Alisol A Suppresses Proliferation, Migration, and Invasion in Human Breast Cancer MDA-MB-231 Cells
Abstract
1. Introduction
2. Material and Methods
2.1. Cell Culture and Reagents
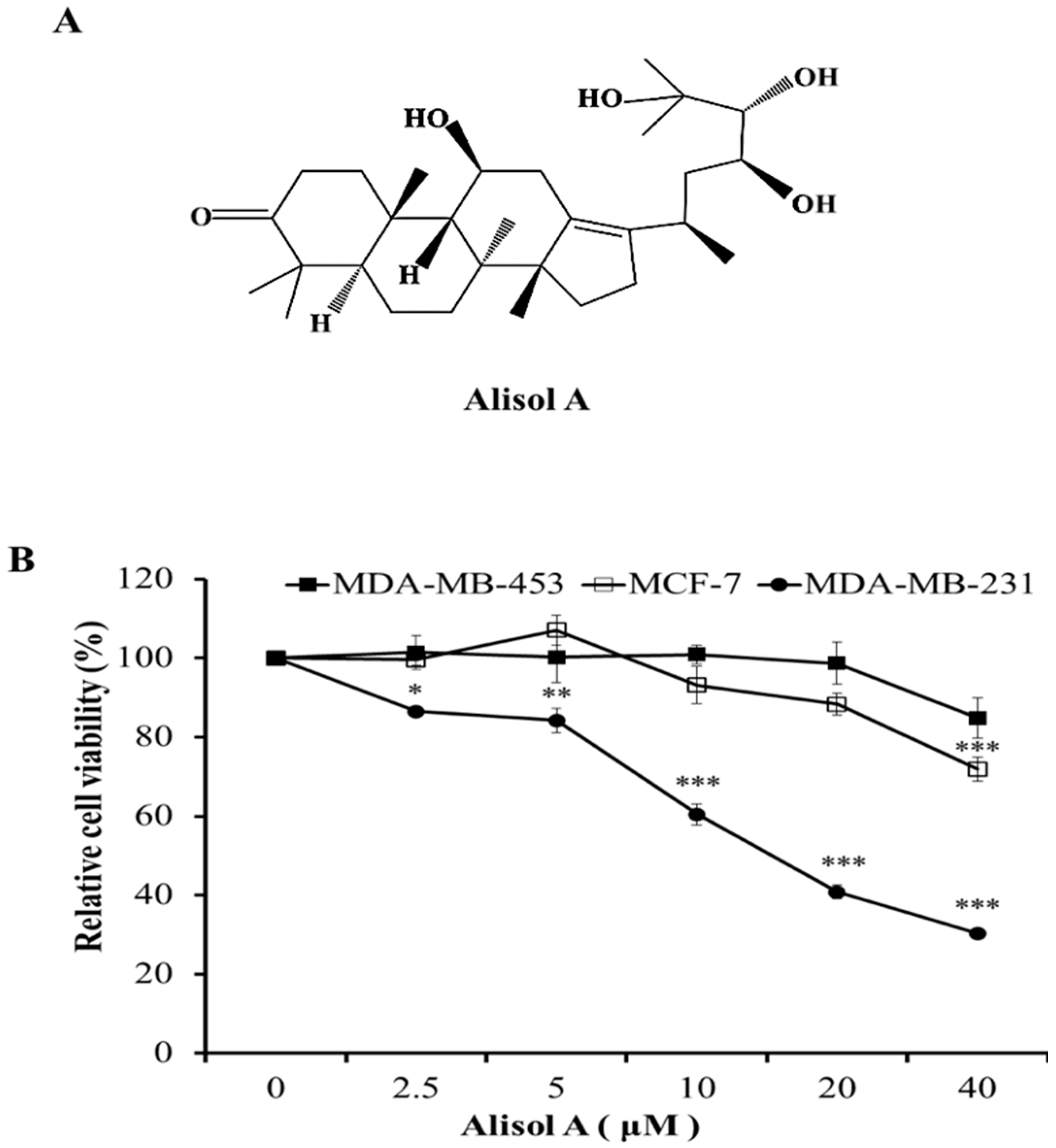
2.2. Cell Viability Assay
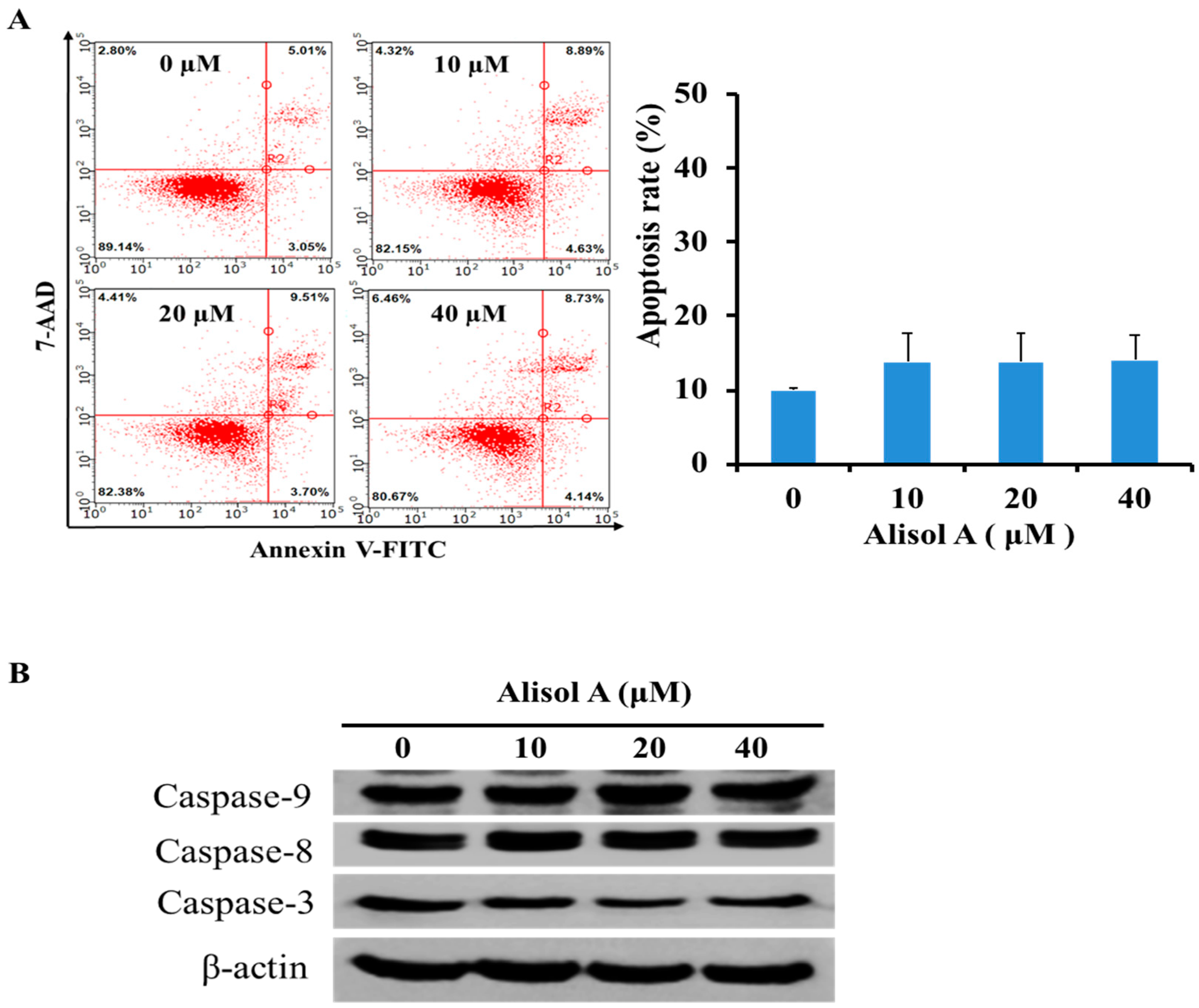
2.3. Annexin V-FITC/PI Double Staining Assay
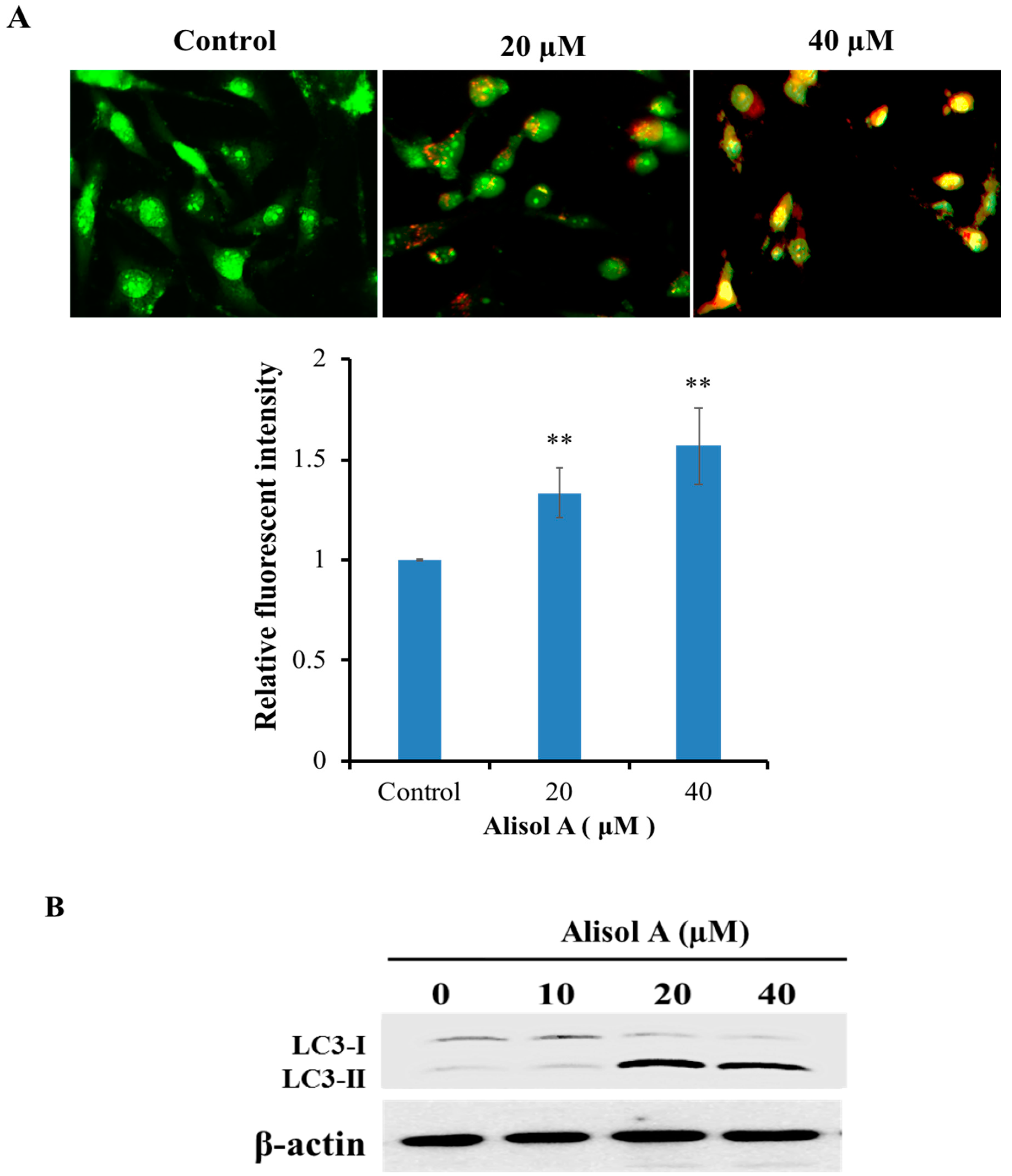
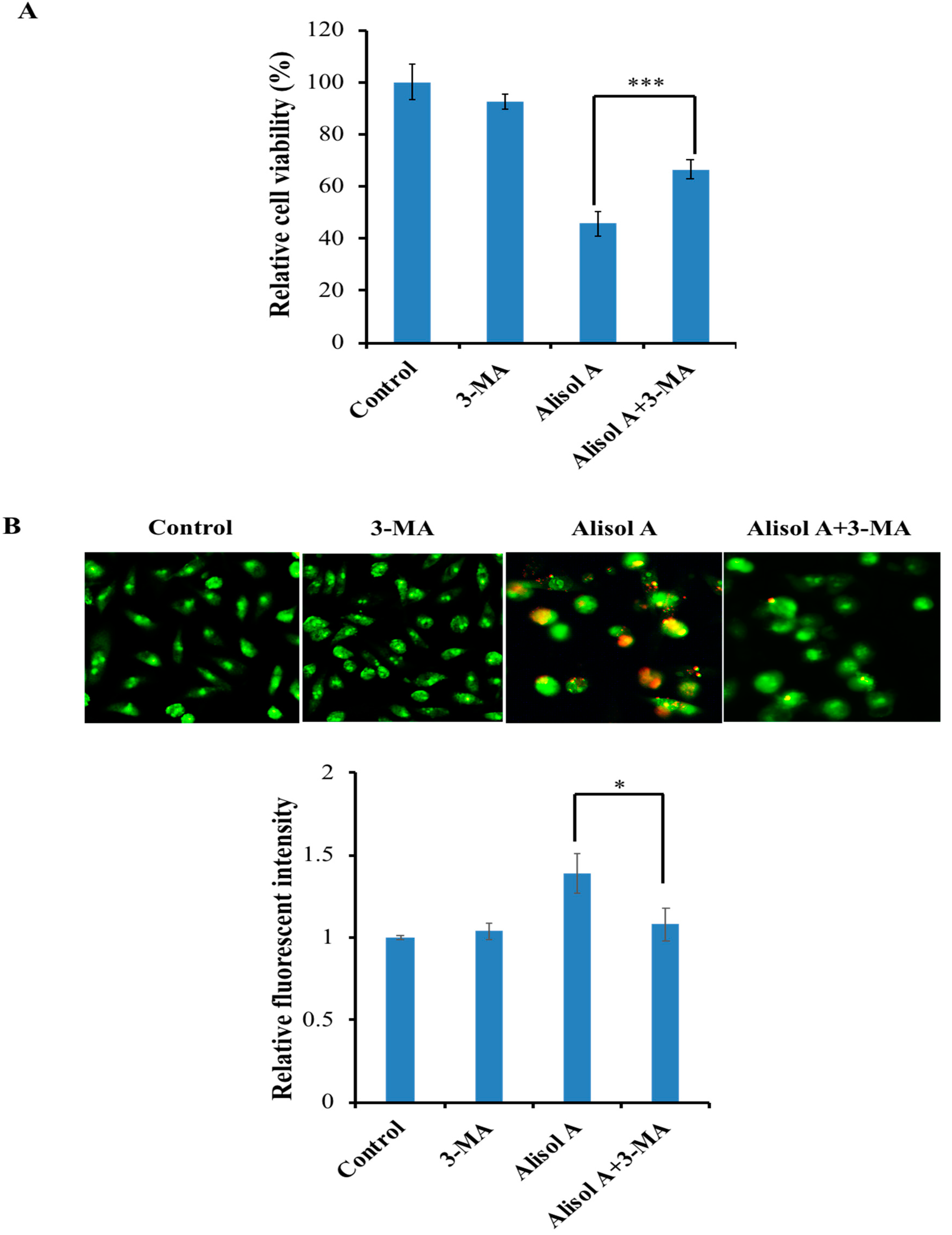
2.4. Acridine Orange Staining
2.5. Cell Cycle Analysis
2.6. Wound Healing Assay
2.7. Transwell Migration and Invasion Assays
2.8. Gelatin Zymography
2.9. Western Blotting
2.10. Statistical Analysis
3. Results
3.1. Effects of Alisol A on Cell Viability in Human Breast Cancer Cells
3.2. Effects of Alisol A on Induction of Cell Apoptosis
3.3. Effects of Alisol A on Induction of Autophagy
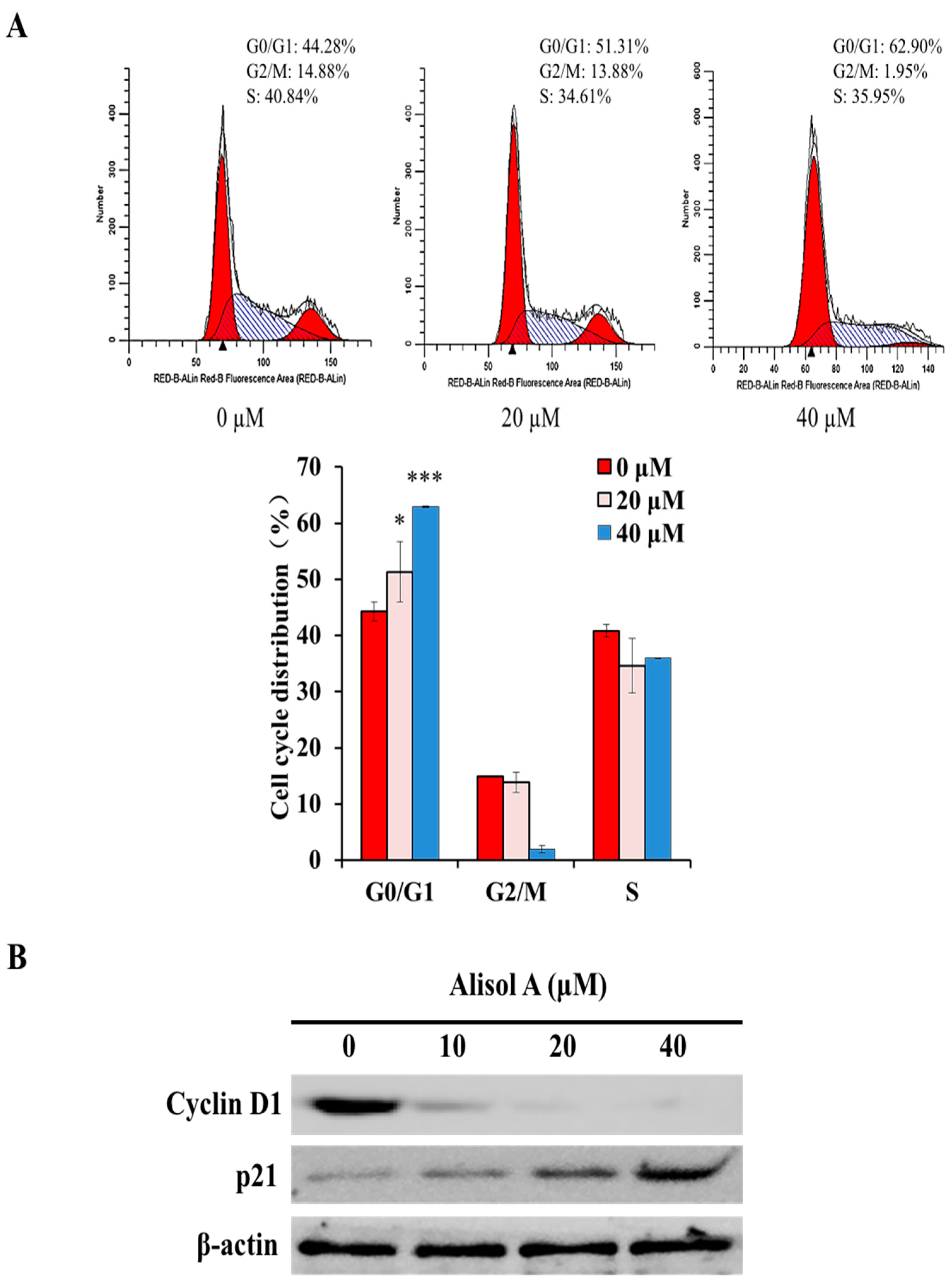
3.4. Effects of Alisol A on Cell Cycle Arrest
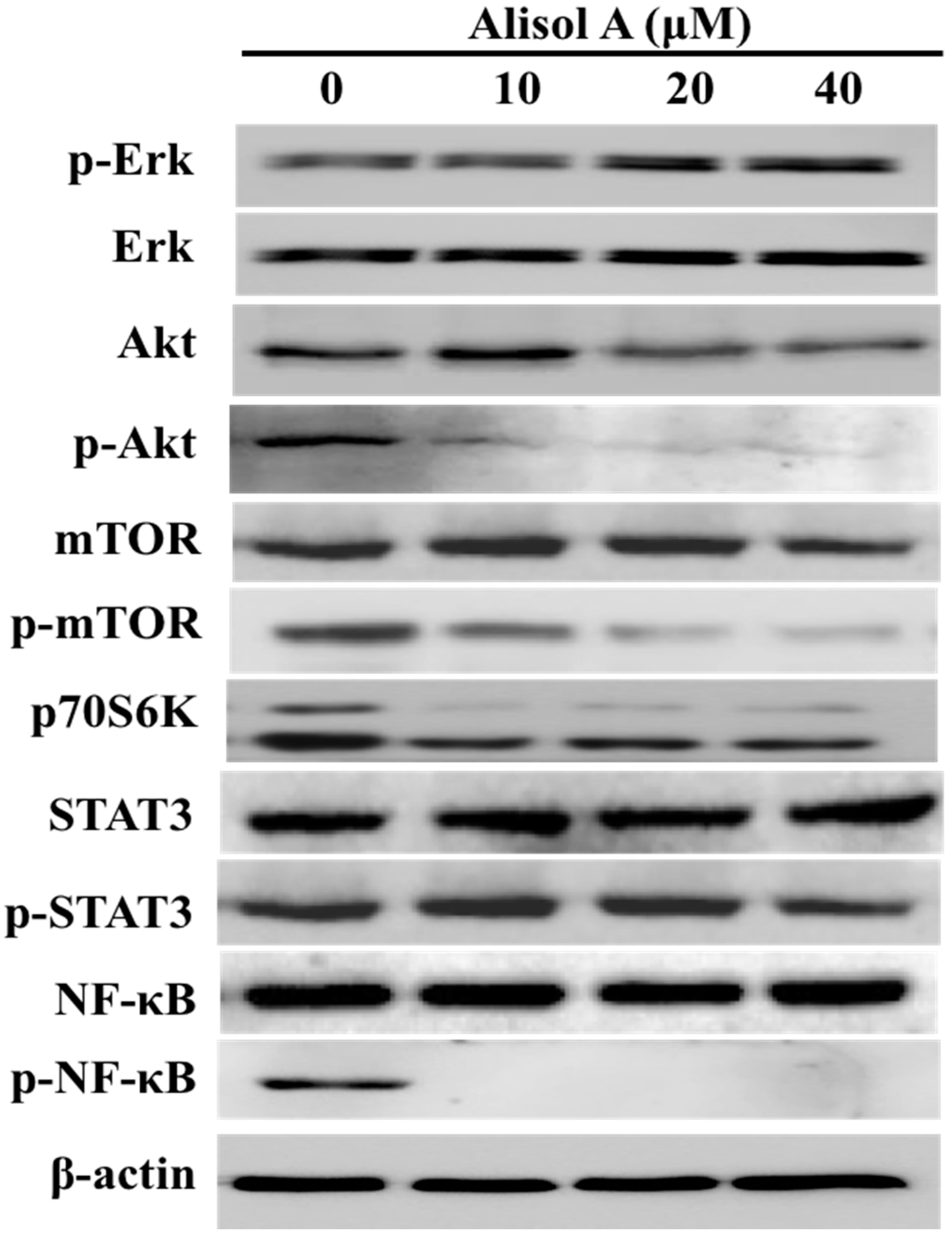
3.5. Effects of Alisol A on Autophagy Related Signaling Pathways
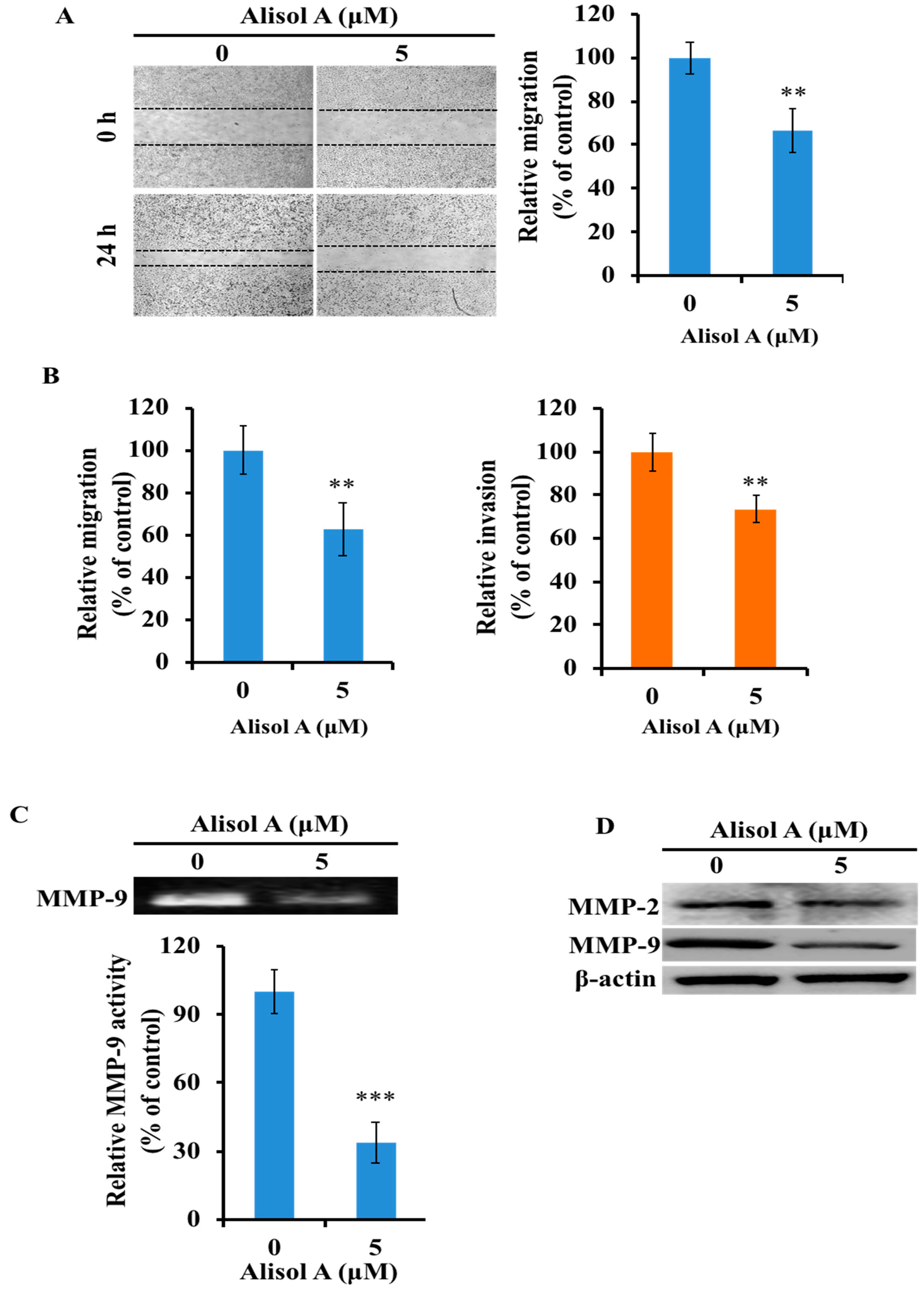
3.6. Effects of Alisol A on Cell Migration and Invasion
3.7. Effects of Alisol A on MMP-2 and MMP-9 in MDA-MB-231 Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| DMSO | Dimethyl sulfoxide |
| MTT | 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide |
| FBS | Fetal bovine serum |
| DMEM | Dulbecco’s Modified Eagle’s Medium |
| BSA | Bovine serum albumin |
| PVDF | Polyvinylidene fluoride |
| PBS | Phosphate buffered saline |
| AO | Acridine orange |
| PI | Propidium iodide |
| 3-MA | 3-methyladenine |
| TBST | Tris-buffered saline-5% Tween 20 |
| SDS | Sodium dodecyl sulfate |
| ECL | Enhanced chemiluminescence |
| TNBC | Triple-negative breast cancer |
| ER | Estrogen receptor |
| PR | Progesterone receptor |
| HER2 | Human epidermal growth factor receptor 2 |
| STAT3 | Signal transducer and activator of transcription 3 |
| NF-κB | Nuclear factor-κB |
