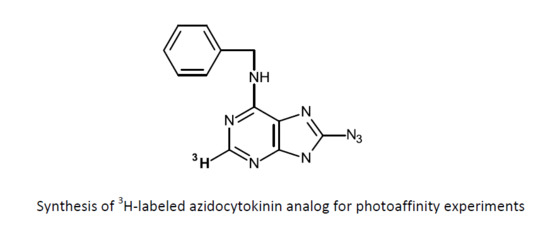
The Synthesis of 3H-Labelled 8-Azido-N6-Benzyladenine and Related Compounds for Photoaffinity Labelling of Cytokinin-Binding Proteins
Abstract
1. Introduction
1.1. Binding of Cytokinins as Plant Regulators
1.2. Cytokinins as Inhibitors of Tumour Cell Growth
1.3. Cytokinins and Microorganisms
1.4. Cytokinins and Photoaffinity Labelling
2. Results and Discussion
2.1. Possible Synthetic Routes to [3H]8-Azido-BA
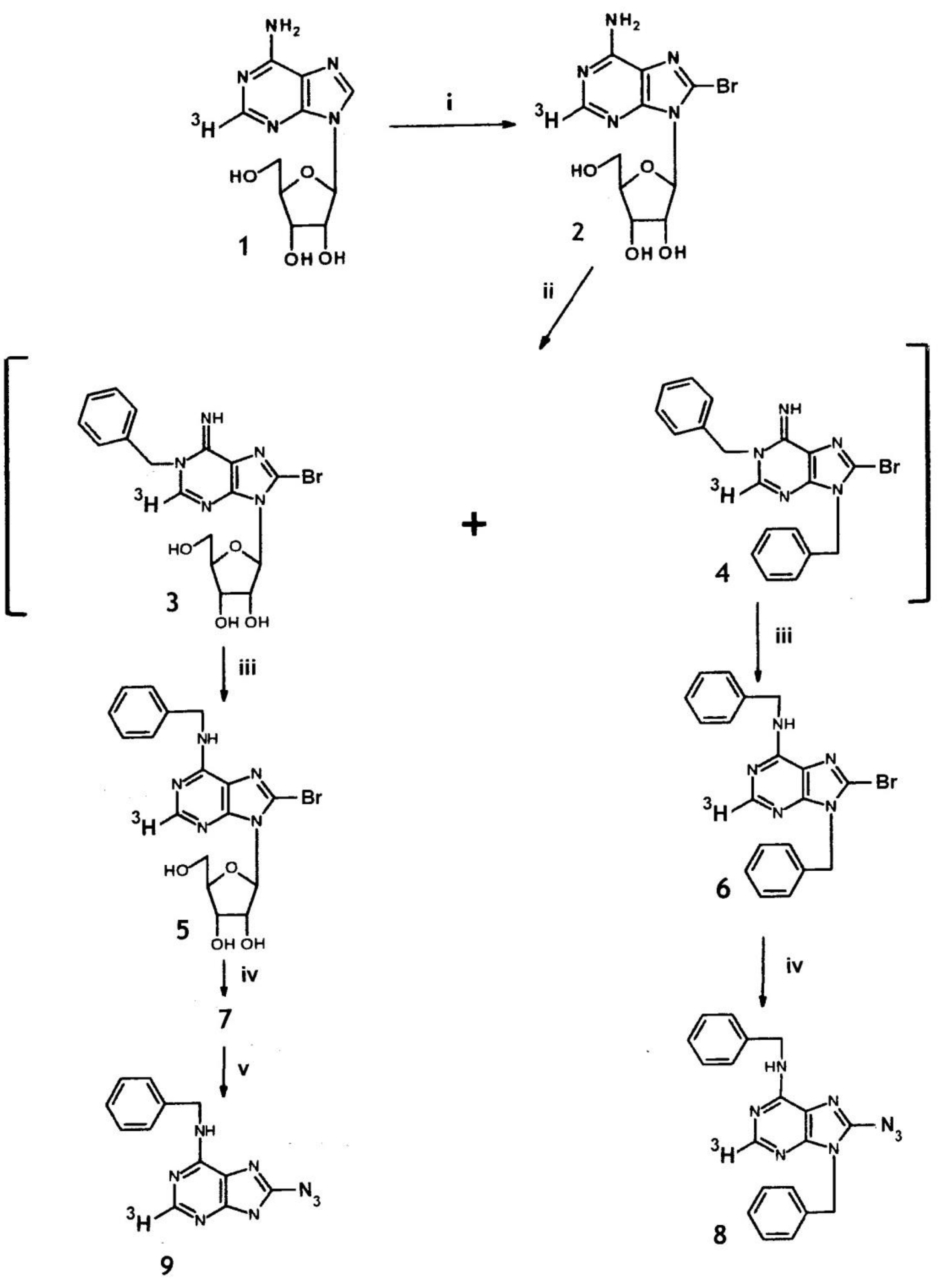
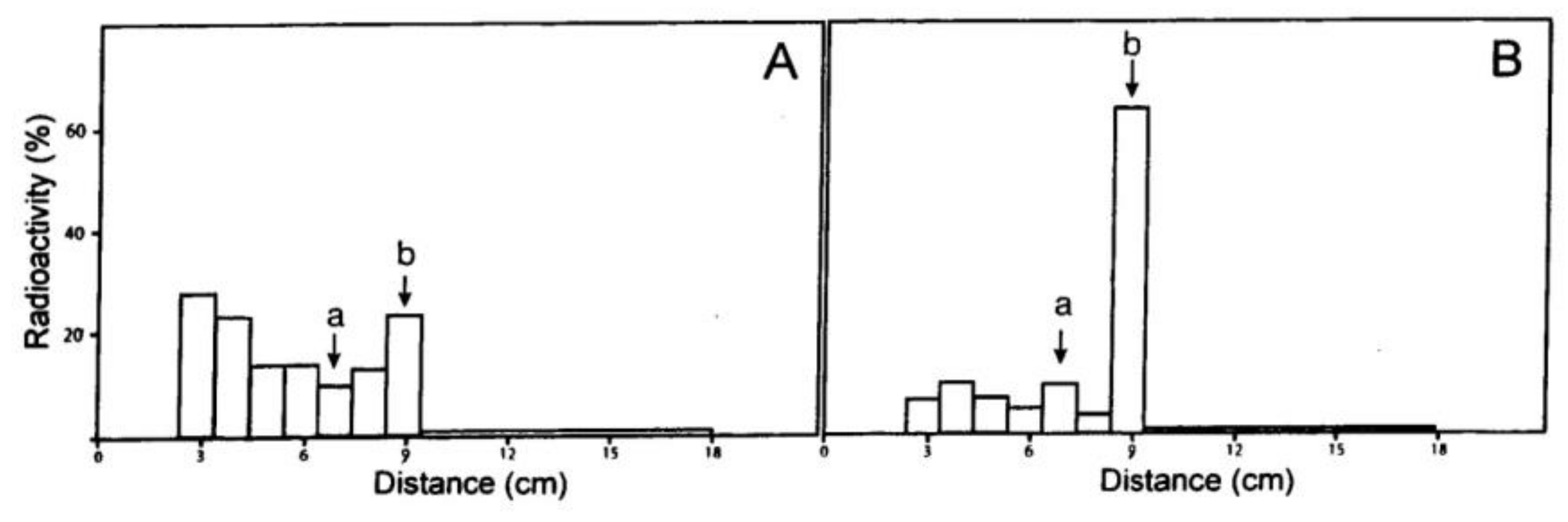
2.2. Synthesis of 8-Azido-N6-benzyl-[2-3H]adenine from [2-3H]Adenosine
