Effectiveness of Prenyl Group on Flavonoids from Epimedium koreanum Nakai on Bacterial Neuraminidase Inhibition
Abstract
1. Introduction
2. Results and Discussion
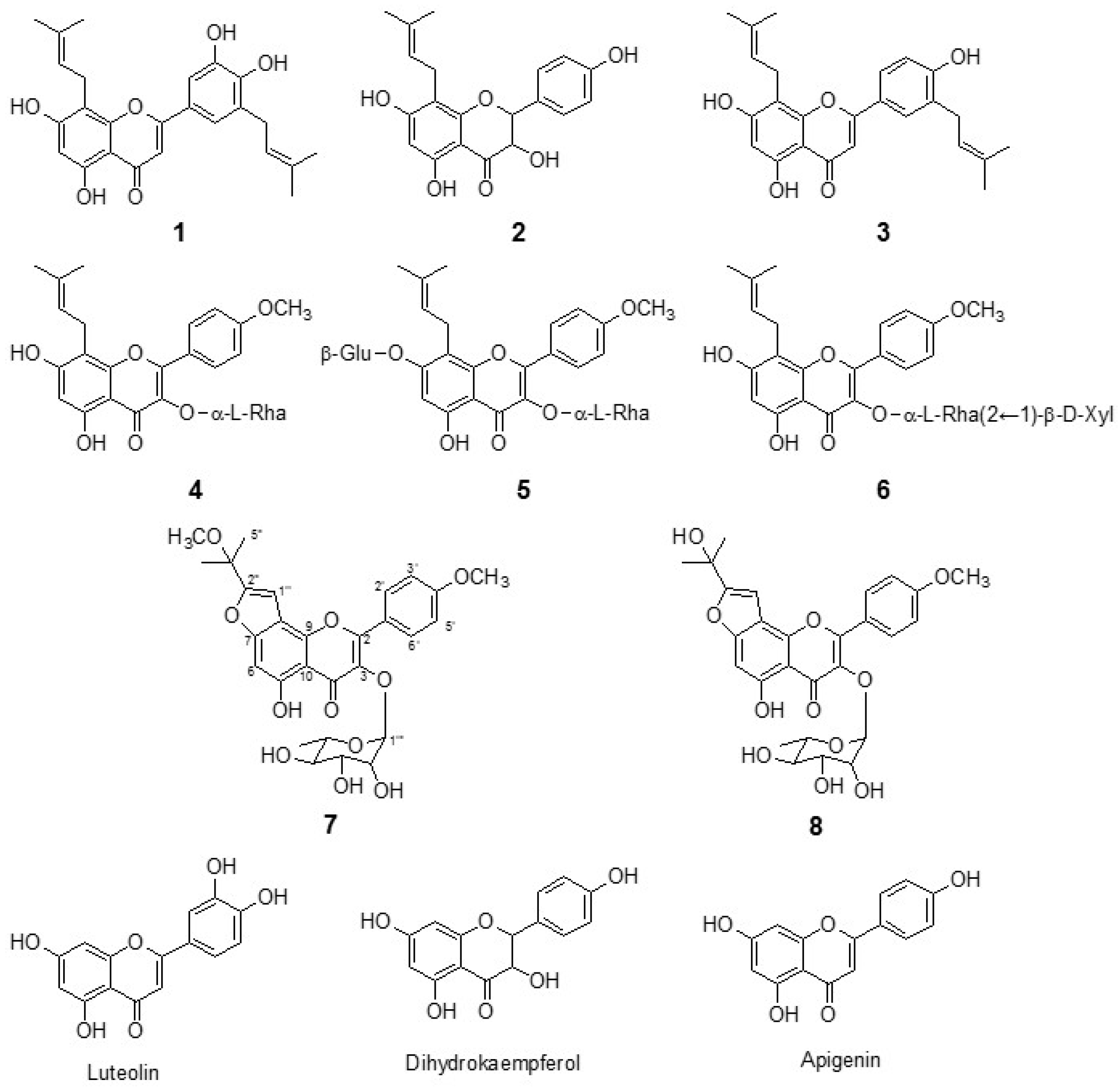
2.1. Isolation of Flavonoids from E. koreanum Nakai
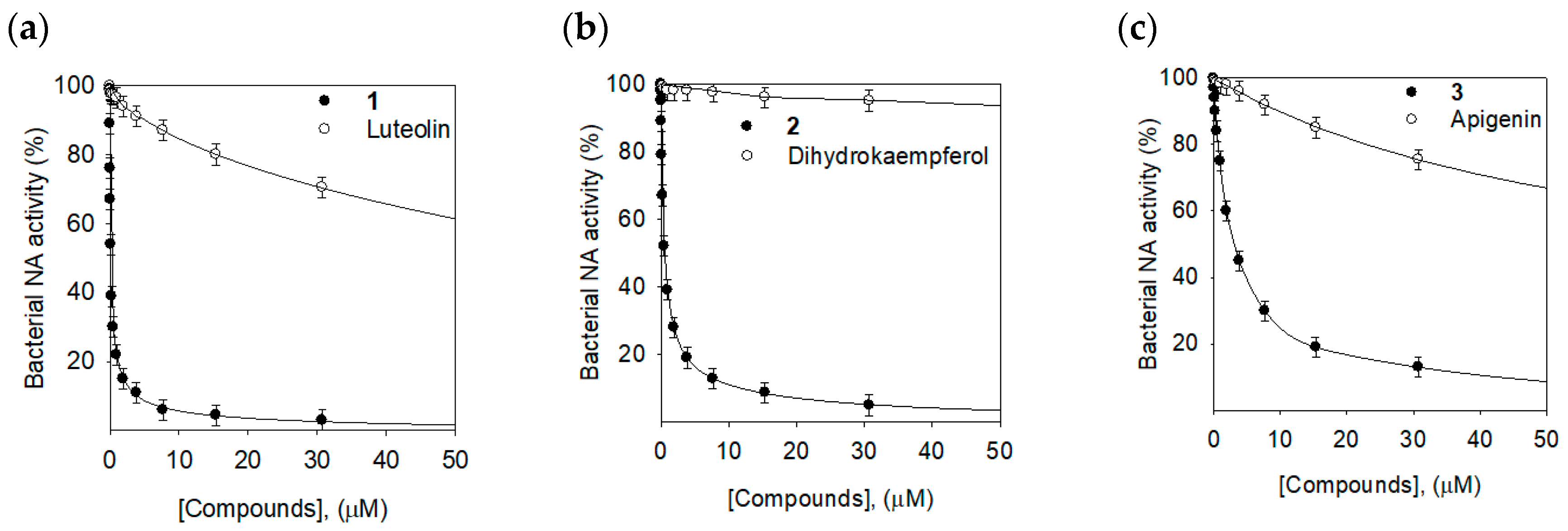
2.2. Bacterial Neuraminidase Inhibitory Activities
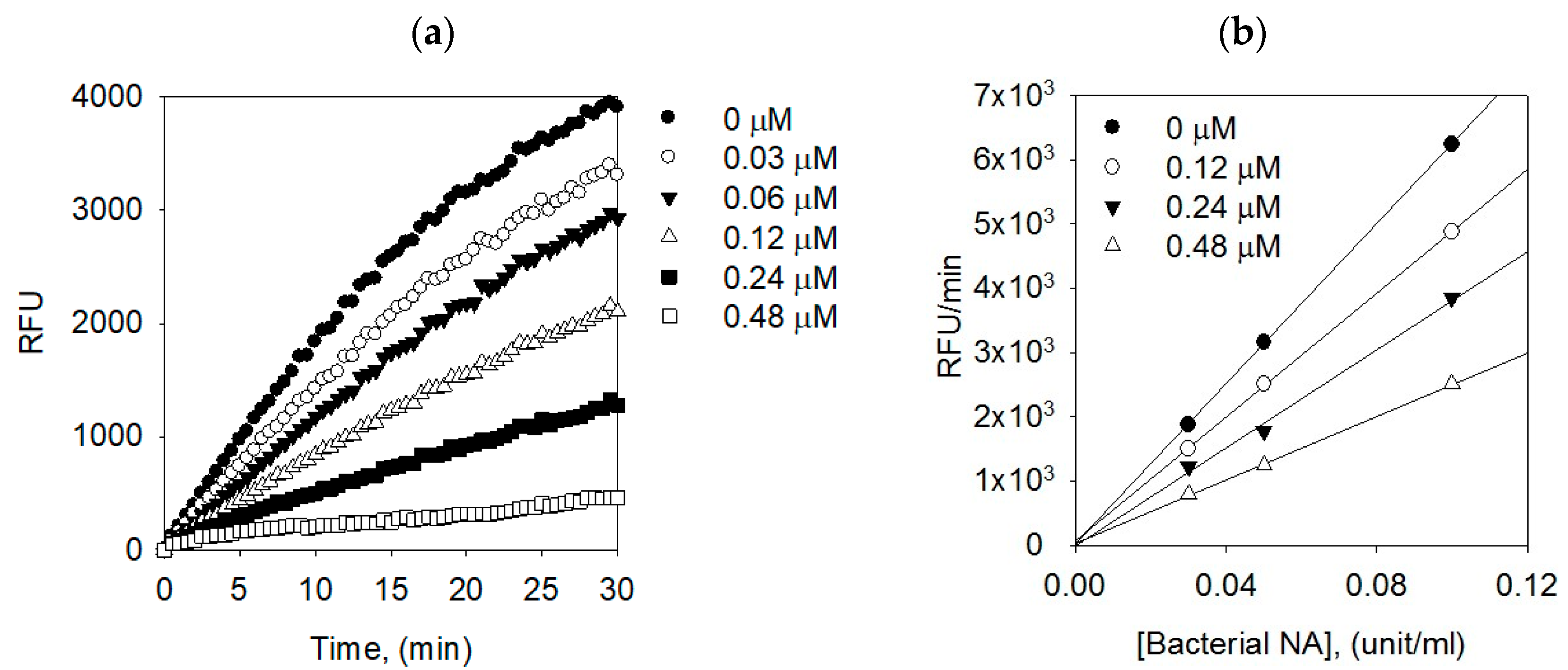
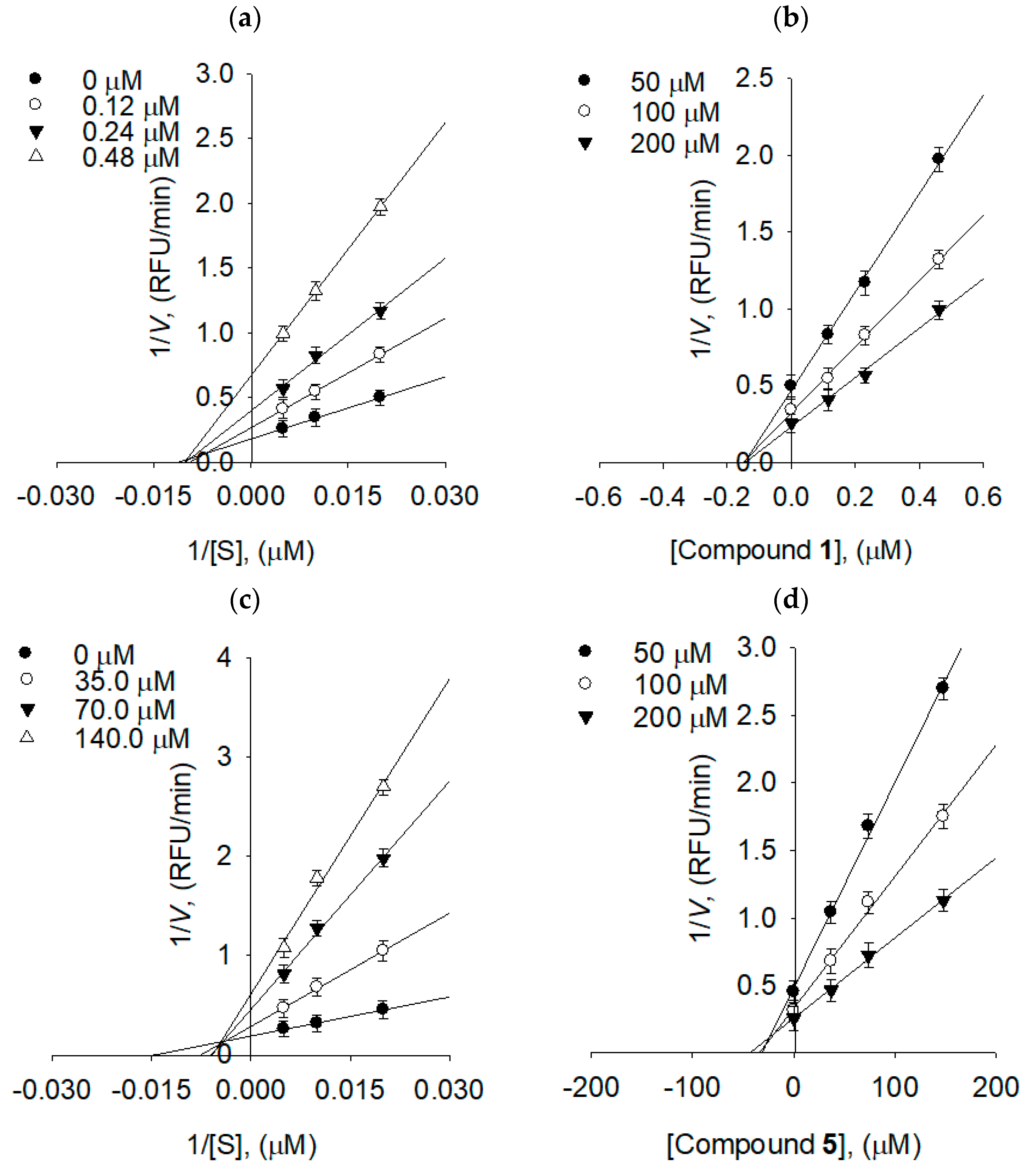
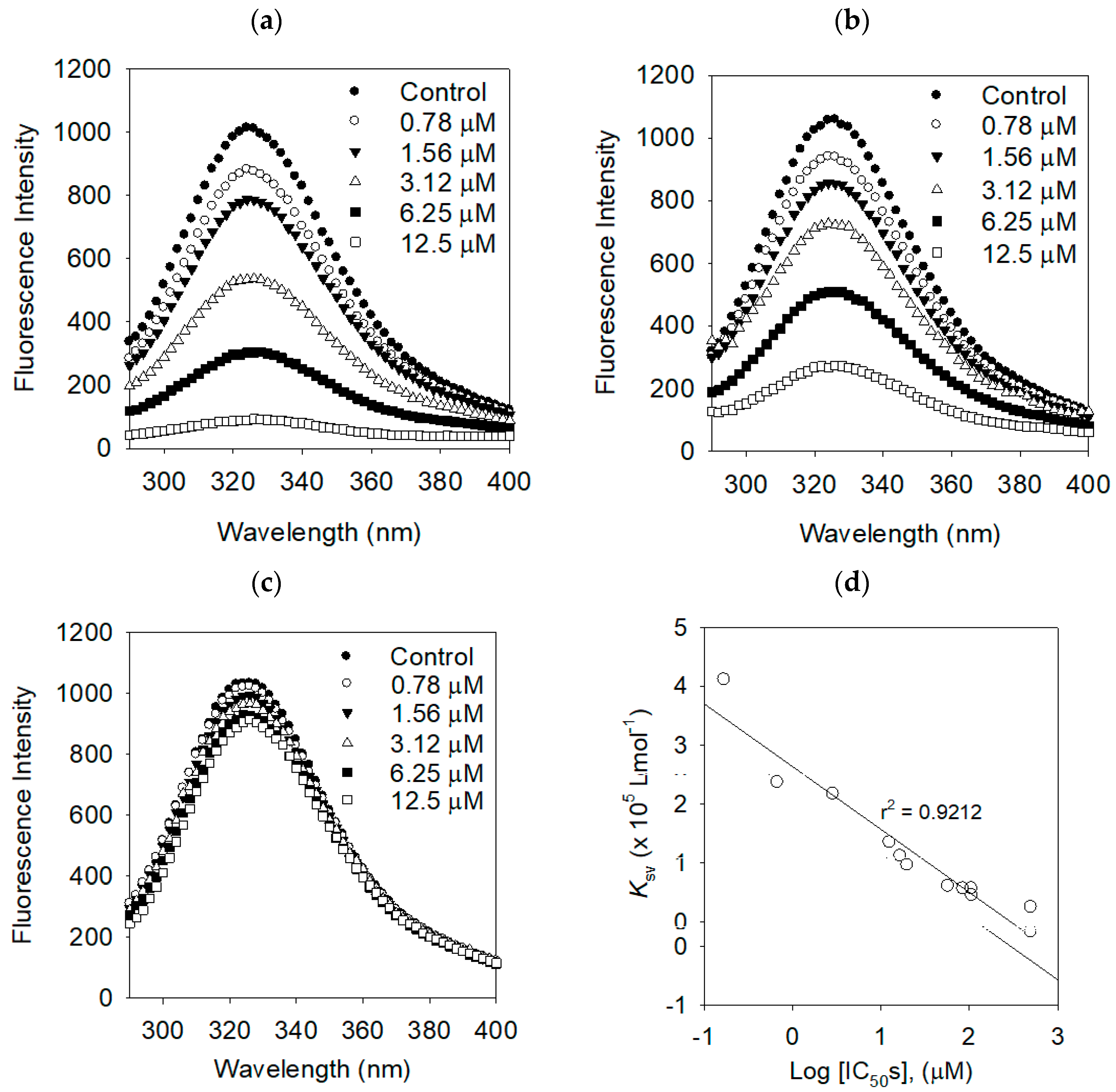
2.3. Binding Affinities between Bacterial Neuraminidase and Compounds
3. Materials and Methods
3.1. Chemicals and Materials
3.2. Instruments
3.3. Extraction and Isolation
3.3.1. Epimedokoreanin B (Compound 1)
3.3.2. 8-(γ,γ-Dimethyl allyl)-5,7,4′-trihydroxydihydroflavonol (Compound 2)
3.3.3. 5,7,4′-Trihydroxy-8,3′-diprenylflavone (Compound 3)
3.3.4. Icariside II (Compound 4)
3.3.5. Icariin (Compound 5)
3.3.6. Sagittatoside B (Compound 6)
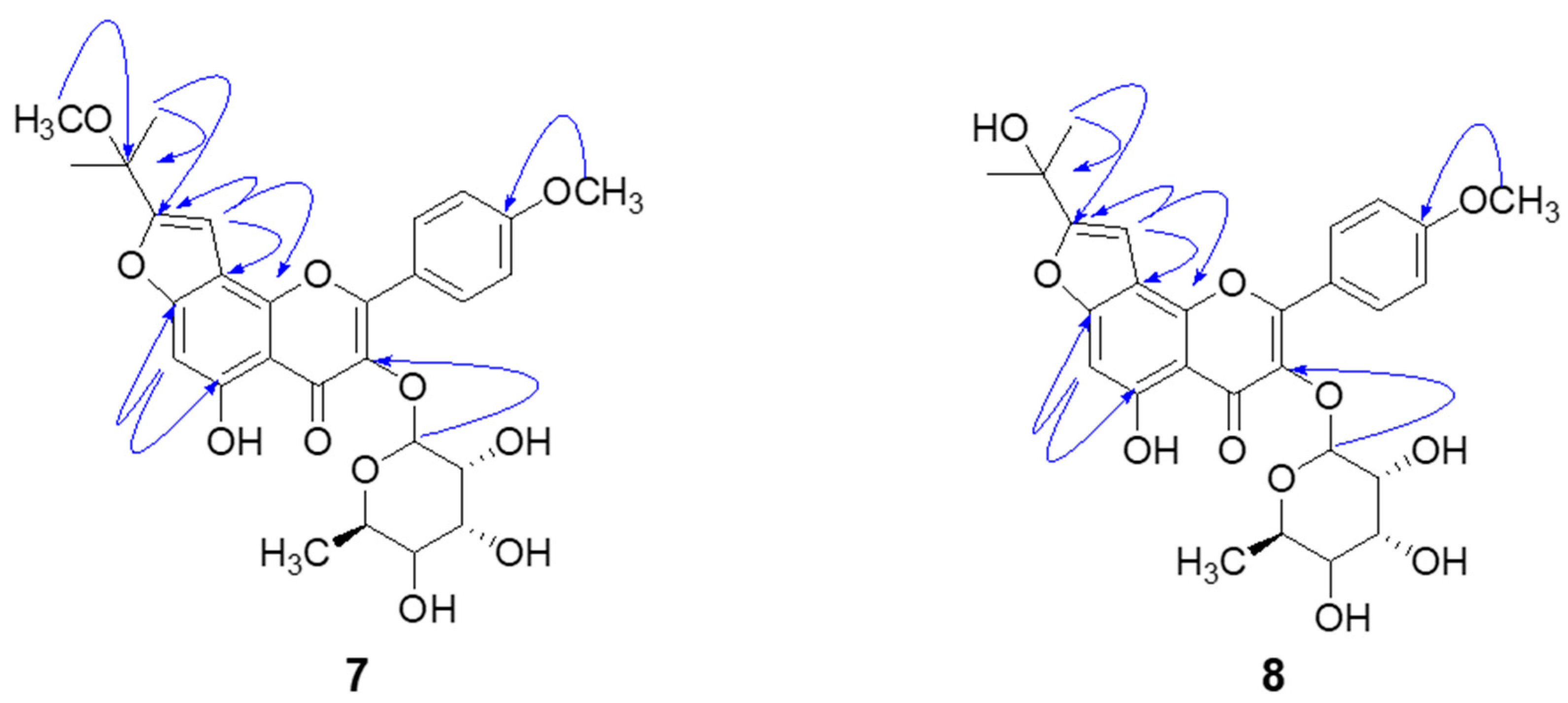
3.3.7. Koreanoside F (Compound 7)
3.3.8. Koreanoside G (Compound 8)
3.4. Bacterial Neuraminidase Inhibitory Activity Assay