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Ginsburg, O.; Bray, F.; Coleman, M.P.; Vanderpuye, V.; Eniu, A.; Kotha, S.R.; Sarker, M.; Huong, T.T.; Allemani, C.; Dvaladze, A.; et al. The global burden of women’s cancers: A grand challenge in global health. Lancet 2017, 389, 847–860. [Google Scholar] [CrossRef]
- Parise, C.A.; Caggiano, V. Breast cancer survival defined by the ER/PR/HER2 subtypes and a surrogate classification according to tumor grade and immunohistochemical biomarkers. J. Cancer Epidemiol. 2014, 2014, 469251. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 2012, 75, 311–335. [Google Scholar] [CrossRef] [PubMed]
- Mann, J. Natural products in cancer chemotherapy: Past, present and future. Nat. Rev. Cancer 2002, 2, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Koehn, F.E.; Carter, G.T. The evolving role of natural products in drug discovery. Nat. Rev. Drug Discov. 2005, 4, 206–220. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Chen, H.; Zhao, Y.Y. Traditional uses, phytochemistry, pharmacology, toxicology and quality control of Alisma orientale (Sam.) Juzep: A review. J. Ethnopharmacol. 2014, 158, 373–387. [Google Scholar] [CrossRef] [PubMed]
- Xiu, R.J. Microcirculation and traditional Chinese medicine. Jama 1988, 260, 1755–1757. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Song, H.H.; Ahn, K.S.; Oh, S.R.; Sadikot, R.T.; Joo, M. Ethanol extract of the tuber of Alisma orientale reduces the pathologic features in a chronic obstructive pulmonary disease mouse model. J. Ethnopharmacol. 2016, 188, 21–30. [Google Scholar] [CrossRef]
- Xu, W.; Li, T.; Qiu, J.F.; Wu, S.S.; Huang, M.Q.; Lin, L.G.; Zhang, Q.W.; Chen, X.P.; Lu, J.J. Anti-proliferative activities of terpenoids isolated from Alisma orientalis and their structure-activity relationships. Anticancer Agents Med. Chem. 2015, 15, 228–235. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.L.; Xu, Y.L.; Tang, Z.H.; Xu, X.H.; Chen, X.; Li, T.; Ding, C.Y.; Huang, M.Q.; Chen, X.P.; Wang, Y.T.; et al. Effects of alisol B 23-acetate on ovarian cancer cells: G1 phase cell cycle arrest, apoptosis, migration and invasion inhibition. Phytomedicine 2016, 23, 800–809. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.H.; Zhao, L.J.; Li, Y. Alisol B acetate induces apoptosis of SGC7901 cells via mitochondrial and phosphatidylinositol 3-kinases/Akt signaling pathways. World J. Gastroenterol. 2009, 15, 2870–2877. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Qu, H. Study on the hypoglycemic activities and metabolism of alcohol extract of Alismatis Rhizoma. Fitoterapia 2012, 83, 1046–1053. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.Q.; Feng, Y.L.; Tian, T.; Chen, H.; Yin, L.; Zhao, Y.Y.; Lin, R.C. Diuretic and anti-diuretic activities of fractions of Alismatis rhizoma. J. Ethnopharmacol. 2014, 157, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Lu, C.; Wu, Q.; Chen, J.; Gu, W.; Du, W.; You, M. Study on antitumor molecular mechanism of Alisols based on p53DNA. Int. J. Biol. Macromol. 2018, 116, 1163–1174. [Google Scholar] [CrossRef]
- Xu, W.; Li, X.; Lin, N.; Zhang, X.; Huang, X.; Wu, T.; Tai, Y.; Chen, S.; Wu, C.H.; Huang, M.; et al. Pharmacokinetics and tissue distribution of five major triterpenoids after oral administration of Rhizoma Alismatis extract to rats using ultra high-performance liquid chromatography-tandem mass spectrometry. J. Pharm. Biomed. Anal. 2017, 146, 314–323. [Google Scholar] [CrossRef] [PubMed]
- Polyak, K. Heterogeneity in breast cancer. J. Clin. Investig. 2011, 121, 3786–3788. [Google Scholar] [CrossRef]
- Dent, R.; Trudeau, M.; Pritchard, K.I.; Hanna, W.M.; Kahn, H.K.; Sawka, C.A.; Lickley, L.A.; Rawlinson, E.; Sun, P.; Narod, S.A. Triple-negative breast cancer: Clinical features and patterns of recurrence. Clin. Cancer Res. 2007, 13, 4429–4434. [Google Scholar] [CrossRef]
