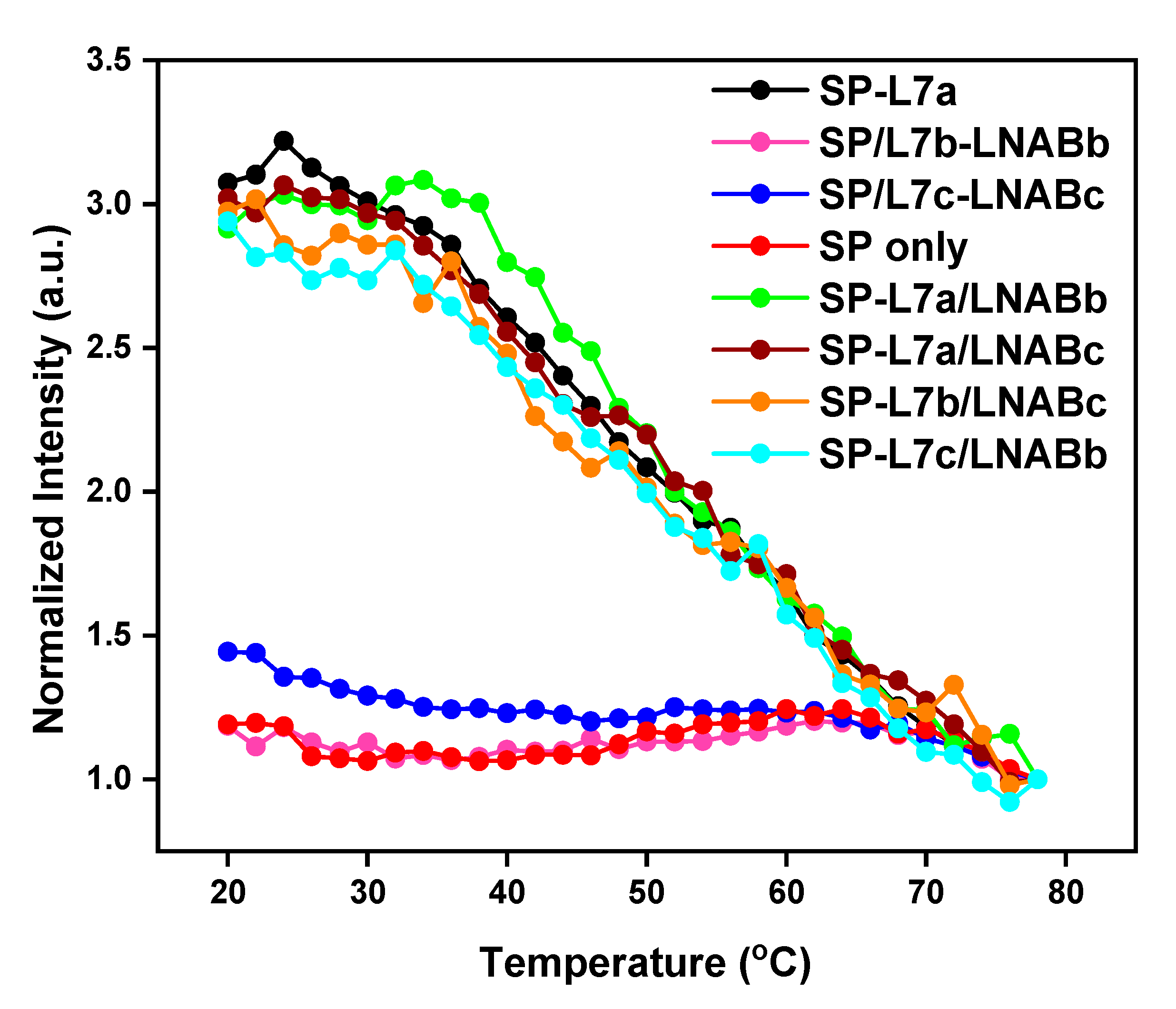
2.2. Melting Profiles
2.3. Recognition of L7b and L7c through Non-Specific Hybridization
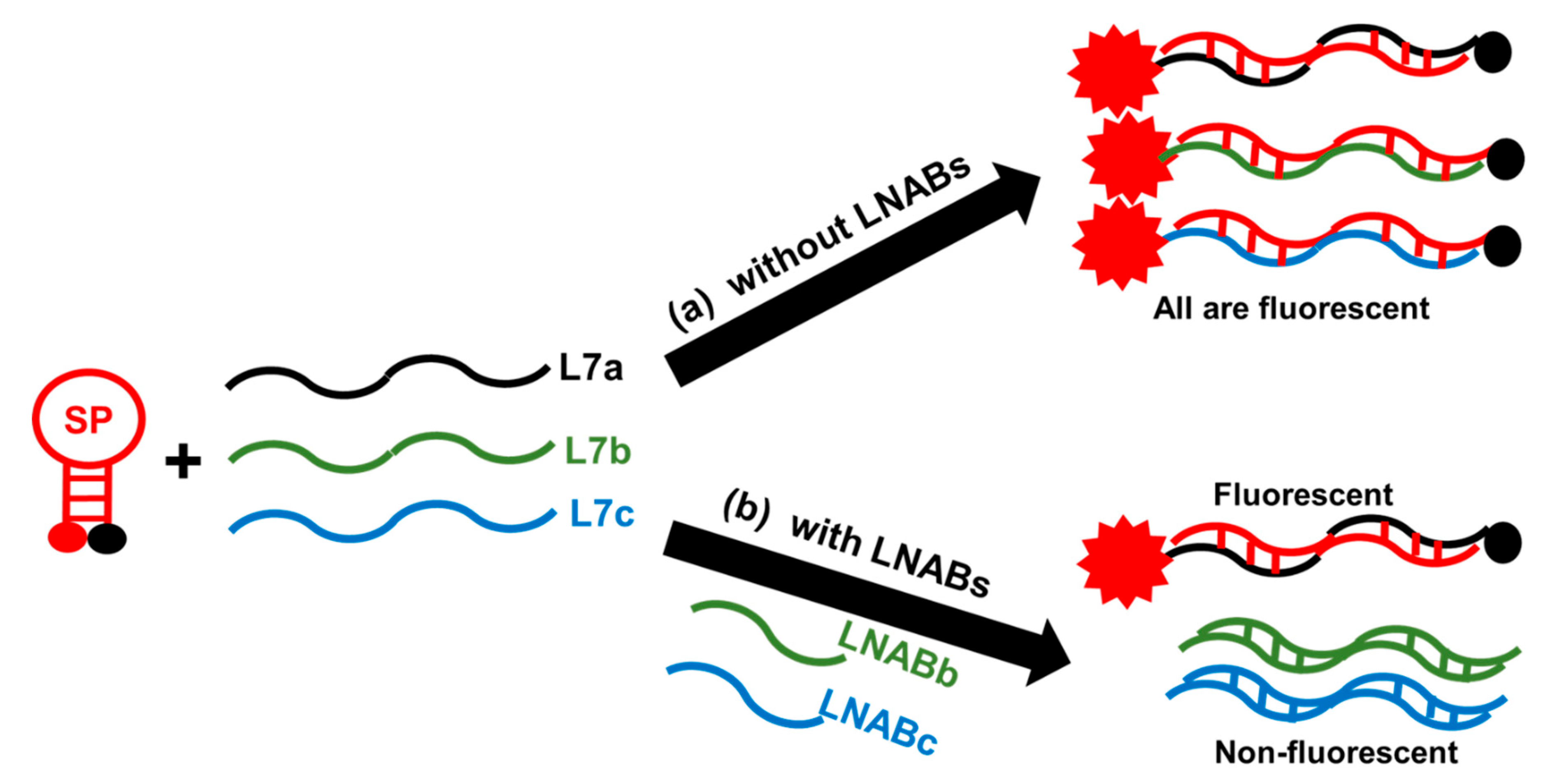
2.4. Sequence-Specificity by Means of Linear Nucleic Acid Blockers (LNABs)
2.5. No Evidence of Target-LNABs Interaction
2.6. Concentration-Dependent Studies
3. Materials and Methods
3.1. Materials
3.2. Absorbance and Fluorescence Measurements
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Knemeyer, J.-P.; Marme, N.; Sauer, M. Probes for detection of specific DNA sequences at the single-molecule level. Anal. Chem. 2000, 72, 3717–3724. [Google Scholar] [CrossRef] [PubMed]
- Stohr, K.; Hafner, B.; Nolte, O.; Wolfrum, J.; Sauer, M.; Herten, D.-P. Species-specific identification of mycobacterial 16s rRNA PCR amplicons using smart probes. Anal. Chem. 2005, 77, 7195–7203. [Google Scholar] [CrossRef] [PubMed]
- Marme, N.; Knemeyer, J.-P. Sensitive bioanalysis—Combining single-molecule spectroscopy with mono-labeled self-quenching probes. Anal. Bioanal. Chem. 2007, 388, 1075–1085. [Google Scholar] [CrossRef] [PubMed]
- Heinlein, T.; Knemeyer, J.-P.; Piestert, O.; Sauer, M. Photoinduced electron transfer between fluorescent dyes and guanosine residues in DNA-hairpins. J. Phys. Chem. B 2003, 107, 7957–7964. [Google Scholar] [CrossRef]
- Misra, A.; Kumar, P.; Gupta, K.C. Synthesis of hairpin probe using deoxyguanosine as a quencher: Fluorescence and hybridization studies. Anal. Biochem. 2007, 364, 86–88. [Google Scholar] [CrossRef] [PubMed]
- Steenken, S.; Jovanovic, S.V. How easily oxidizable is DNA? One-electron reduction potentials of adenosine and guanosine radicals in aqueous solution. J. Am. Chem. Soc. 1997, 119, 617–618. [Google Scholar] [CrossRef]
- Seidel, C.A.M.; Schulz, A.; Sauer, M. Nucleobase-specific quenching of fluorescent dyes. 1. Nucleobase one-electron redox potentials and their correlation with static and dynamic quenching efficiencies. J. Phys. Chem. 1996, 100, 5541–5553. [Google Scholar] [CrossRef]
- Song, C.; Zhang, C.; Zhao, M. Rapid and sensitive detection of DNA polymerase fidelity by singly labeled smart fluorescent probes. Biosens. Bioelectron. 2011, 26, 2699–2702. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Zhao, M. Real-time monitoring of the activity and kinetics of T4 polynucleotide kinase by a singly labeled DNA-hairpin smart probe coupled with lambda exonuclease cleavage. Anal. Chem. 2009, 81, 1383–1388. [Google Scholar] [CrossRef]