3.5. Enzyme Kinetics and Progress Linear Determinations
3.6. Binding Affinity between Bacterial Neuraminidase and Compounds
3.7. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Matrosovich, M.N.; Matrosovich, T.Y.; Gray, T.; Roberts, N.A.; Klenk, H.-D. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc. Natl. Acad. Sci. USA 2004, 10113, 4620–4624. [Google Scholar] [CrossRef] [PubMed]
- Shinya, K.; Ebina, M.; Yamada, S.; Ono, M.; Kasai, N.; Kawaoka, Y. Influenza virus receptors in the human airway. Nature 2006, 56, 85–89. [Google Scholar] [CrossRef] [PubMed]
- Couceiro, J.N.S.S.; Paulson, J.C.; Baum, L.G. Influenza virus strains selectively recognize sialyloligosaccharides on human respiratory epithelium; the role of the host cell in selection of hemagglutinin receptor specificity. Virus Res. 1993, 29, 155–165. [Google Scholar] [CrossRef]
- Soong, G.; Muir, A.; Gomez, M.I.; Waks, J.; Reddy, B.; Planet, P.; Singh, P.K.; Kanetko, Y.; Wolfgang, M.C.; Hsiao, Y.S.; et al. Bacterial neuraminidase facilitates mucosal infection by participating in biofilm production. J. Clin. Invest. 