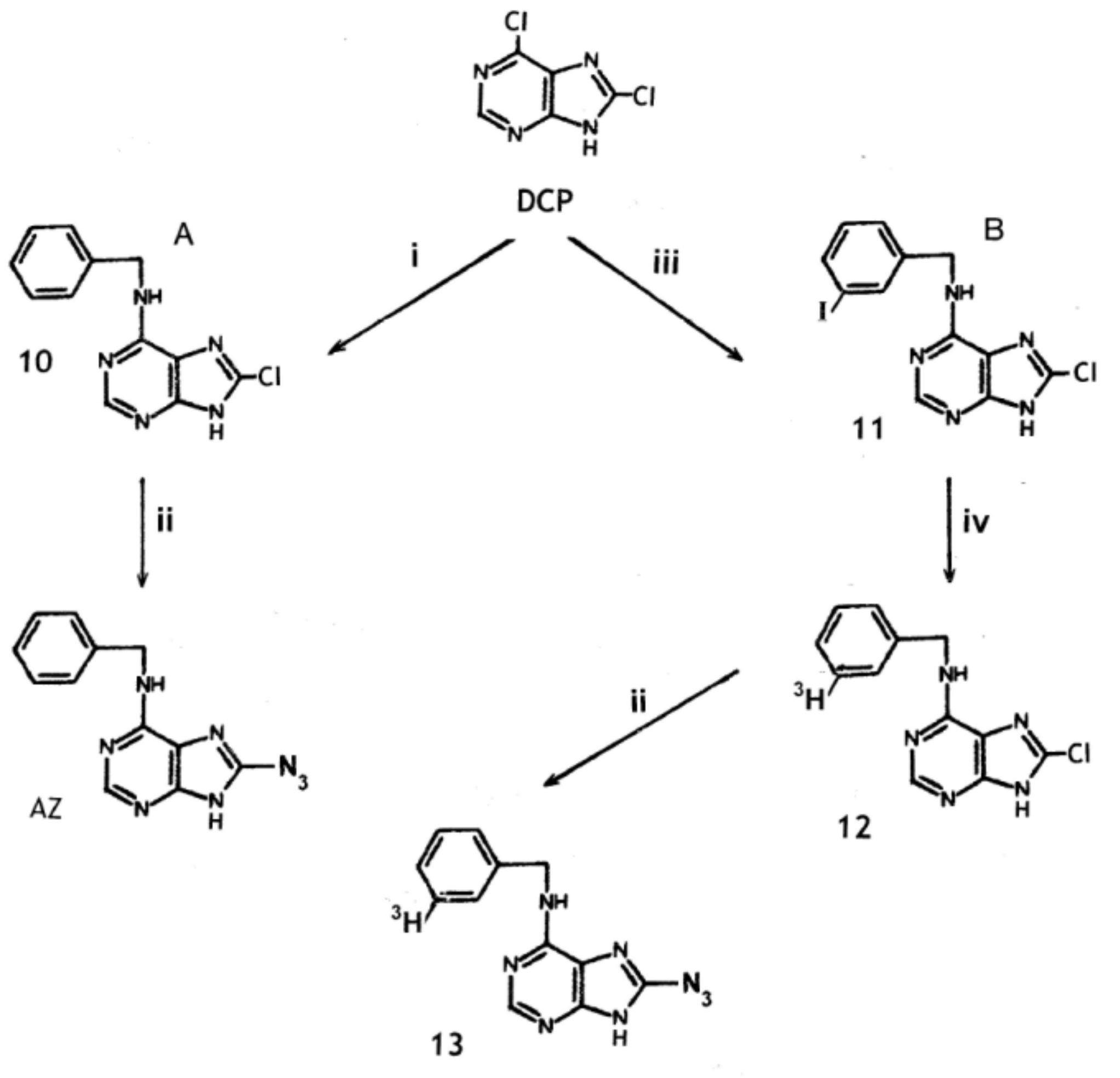
2.3. Synthesis of 8-Azido-N6-Benzyladenine from 6,8-Dichloropurine
2.4. Assessment of PAL
3. Experimental Section
3.1. Chromatographic Methods
3.2. UV and Mass Spectra
3.3. Synthesis of 8-Azido-N6-benzyl-[2-3H]adenine from [2-3H]adenosine
3.3.1. 8-Bromo-[2-3H]adenosine (2)