- Ma, C.; Chen, H.; Han, R.; He, H.; Zeng, W. Fluorescence detection of adenosine triphosphate using smart probe. Anal. Biochem. 2012, 429, 8–10. [Google Scholar] [CrossRef]
- Oladepo, S.A.; Loppnow, G.R. Self-quenching smart probes as a platform for the detection of sequence-specific UV-induced DNA photodamage. Anal. Bioanal. Chem. 2010, 397, 2949–2957. [Google Scholar] [CrossRef] [PubMed]
- Nair, S.G.; Loppnow, G.R. Multiplexed, UVC-induced, sequence-dependent DNA damage detection. Photochem. Photobiol. 2013, 89, 884–890. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, A.; Habl, G.; Sauer, M.; Wolfrum, J.; Hoheisel, J.; Marmé, N.; Knemeyer, J.-P. New hairpin-structured DNA probes: Alternatives to classical molecular beacons. In Proceedings of the SPIE, San Jose, CA, USA, 14 February 2007; Volume 6444, pp. 64440M-1–64440M-7. [Google Scholar]
- Tyagi, S.; Kramer, F.R. Molecular beacons: Probes that fluoresce upon hybridization. Nat. Biotechnol. 1996, 14, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, G.; Tyagi, S.; Libchaber, A.; Kramer, F.R. Thermodynamic basis of the enhanced specificity of structured DNA probes. Proc. Natl. Acad. Sci. USA 1999, 96, 6171–6176. [Google Scholar] [CrossRef] [PubMed]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Si, M.-L.; Wu, H.; Mo, Y.-Y. MicroRNA-21 targets the tumor suppressor gene Tropomyosin 1 (TPM1). J. Biol. Chem. 2007, 282, 14328–14336. [Google Scholar] [CrossRef] [PubMed]
- Cissell, K.A.; Rahimi, Y.; Shrestha, S.; Hunt, E.A.; Deo, S.K. Bioluminescence-based detection of microRNA, miR21 in breast cancer cells. Anal. Chem. 2008, 80, 2319–2325. [Google Scholar] [CrossRef] [PubMed]
- Croce, C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009, 10, 704–714. [Google Scholar] [CrossRef] [PubMed]
- Catuogno, S.; Esposito, C.L.; Quintavalle, C.; Cerchia, L.; Condorelli, G.; de Franciscis, V. Recent advance in biosensors for microRNAs detection in cancer. Cancers 2011, 3, 1877–1898. [Google Scholar] [CrossRef]
- Lee, Y.S.; Dutta, A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007, 21, 1025–1030. [Google Scholar] [CrossRef] [PubMed]
- Yanaihara, N.; Caplen, N.; Bowman, E.; Seike, M.; Kumamoto, K.; Yi, M.; Stephens, R.M.; Okamoto, A.; Yokota, J.; Tanaka, T.; et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006, 9, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.M.; Grosshans, H.; Shingara, J.; Byrom, M.; Jarvis, R.; Cheng, A.; Labourier, E.; Reinert, K.L.; Brown, D.; Slack, F.J. RAS is regulated by the let-7 microRNA family. Cell 2005, 120, 635–647. [Google Scholar] [CrossRef] [PubMed]