2006, 116, 2297–2305. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.Y.; Chen, X.; King, S.; Cavassani, K.A.; Cheng, J.; Zheng, X.; Cao, H.; Yu, H.; Qu, J.; Fang, D.; et al. Amelioration of sepsis by inhibiting sialidase-mediated disruption of the CD24-SiglecG interaction. Nat. Biotechnol. 2011, 29, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Hiramatsu, K. Molecular Evolution of MRSA. Microbiol. Immunol. 1995, 39, 531–543. [Google Scholar] [CrossRef] [PubMed]
- Ryu, H.W.; Curtis-Long, M.J.; Jung, S.; Jin, Y.M.; Cho, J.K.; Ryu, Y.B.; Lee, W.S.; Park, K.H. Xanthones with neuraminidase inhibitory activity from the seedcases of Garcinia mangostana. Bioorganic Med. Chem. 2010, 18, 6258–6264. [Google Scholar] [CrossRef] [PubMed]
- Ryu, Y.B.; Curtis-Long, M.J.; Kim, J.H.; Jeong, S.H.; Yang, M.S.; Lee, K.W.; Lee, W.S.; Park, K.H. Pterocarpans and flavanones from Sophora flavescens displaying potent neuraminidase inhibition. Bioorganic Med. Chem. Lett. 2008, 18, 6046–6049. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Ryu, Y.B.; Youn, H.S.; Cho, J.K.; Kim, Y.M.; Park, J.Y.; Lee, W.S.; Park, K.H.; Eom, S.H. Structural basis of sialidase in complex with geranylated flavonoids as potent natural inhibitors. Acta Crystallogr. Sect. D Biol. Crystallogr. 2014, 70, 1357–1365. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; He, X.; Yang, Y.; Li, M.; Hao, D.; Jia, Z. The genus Epimedium: An ethnopharmacological and phytochemical review. J. Ethnopharmacol. 2011, 134, 519–541. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.-N. An illustrated Chinese Materia Medica; Oxford University Press, Inc.: New York, NY, USA, 1932; ISBN 9780195140170. [Google Scholar]
- Eddouks, M. Handbook of Ethnopharmacology; Research Signpost: Trivandrum, India, 2008; ISBN 9788130802138. [Google Scholar]
- Zhang, W.; Chen, H.; Wang, Z.; Lan, G.; Zhang, L. Comparative studies on antioxidant activities of extracts and fractions from the leaves and stem of Epimedium koreanum Nakai. J. Food Sci. Technol. 2013, 50, 1122–1129. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Du, Z.R.; Xu, A.L.; Yan, Z.; Xiao, H.H.; Wong, M.S.; Yao, X.S.; Chen, W.F. Neuroprotective effects of total flavonoid fraction of the Epimedium koreanum Nakai extract on dopaminergic neurons: In vivo and in vitro. Biomed. Pharmacother. 2017, 91, 656–663. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Du, Z.; Pu, J.; Zhao, H.; Chen, H.; Liu, Y.; Li, Z.; Cheng, Z.; Zhong, H.; Liao, F. Classification of difference between inhibition constants of an inhibitor to facilitate identifying the inhibition type. J. Enzyme Inhib. Med. Chem. 2013, 28, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Eftink, M.R.; Ghiron, C.A. Fluorescence quenching studies with proteins. Anal. Biochem. 1981, 114, 199–227. [Google Scholar] [CrossRef]
- Uddin, Z.; Song, Y.H.; Curtis-Long, M.J.; Kim, J.Y.; Yuk, H.J.; Park, K.H. Potent bacterial neuraminidase inhibitors, anthraquinone glucosides from Polygonum cuspidatum and their inhibitory mechanism. J. Ethnopharmacol. 2016, 193, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Zhang, D.-W.; Zhang, J.