- Huang, Y.T.; Huang, D.M.; Chueh, S.C.; Teng, C.M.; Guh, J.H. Alisol B acetate, a triterpene from Alismatis rhizoma, induces Bax nuclear translocation and apoptosis in human hormone-resistant prostate cancer PC-3 cells. Cancer Lett. 2006, 231, 270–278. [Google Scholar] [CrossRef]
- Zhang, A.; Sheng, Y.; Zou, M. Antiproliferative activity of Alisol B in MDA-MB-231 cells is mediated by apoptosis, dysregulation of mitochondrial functions, cell cycle arrest and generation of reactive oxygen species. Biomed. Pharmacother. 2017, 87, 110–117. [Google Scholar] [CrossRef]
- Fong, W.F.; Wang, C.; Zhu, G.Y.; Leung, C.H.; Yang, M.S.; Cheung, H.Y. Reversal of multidrug resistance in cancer cells by Rhizoma Alismatis extract. Phytomedicine 2007, 14, 160–165. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, J.X.; Shen, X.L.; Wan, C.K.; Tse, A.K.; Fong, W.F. Reversal of P-glycoprotein-mediated multidrug resistance by Alisol B 23-acetate. Biochem. Pharmacol. 2004, 68, 843–855. [Google Scholar] [CrossRef] [PubMed]
- Knickle, A.; Fernando, W.; Greenshields, A.L.; Rupasinghe, H.P.V.; Hoskin, D.W. Myricetin-induced apoptosis of triple-negative breast cancer cells is mediated by the iron-dependent generation of reactive oxygen species from hydrogen peroxide. Food Chem. Toxicol. 2018, 118, 154–167. [Google Scholar] [CrossRef] [PubMed]
- Moeinifard, M.; Hassan, Z.M.; Fallahian, F.; Hamzeloo-Moghadam, M.; Taghikhani, M. Britannin induces apoptosis through AKT-FOXO1 pathway in human pancreatic cancer cells. Biomed. Pharmacother. 2017, 94, 1101–1110. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Emr, S.D. Autophagy as a regulated pathway of cellular degradation. Science 2000, 290, 1717–1721. [Google Scholar] [CrossRef] [PubMed]