- Crockett, A.O.; Wittwer, C.T. Fluorescein-labelled oligonucleotides for real-time PCR: Using inherent quenching of deoxyguanosine nucleotides. Anal. Biochem. 2001, 290, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.-X.; Liang, Z.Z.; Ma, Y.-H.; Kong, D.-M.; Hong, Z.-Y. G-quadruplex fluorescent probe-mediated real-time rolling circle amplification strategy for highly sensitive microRNA detection. Anal. Chim. Acta 2016, 943, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-Q.; Zhang, M.; Yin, B.-C.; Ye, B.-C. Attomolar ultrasensitive microrna detection by DNA-scaffolded silver-nanocluster probe based on isothermal amplification. Anal. Chem. 2012, 84, 5165–5169. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Song, Y.; Zhang, C.; Zhi, X.; Fu, H.; Ma, Y.; Chen, Y.; Pan, F.; Wang, K.; Ni, J.; et al. Circulating miR-16-5p and miR-19b-3p as two novel potential biomarkers to indicate progression of gastric cancer. Theranostics 2015, 5, 733–745. [Google Scholar] [CrossRef] [PubMed]
- Kilic, T.; Topkaya, S.N.; Ariksoysal, D.O.; Ozsoz, M.; Ballar, P.; Erac, Y.; Gozen, O. Electrochemical based detection of microRNA, miR21 in breast cancer cells. Biosens. Bioelectron. 2012, 38, 195–201. [Google Scholar] [CrossRef]
- Park, J.; Yeo, J.-S. Colorimetric detection of microRNA miR-21 based on nanoplasmonic core-satellite assembly. Chem. Commun. 2014, 50, 1366–1368. [Google Scholar] [CrossRef]
- Guven, B.; Dudak, F.C.; Boyaci, I.H.; Tamer, U.; Ozsoz, M. SERS-based direct and sandwich assay methods from miR-21 detection. Analyst 2014, 139, 1141–1147. [Google Scholar] [CrossRef]
- Du, W.; Lv, M.; Li, J.; Yu, R.; Jiang, J. A ligation-based loop-mediated isothermal amplification (ligation-lamp) strategy for highly selective microRNA detection. Chem. Commun. 2016, 52, 12721–12724. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.-M.; Du, W.-F.; Xi, Q.; Ge, J.; Jiang, J.-H. Isothermal Nucleic Acid Amplification Strategy by Cyclic Enzymatic Repairing for Highly Sensitive MicroRNA Detection. Anal. Chem. 2014, 86, 6763–6767. [Google Scholar] [CrossRef] [PubMed]
- Duan, R.; Zuo, X.; Wang, S.; Quan, X.; Chen, D.; Chen, Z.; Jiang, L.; Fan, C.; Xian, F. Lab in a tube. ultrasensitive detection of microRNAs at the single-cell level and in breast cancer patients using quadratic isothermal amplification. J. Am. Chem. Soc. 2013, 135, 4604–4607. [Google Scholar] [CrossRef]