-C.; Yang, M.-S.; Xiao, P.-G. Isolation and identification of flavonoids from Epimedium koreanum and their effects on proliferation of RAW 264.7 cell line. Chinese Tradit. Herb. Drugs 2006, 37, 1458–1462. [Google Scholar]
- Li, J.Y.; Li, H.M.; Liu, D.; Chen, X.Q.; Chen, C.H.; Li, R.T. Three new acylated prenylflavonol glycosides from Epimedium koreanum. Phytochem. Lett. 2016, 17, 206–212. [Google Scholar] [CrossRef]
- Li, H.M.; Zhou, C.; Chen, C.H.; Li, R.T.; Lee, K.H. Flavonoids isolated from heat-processed Epimedium koreanum and their anti-HIV-1 activities. Helv. Chim. Acta 2015, 98, 1177–1187. [Google Scholar] [CrossRef]
- Potier, M.; Mameli, L.; Bélisle, M.; Dallaire, L.; Melançon, S.B. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-α-d-N-acetylneuraminate) substrate. Anal. Biochem. 1979, 2697, 90362. [Google Scholar] [CrossRef]
- Boaz, H.; Rollefson, G.K. The Quenching of Fluorescence. Deviations from the Stern-Volmer Law. J. Am. Chem. Soc. 1950, 72, 3435–3443. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are available from the authors. |