- Mathew, R.; Karantza-Wadsworth, V.; White, E. Role of autophagy in cancer. Nat. Rev. Cancer 2007, 7, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Rubinsztein, D.C.; Codogno, P.; Levine, B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat. Rev. Drug Discov. 2012, 11, 709–730. [Google Scholar] [CrossRef]
- Amaravadi, R.K.; Lippincott-Schwartz, J.; Yin, X.M.; Weiss, W.A.; Takebe, N.; Timmer, W.; DiPaola, R.S.; Lotze, M.T.; White, E. Principles and current strategies for targeting autophagy for cancer treatment. Clin. Cancer Res. 2011, 17, 654–666. [Google Scholar] [CrossRef]
- Li, B.; Wang, W.; Li, Z.; Chen, Z.; Zhi, X.; Xu, J.; Li, Q.; Wang, L.; Huang, X.; Wang, L.; et al. MicroRNA-148a-3p enhances cisplatin cytotoxicity in gastric cancer through mitochondrial fission induction and cyto-protective autophagy suppression. Cancer Lett. 2017, 410, 212–227. [Google Scholar] [CrossRef]
- Peng, Y.F.; Shi, Y.H.; Ding, Z.B.; Ke, A.W.; Gu, C.Y.; Hui, B.; Zhou, J.; Qiu, S.J.; Dai, Z.; Fan, J. Autophagy inhibition suppresses pulmonary metastasis of HCC in mice via impairing anoikis resistance and colonization of HCC cells. Autophagy 2013, 9, 2056–2068. [Google Scholar] [CrossRef]
- Hsin, M.C.; Hsieh, Y.H.; Wang, P.H.; Ko, J.L.; Hsin, I.L.; Yang, S.F. Hispolon suppresses metastasis via autophagic degradation of cathepsin S in cervical cancer cells. Cell Death Dis. 2017, 8, e3089. [Google Scholar] [CrossRef] [PubMed]
- Kenific, C.M.; Thorburn, A.; Debnath, J. Autophagy and metastasis: Another double-edged sword. Curr. Opin. Cell Biol. 2010, 22, 241–245. [Google Scholar] [CrossRef]
- Washington, M.N.; Suh, G.; Orozco, A.F.; Sutton, M.N.; Yang, H.; Wang, Y.; Mao, W.; Millward, S.; Ornelas, A.; Atkinson, N.; et al. ARHI (DIRAS3)-mediated autophagy-associated cell death enhances chemosensitivity to cisplatin in ovarian cancer cell lines and xenografts. Cell Death Dis. 2015, 6, e1836. [Google Scholar] [CrossRef] [PubMed]