- Cheng, F.-F.; Jiang, N.; Li, X.; Zhang, L.; Hu, L.; Chen, X.; Jiang, L.-P.; Abdel-Halim, E.S.; Zhu, J.-J. Target-triggered triple isothermal cascade amplification strategy for ultrasensitive microRNA-21 detection at sub-attomole level. Biosens. Bioelectron. 2016, 85, 891–896. [Google Scholar] [CrossRef] [PubMed]
- Duan, R.; Zuo, X.; Wang, S.; Quan, X.; Chen, D.; Chen, Z.; Jiang, L.; Fan, C.; Xian, F. Quadratic isothermal amplification for the detection of microRNA. Nat. Protoc. 2014, 9, 597–607. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, C.; Zhi, X.; Ramon, G.A.; Liu, Y.; Zhang, C.; Pan, F.; Cui, D. Hairpin DNA-templated silver nanoclusters as novel beacons in strand displacement amplification for microRNA detection. Anal. Chem. 2016, 88, 1294–1302. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.-L.; Zhang, C.-Y. Sensitive detection of microRNAs with hairpin probe-based circular exponential amplification assay. Anal. Chem. 2012, 84, 7037–7042. [Google Scholar] [CrossRef]
- Oladepo, S.A. Design and characterization of a singly labelled fluorescent smart probe for in vitro detection of miR-21. Appl. Spectrosc. 2018, 72, 79–88. [Google Scholar] [CrossRef]
- Zuo, X.; Xia, F.; Xiao, Y.; Plaxco, K.W. Sensitive and selective amplified fluorescence DNA detection based on exonuclease III-aided target recycling. J. Am. Chem. Soc. 2010, 132, 1816–1818. [Google Scholar] [CrossRef]
- Oladepo, S.A.; Yusuf, B.O. Simple protocol for sequence-specific detection of mixed-base nucleic acids using a smart probe with NABs. Anal. Biochem. 2019, 568, 53–56. [Google Scholar] [CrossRef]
- Friedrich, A.; Hoheisel, J.D.; Marme, N.; Knemeyer, J.-P. DNA-probes for the highly sensitive identification of single nucleotide polymorphism using single-molecule spectroscopy. FEBS Lett. 2007, 581, 1644–1648. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Tang, W.; Zhao, Y.; Li, N.; Liu, F. Ultra-specific discrimination of single-nucleotide mutations using sequestration-assisted molecular beacons. Chem. Sci. 2017, 8, 1021–1026. [Google Scholar] [CrossRef] [PubMed]
- Poon, K.; MacGregor, R.B., Jr. Unusual behavior exhibited by multistranded guanine-rich DNA complexes. Biopolymers 1998, 45, 427–434. [Google Scholar] [CrossRef]