3.3.2. N6-Benzyl-8-bromo-[2-3H]adenosine (5) and N,9-dibenzyl-8-bromo-[2-3H]adenine (6).
3.3.3. 8-Azido-N6-benzyl-[2-3H]adenosine (7) and 8-azido-N6, 9-dibenzyl-[2-3H]adenine (8)
3.3.4. 8-Azido-N6-benzyl-[2-3H] adenine (9)
3.4. N6-Benzyl-8-Chloro-Adenine (10), Ddirect Azido Insertion
3.5. Additional Compounds
3.5.1. 2,6,8-Triazidopurine
3.5.2. 2,6,8-Triazido-9-(tetrahydropyran-2-yl)purine
3.5.3. 6-Benzylamino-2,8-Dichloro-9-(tetrahydropyran-2-yl)purine
3.5.4. 2-Azido-N6-(3,4-dimethoxybenzyl)adenine
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kaminek, M. Tracking the story of cytokinin research. J. Plant Growth Regul. 2015, 34, 723–739. [Google Scholar] [CrossRef]
- Letham, D.S. Cytokinins as Phytohormones—Sites of biosynthesis, translocation, and function of translocated cytokinin. In Cytokinins: Chemistry, Activity and Function; Mok, D.W.S., Mok, M.C., Eds.; CRC Press: Boca Raton, FL, USA, 1994; pp. 57–80. [Google Scholar]
- Spichal, L. Cytokinins—Recent news and views of evolutionally old molecules. Funct. Plant Biol. 2012, 39, 267–284. [Google Scholar] [CrossRef]
- Lomin, S.N.; Krivosheev, D.M.; Steklov, M.; Arkhipov, D.V.; Osolodkin, D.I.; Schmulling, T.; Romanov, G.A. Plant membrane assays with cytokinin receptors underpin the unique role of free cytokinin bases as biologically active ligands. J. Exp. Bot. 2015, 66, 1851–1863. [Google Scholar] [CrossRef] [PubMed]
- Wulfetange, K.; Lomin, S.N.; Romanov, G.A.; Stolz, A.; Heyl, A. The cytokinin receptors of Arabidopsis are located mainly to the endoplasmic reticulum. Plant Physiol. 2011, 156, 1808–1818. [Google Scholar] [CrossRef] [PubMed]
- Lomin, S.N.; Yonekura-Sakakibara, K.; Romanov, G.A.; Sakakibara, H. Ligand-binding properties and subcellular localization of maize cytokinin receptors. J. Exp. Bot. 2011, 62, 5149–5159. [Google Scholar] [CrossRef] [PubMed]
- Lomin, S.N.; Myakushina, Y.A.; Arkhipov, D.V.; Leonova, O.G.; Popenko, V.I.; Schmulling, T.; Romanov, G.A. Studies of cytokinin receptor-phosphotransmitter interaction provide evidences for the initiation of cytokinin signaling in the endoplasmic reticulum. Funct. Plant Biol. 2018, 45, 192–202. [Google Scholar] [CrossRef]
- Gruhn, N.; Halawa, M.; Snel, B.; Seidl, M.F.; Heyl, A. A subfamily of putative cytokinin receptors is revealed by an analysis of the evolution of the two-component signaling system of plants. Plant Physiol. 2014, 165, 227–237. [Google Scholar] [CrossRef]
- Doležal, K.; Popa, I.; Hauserová, E.; Spíchal, L.; Chakrabarty, K.; Novák, O.; Kryštof, V.; Voller, J.; Holub, J.; Strnad, M. Preparation, biological activity and endogenous occurrence of N6-benzyladenosines. Bioorg. Med. Chem. 2007, 15, 3737–3747. [Google Scholar] [CrossRef]
- Brinegar, C. Cytokinin binding proteins and receptors. In Cytokinins: Chemistry, Activity and Function; Mok, D.W.S., Mok, M.C., Eds.; CRC Press: Boca Raton, FL, USA, 1994; pp. 217–232. [Google Scholar]
- Penny, P.; Penny, D. Rapid Responses to Phytohormones. In Phytohormones and Related Compounds—A Comprehensive Treatise; Letham, D.S., Goodwin, P.B., Higgins, T.J.V., Eds.; Elsevier/North Holland Biomedical Press: Amsterdam, The Netherlands, 1978; Volume 2, pp. 537–597, See also: Kuraishi, S.; Hashimoto, Y.; Shiraishi, M. Plant Cell Physiol. 1981, 22, 911–916. [Google Scholar]
- Yong, J.W.H.; Letham, D.S.; Wong, S.C.; Farquhar, G.D. Rhizobium-induced elevation in xylem cytokinin delivery in pigeonpea induces changes in shoot development and leaf physiology. Funct. Plant Biol. 2014, 41, 1323–1335. [Google Scholar] [CrossRef]