- Glick, D.; Barth, S.; Macleod, K.F. Autophagy: Cellular and molecular mechanisms. J. Pathol. 2010, 221, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Tanida, I.; Ueno, T.; Kominami, E. LC3 and Autophagy. Methods Mol. Biol. 2008, 445, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.E.; Johansen, T. Following autophagy step by step. BMC Biol. 2011, 9, 39. [Google Scholar] [CrossRef]
- Schaaf, M.B.; Keulers, T.G.; Vooijs, M.A.; Rouschop, K.M. LC3/GABARAP family proteins: Autophagy-(un)related functions. FASEB J. 2016, 30, 3961–3978. [Google Scholar] [CrossRef]
- Jung, C.H.; Ro, S.H.; Cao, J.; Otto, N.M.; Kim, D.H. mTOR regulation of autophagy. FEBS Lett. 2010, 584, 1287–1295. [Google Scholar] [CrossRef]
- He, C.; Klionsky, D.J. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 2009, 43, 67–93. [Google Scholar] [CrossRef]
- Tapia, O.; Riquelme, I.; Leal, P.; Sandoval, A.; Aedo, S.; Weber, H.; Letelier, P.; Bellolio, E.; Villaseca, M.; Garcia, P.; et al. The PI3K/AKT/mTOR pathway is activated in gastric cancer with potential prognostic and predictive significance. Virchows Arch. 2014, 465, 25–33. [Google Scholar] [CrossRef]
- Basseres, D.S.; Baldwin, A.S. Nuclear factor-kappaB and inhibitor of kappaB kinase pathways in oncogenic initiation and progression. Oncogene 2006, 25, 6817–6830. [Google Scholar] [CrossRef] [PubMed]
- Bours, V.; Bonizzi, G.; Bentires-Alj, M.; Bureau, F.; Piette, J.; Lekeux, P.; Merville, M. NF-kappaB activation in response to toxical and therapeutical agents: Role in inflammation and cancer treatment. Toxicology 2000, 153, 27–38. [Google Scholar] [CrossRef]
- Hu, W.; Chen, S.S.; Zhang, J.L.; Lou, X.E.; Zhou, H.J. Dihydroartemisinin induces autophagy by suppressing NF-kappaB activation. Cancer Lett. 2014, 343, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Xavier, C.P.; Lima, C.F.; Preto, A.; Seruca, R.; Fernandes-Ferreira, M.; Pereira-Wilson, C. Luteolin, quercetin and ursolic acid are potent inhibitors of proliferation and inducers of apoptosis in both KRAS and BRAF mutated human colorectal cancer cells. Cancer Lett. 2009, 281, 162–170. [Google Scholar] [CrossRef] [PubMed]