| Position | 7 | 8 | ||
|---|---|---|---|---|
| δH, mult, (J, Hz) | δc | δH, mult, (J, Hz) | δc | |
| 2 | - | 157.7 | - | 157.5 |
| 3 | - | 135.9 | - | 135.8 |
| 4 | - | 179.1 | - | 179.2 |
| 5 | - | 159.5 | - | 157.8 |
| 6 | 6.78, s | 94.2 | 6.90, s | 94.2 |
| 7 | - | 148.9 | - | 148.9 |
| 8 | - | 108.9 | - | 109.2 |
| 9 | - | 158.2 | - | 158.9 |
| 10 | - | 107.4 | - | 107.3 |
| 1′ | - | 122.2 | - | 122.2 |
| 2′,6′ | 7.88, d, (J = 8.7 Hz) | 130.6 | 7.98, d, (J = 8.8 Hz) | 130.5 |
| 3′,5′ | 7.04, d, (J = 8.7 Hz) | 113.9 | 7.15, d, (J = 8.8 Hz) | 113.9 |
| 4′ | - | 162.3 | - | 162.3 |
| 1″ | 6.93, s | 100.6 | 6.91, s | 96.9 |
| 2″ | - | 159.1 | - | 163.5 |
| 3″ | - | 73.3 | - | 68.2 |
| 4″ | 1.53, s | 24.4 | 1.66, s | 27.5 |
| 5″ | 1.53, s | 24.1 | 1.66, s | 27.5 |
| 1‴ | 5.38, s | 102.2 | 5.48, s | 102.2 |
| 2‴ | 3.19, overlap | 70.5 | 3.30, overlap | 70.5 |
| 3‴ | 4.16, m | 70.7 | 4.27, d, (J = 1.7 Hz) | 70.7 |
| 4‴ | 3.63, m | 70.8 | 3.75, m | 70.8 |
| 5‴ | 3.25, overlap | 71.7 | 3.37, overlap | 71.7 |
| 6‴ | 0.82, d, (J = 5.8 Hz) | 16.3 | 0.93, d, (J = 5.90 Hz) | 16.3 |