- Badenoch-Jones, B.; Parker, C.W.; Letham, D.S.; Singh, S. Effect of cytokinins supplied via the xylem at multiples of endogenous concentrations on transpiration and serescence in derooted seedlings of oat and wheat. Plant Cell Environ. 1996, 19, 504–516. [Google Scholar] [CrossRef]
- Matsumoto-Kitano, M.; Kusumoto, T.; Tarkowski, P.; Kinoshita-Tsujimura, K.; Vaclavikova, K.; Miyawaki, K.; Kakimoto, T. Cytokinins are central regulators of cambial activity. Proc. Natl. Acad. Sci. USA 2008, 105, 20027–20031. [Google Scholar] [CrossRef] [PubMed]
- Kiba, T.; Takei, K.; Kojima, M.; Sakakibara, H. Scale-chain modification of cytokinins controls shoot growth in Arabidopsis. Dev. Cell 2013, 27, 452–461. [Google Scholar] [CrossRef] [PubMed]
- Bishopp, A.; Lehesranta, S.; Vatén, A.; Help, H.; El-Showk, S.; Scheres, B.; Helariutta, K.; Mähönen, A.P.; Sakakibara, H.; Helariutta, Y. Phloem-transported cytokinin regulates polar auxin transport and maintains vascular pattern in the root meristem. Curr. Biol. 2011, 21, 927–932. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Lee, Y.; Sakakibara, H.; Martinoia, E. Cytokinin transporters: GO and STOP in signaling. Trends Plant Sci. 2017, 22, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Knypl, J.S.; Letham, D.S.; Palni, L.M.S. Cytokinins in maturing and germinating Lupinus luteus L. seeds. Biol. Plant. 1985, 27, 188–194. [Google Scholar] [CrossRef]
- Nandi, S.K.; Palni, L.M.S.; Letham, D.S.; Knypl, J.S. The biosynthesis of cytokinins in germinating lupin seeds. J. Expl. Bot. 1988, 39, 1649–1665. [Google Scholar] [CrossRef]
- Nandi, S.K.; Palni, L.M.S. Transport and metabolism of dihydrozeatin riboside in germinating lupin seeds. J. Expl. Bot. 1989, 40, 615–621. [Google Scholar] [CrossRef]
- Nandi, S.K.; Palni, L.M.S.; Letham, D.S. Axial control of cotyledon expansion and chlorophyll formation in germinating lupin seeds. Plant Sci. 1989, 60, 181–188. [Google Scholar] [CrossRef]
- Nandi, S.K.; Palni, L.M.S.; de Klerk, G.J.M. The influence of the embryonic axis and cytokinins on reserve mobilization in germinating lupin seeds. J. Expl. Bot. 1995, 46, 329–336. [Google Scholar] [CrossRef]
- Wang, Y.; Letham, D.S.; John, P.C.L.; Zhang, R. Synthesis of a cytokinin linked by a spacer to dexamethasone and biotin: Conjugates to detect cytokinin-binding proteins. Molecules 2016, 21, 576. [Google Scholar] [CrossRef]
- Spinola, M.; Colombo, F.; Falvella, F.S.; Dragani, T.A. N6-isopentenyladenosine: A potential therapeutic agent for a variety of epithelial cancers. Int. J. Cancer 2007, 120, 2744–2748. [Google Scholar] [CrossRef] [PubMed]
- Voller, J.; Zatloukal, M.; Lenobel, R.; Doležal, K.; Béreš, T.; Kryštof, V.; Spíchal, L.; Niemann, P.; Džubák, P.; Hajdúch, M.; et al. Anticancer activity of natural cytokinins: A structure-activity relationship study. Phytochemistry 2010, 71, 1350–1359. [Google Scholar] [CrossRef] [PubMed]
- Parker, C.W.; Entsch, B.; Letham, D.S. Inhibitors of two enzymes which metabolize cytokinins. Phytochemistry 1986, 25, 303–310. [Google Scholar] [CrossRef]
- Entsch, B.; Parker, C.W.; Letham, D.S.; Summons, R.E. Preparation and characterization, using HPLC, of an enzyme forming glucosides of cytokinins. Biochim. Biophys. Acta 1979, 570, 124–139. [Google Scholar] [CrossRef]