- Jarnicki, A.; Putoczki, T.; Ernst, M. Stat3: Linking inflammation to epithelial cancer—More than a "gut" feeling? Cell Div. 2010, 5, 14. [Google Scholar] [CrossRef] [PubMed]
- Sinibaldi, D.; Wharton, W.; Turkson, J.; Bowman, T.; Pledger, W.J.; Jove, R. Induction of p21WAF1/CIP1 and cyclin D1 expression by the Src oncoprotein in mouse fibroblasts: Role of activated STAT3 signaling. Oncogene 2000, 19, 5419–5427. [Google Scholar] [CrossRef]
- Wohlbold, L.; Fisher, R.P. Behind the wheel and under the hood: Functions of cyclin-dependent kinases in response to DNA damage. DNA Repair 2009, 8, 1018–1024. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Stewart, Z.A.; Pietenpol, J.A. p53 Signaling and cell cycle checkpoints. Chem. Res. Toxicol. 2001, 14, 243–263. [Google Scholar] [CrossRef]
- Overall, C.M.; Lopez-Otin, C. Strategies for MMP inhibition in cancer: Innovations for the post-trial era. Nat. Rev. Cancer 2002, 2, 657–672. [Google Scholar] [CrossRef]
- Kong, C.S.; Kim, Y.A.; Kim, M.M.; Park, J.S.; Kim, J.A.; Kim, S.K.; Lee, B.J.; Nam, T.J.; Seo, Y. Flavonoid glycosides isolated from Salicornia herbacea inhibit matrix metalloproteinase in HT1080 cells. Toxicol. in Vitro 2008, 22, 1742–1748. [Google Scholar] [CrossRef]
- Wu, C.; Qiu, S.; Liu, P.; Ge, Y.; Gao, X. Rhizoma Amorphophalli inhibits TNBC cell proliferation, migration, invasion and metastasis through the PI3K/Akt/mTOR pathway. J. Ethnopharmacol. 2018, 211, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Y.; Lu, H.F.; Hsu, S.C.; Kuo, C.L.; Chang, S.J.; Lin, J.J.; Wu, P.P.; Liu, J.Y.; Lee, C.H.; Chung, J.G.; et al. Bufalin inhibits migration and invasion in human hepatocellular carcinoma SK-Hep1 cells through the inhibitions of NF-kB and matrix metalloproteinase-2/-9-signaling pathways. Environ. Toxicol. 