- Ong, A.A.L.; Toh, D.-F.K.; Krishna, M.S.; Patil, K.M.; Okamura, K.; Chen, G. Incorporating 2-thiouracil into short double-stranded RNA-binding peptide nucleic acids for enhanced recognition of A-U pairs and for targeting a microRNA hairpin precursor. Biochemistry 2019, 58, 3444–3453. [Google Scholar] [CrossRef] [PubMed]
- Patil, K.M.; Chen, G. Recognition of RNA sequence and structure by duplex and triplet formation: Targeting miRNA and pre-miRNA. In Modified Nucleic Acids in Biology and Medicine, 1st ed.; Jurga, S., Erdmann, V., Barciszewski, J., Eds.; Springer: Cham, Switzerland, 2016; pp. 299–317. [Google Scholar]
- El-Yazbi, A.F.; Loppnow, G.R. Chimeric RNA–DNA molecular beacons for quantification of nucleic acids, single nucleotide polymophisms, and nucleic acid damage. Anal. Chem. 2013, 85, 4321–4327. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds SP, L7a, L7b, L7c, LNABb and LNABc may not be available from the authors, interested parties may obtain them from a reliable oligo manufacturer. |


| Sequence | Description (Label) |
|---|---|
| /56-FAM/-ACCCGGAACTATACAACCTACTACCTCACCGGGTGGG-3′ | Smart Probe (SP) a |
| 5′-rUrGrArGrGrUrArGrUrArGrGrUrUrGrUrArUrArGrUrU-3′ | Let-7a (L7a) |
| 5′-rUrGrArGrGrUrArGrUrArGrGrUrUrGrUrGrUrGrGrUrU-3′ | Let-7b (L7b) b |
| 5′-rUrGrArGrGrUrArGrUrArGrGrUrUrGrUrArUrGrGrUrU-3′ | Let-7c (L7c) b |
| 5′-AACCACACAACCTACTACCTCA-3′ | Linear nucleic acid blocker for L7b (LNABb) c |
| 5′-AACCATACAACCTACTACCTCA-3′ | Linear nucleic acid blocker for L7c (LNABc) d |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oladepo, S.A.; Yusuf, B.O. Detection of Several Homologous MicroRNAs by a Single Smart Probe System Consisting of Linear Nucleic Acid Blockers. Molecules 2019, 24, 3691. https://doi.org/10.3390/molecules24203691
Oladepo SA, Yusuf BO. Detection of Several Homologous MicroRNAs by a Single Smart Probe System Consisting of Linear Nucleic Acid Blockers. Molecules. 2019; 24(20):3691. https://doi.org/10.3390/molecules24203691
Chicago/Turabian StyleOladepo, Sulayman A., and Basiru O. Yusuf. 2019. "Detection of Several Homologous MicroRNAs by a Single Smart Probe System Consisting of Linear Nucleic Acid Blockers" Molecules 24, no. 20: 3691. https://doi.org/10.3390/molecules24203691
APA StyleOladepo, S. A., & Yusuf, B. O. (2019). Detection of Several Homologous MicroRNAs by a Single Smart Probe System Consisting of Linear Nucleic Acid Blockers. Molecules, 24(20), 3691. https://doi.org/10.3390/molecules24203691