- Tao, G.Q.; Letham, D.S.; Hocart, C.H.; Summons, R.E. Inhibitors of cytokinin metabolism III. The inhibition of cytokinin N-glucosylation in radish cotyledons. J. Plant Growth Regul. 1991, 10, 179–185. [Google Scholar] [CrossRef]
- Veselý, J.; Havliček, L.; Strnad, M.; Blow, J.J.; Donella-Deana, A.; Pinna, L.; Letham, D.S.; Kato, J.Y.; Detivaud, L.; Leclerc, S.; et al. Inhibition of cyclin-dependent kinases by purine analogues. Eur. J. Biochem. 1994, 224, 771–786. [Google Scholar] [CrossRef] [PubMed]
- Spellek, T.; Gan, P.; Kadota, Y.; Shirasu, K. Same tune, different song—Cytokinins as virulence factors in plant-pathogen interactions. Curr. Opin. Plant Biol. 2018, 44, 82–87. [Google Scholar] [CrossRef]
- Kabbara, S.; Schmulling, T.; Papon, N. CHASEing cytokinin receptors in plants, bacteria, fungi, and beyond. Trends Plant Sci. 2018, 23, 1–3. [Google Scholar] [CrossRef]
- Andrabi, S.B.; Tahara, M.; Matsubara, R.; Toyama, T.; Aonuma, H.; Sakakibara, H.; Suematsu, M.; Tanabe, K.; Nozaki, T.; Nagamune, K. Plant hormone cytokinins control cell cycle progression and plastid replication in apicomplexan parasites. Parasitol. Int. 2018, 67, 47–58. [Google Scholar] [CrossRef]
- Samanovic, M.I.; Darwin, K.H. Cytokinins beyond plants: Synthesis by Mycobacterium tuberculosis. Microb. Cell 2015, 2, 168–170. [Google Scholar] [CrossRef]
- Samanovic, M.I.; Hsu, H.C.; Jones, M.B.; Jones, V.; McNeil, M.R.; Becker, S.H.; Jordan, A.T.; Strnad, M.; Xu, C.; Jackson, M.; et al. Cytokinin signaling in Mycobacterium tuberculosis. Mbio 2018, 9, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.; Collins, I. Photoaffinity labelling in target- and binding-site identification. Future Med. Chem. 2015, 7, 159–183. [Google Scholar] [CrossRef]
- Sumranjit, J.; Chung, S.J. Recent advances in target characterization and identification by photoaffinity probes. Molecules 2013, 18, 10425–10451. [Google Scholar] [CrossRef]
- Gonneau, M.; Mornet, R.; Laloue, M. A Nicotiana plumbaginifolia protein labelled with an azido cytokinin agonist is a glutathione S-transferase. Physiol. Plant. 1998, 103, 114–124. [Google Scholar] [CrossRef]
- Nogué, F.; Mornet, R.; Laloue, M. Specific photoaffinity labelling of a thylakoid membrane protein with an azido-cytokinin agonist. Plant Growth Regul. 1996, 18, 51–58. [Google Scholar] [CrossRef]
- Brinegar, A.C.; Cooper, G.; Stevens, A.; Hauer, C.R.; Shabanowitz, J.; Hunt, D.F.; Fox, J.E. Characterization of a benzyladenine binding-site peptide isolated from a wheat cytokinin-binding protein: Sequence analysis and identification of a single affinity-labelled histidine residue by mass spectrometry. Proc. Natl. Acad. Sci. USA 1988, 85, 5927–5931. [Google Scholar] [CrossRef] [PubMed]
- Brinegar, C.; Shah, G.; Cooper, G. Photoaffinity labelling of a cytokinin-binding integral membrane protein in plant mitochondria. Plant Growth Regul. 1996, 18, 45–50. [Google Scholar] [CrossRef]
- Mornet, R.; Theiler, J.B.; Leonard, N.J. Photoaffinity labelling reagents to detect binding of cytokinins: Chemical limitations. In Metabolism and Molecular Activities of Cytokinins; Guern, J., Peaud-Lenoel, C., Eds.; Springer: Berlin, Germany, 1981; pp. 153–161. [Google Scholar]
- Mornet, R.; Theiler, J.B.; Leonard, N.J.; Schmitz, R.Y.; Moore, F.H.; Skoog, F. Active cytokinins. Plant Physiol. 1979, 64, 600–610. [Google Scholar] [CrossRef]