2015, 30, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.R.; Jin, L.; Tian, M.X.; Jiang, X.F.; Yang, L.X.; Ding, Z.B.; Shen, Y.H.; Peng, Y.F.; Gao, D.M.; Zhou, J.; et al. Caveolin-1 promotes tumor growth and metastasis via autophagy inhibition in hepatocellular carcinoma. Clin. Res. Hepatol. Gastroenterol. 2016, 40, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Su, Z.; DeWitt, J.P.; Xie, L.; Chen, Y.; Li, X.; Han, L.; Li, D.; Xia, J.; Zhang, Y.; et al. Fluvastatin prevents lung adenocarcinoma bone metastasis by triggering autophagy. EBioMedicine 2017, 19, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Catalano, M.; D’Alessandro, G.; Lepore, F.; Corazzari, M.; Caldarola, S.; Valacca, C.; Faienza, F.; Esposito, V.; Limatola, C.; Cecconi, F.; et al. Autophagy induction impairs migration and invasion by reversing EMT in glioblastoma cells. Mol. Oncol. 2015, 9, 1612–1625. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lou, C.; Xu, X.; Chen, Y.; Zhao, H. Alisol A Suppresses Proliferation, Migration, and Invasion in Human Breast Cancer MDA-MB-231 Cells. Molecules 2019, 24, 3651. https://doi.org/10.3390/molecules24203651
Lou C, Xu X, Chen Y, Zhao H. Alisol A Suppresses Proliferation, Migration, and Invasion in Human Breast Cancer MDA-MB-231 Cells. Molecules. 2019; 24(20):3651. https://doi.org/10.3390/molecules24203651
Chicago/Turabian StyleLou, Chenghua, Xintong Xu, Yan Chen, and Huajun Zhao. 2019. "Alisol A Suppresses Proliferation, Migration, and Invasion in Human Breast Cancer MDA-MB-231 Cells" Molecules 24, no. 20: 3651. https://doi.org/10.3390/molecules24203651
APA StyleLou, C., Xu, X., Chen, Y., & Zhao, H. (2019). Alisol A Suppresses Proliferation, Migration, and Invasion in Human Breast Cancer MDA-MB-231 Cells. Molecules, 24(20), 3651. https://doi.org/10.3390/molecules24203651