- Sussman, M.R.; Kende, H. The synthesis and biological properties of 8-azido-N6-benzyladenine, a potential photoaffinity reagent for cyotokinin. Planta 1977, 137, 91–96. [Google Scholar] [CrossRef]
- Steklov, M.Y.; Tararov, V.I.; Romanov, G.A.; Mikhailov, S.N. Facile synthesis of 8-azido-6-benzyl-aminopurine. Nucleosides Nucleotides Nucleic Acids 2011, 30, 503–511. [Google Scholar] [CrossRef]
- Leonard, N.J.; Fujii, T. The use of a blocking group at the 9-position of adenine for the synthesis of 1-substitutedadenines. Proc. Natl. Acad. Sci. USA 1964, 51, 73–75. [Google Scholar] [CrossRef] [PubMed]
- Macon, J.B.; Wolfenden, R. 1-methyl-adenosine. Dimroth rearrangement and reversible reduction. Biochemistry 1968, 7, 3453–3458. [Google Scholar] [CrossRef] [PubMed]
- Brown, H.C.; Subba Rao, B.C. A new powerful reducing agent—Sodium borohydride in the presence of aluminium chloride and other polyvalent metal halides. J. Am. Chem. Soc. 1956, 78, 2582–2588. [Google Scholar] [CrossRef]
- Cooper, G.; Bourell, J.; Kaminek, M.; Fox, J.E. Method for synthesis of 2-azido-N6-m-tritiobenzylaminopurine, a photoaffinity label for cytokinin-binding proteins in plants. J. Label. Compd. Radiopharm. 1988, 25, 957–962. [Google Scholar] [CrossRef]
- Sidorov, G.V.; Myasoedov, N.F.; Lomin, S.N.; Romanov, G.A. Synthesis of tritium- and deuterium-labelled isopentenyladenines. Radiochemistry 2015, 57, 108–110. [Google Scholar] [CrossRef]
- Letham, D.S.; Tao, G.Q.; Parker, C.W. An overview of cytokinin metabolism. In Plant Growth Substances 1982; Wareing, P.F., Ed.; Academic Press: London, UK, 1982; pp. 143–153. [Google Scholar]
- Doležal, K.; Popa, I.; Kryštof, V.; Spíchal, L.; Fojtíková, M.; Holub, J.; Lenobel, R.; Schmülling, T.; Strnad, M. Preparation and biological activity of 6-benzylaminopurine derivatives in plants and human cancer cells. Bioorganic. Med. Chem. 2006, 14, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Theiler, J.B.; Leonard, N.J.; Schmitz, R.Y.; Skoog, F. Photoaffinity-labelled cytokinins. Plant Physiol. 1976, 58, 803–805. [Google Scholar] [CrossRef]
- Shelton, K.R.; Clark, J.M. A proton exchange between purines and water and its application to biochemistry. Biochemistry 1967, 6, 2735–2739. [Google Scholar] [CrossRef]
- Letham, D.S.; Singh, S.; Willcocks, D.A. Reversed phase thin-layer chromatographic methods for separation of cytokinins. Phytochem. Anal. 1992, 3, 218–222. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Letham, D.S.; Zhang, X.-D.; Hocart, C.H. The Synthesis of 3H-Labelled 8-Azido-N6-Benzyladenine and Related Compounds for Photoaffinity Labelling of Cytokinin-Binding Proteins. Molecules 2019, 24, 349. https://doi.org/10.3390/molecules24020349
Letham DS, Zhang X-D, Hocart CH. The Synthesis of 3H-Labelled 8-Azido-N6-Benzyladenine and Related Compounds for Photoaffinity Labelling of Cytokinin-Binding Proteins. Molecules. 2019; 24(2):349. https://doi.org/10.3390/molecules24020349
Chicago/Turabian StyleLetham, David. S., Xue-Dong Zhang, and Charles H. Hocart. 2019. "The Synthesis of 3H-Labelled 8-Azido-N6-Benzyladenine and Related Compounds for Photoaffinity Labelling of Cytokinin-Binding Proteins" Molecules 24, no. 2: 349. https://doi.org/10.3390/molecules24020349
APA StyleLetham, D. S., Zhang, X.-D., & Hocart, C. H. (2019). The Synthesis of 3H-Labelled 8-Azido-N6-Benzyladenine and Related Compounds for Photoaffinity Labelling of Cytokinin-Binding Proteins. Molecules, 24(2), 349. https://doi.org/10.3390/molecules24020349