| 3″-OCH3 | 3.03, s | 49.9 | - | - |
| 4′-OCH3 | 3.81, s | 54.6 | 3.93, s | 54.6 |
| Compounds | Bacterial Neuraminidase | |
|---|---|---|
| IC50 1 (µM) | Kinetic Mode (Ki 2, µM) | |
| 1 | 0.17 ± 0.02 | Noncompetitive (0.15 ± 0.01) |
| 2 | 0.68 ± 0.03 | Noncompetitive (0.73 ± 0.03) |
| 3 | 12.6 ± 0.2 | Noncompetitive (12.2 ± 0.3) |
| 4 | 20.0 ± 0.5 | Noncompetitive (21.1 ± 0.8) |
| 5 | 57.8 ± 1.1 | Mixed type I (36.8 ± 0.7) |
| 6 | 106.3 ± 3.2 | Noncompetitive (103.2 ± 1.4) |
| 7 | 2.90 ± 0.7 | Noncompetitive (2.73 ± 0.6) |
| 8 | 16.7 ± 0.9 | Noncompetitive (16.4 ± 0.9) |
| Luteolin | 85.6 ± 2.1 | NT 3 |
| Dihydrokaempferol | 500.4 ± 3.8 | NT |
| Apigenin | 107.5 ± 1.8 | NT |
| Quercetin 4 | 20.2 ± 0.8 | NT |
| Compounds | [I], (µM) | Vmax1 | Km2 | Kik/Kiv3 |
|---|---|---|---|---|
| 1 | 0 | 5.531 | 107.4564 | - |
| - | 0.12 | 2.738 | 105.2853 | 16.0415 |
| - | 0.24 | 2.513 | 98.6856 | 6.6846 |
| - | 0.48 | 1.494 | 97.3510 | 7.7610 |
| 5 | 0 | 5.1813 | 120.1982 | - |
| - | 35.0 | 3.4578 | 131.0961 | 3.6688 |
| - | 70.0 | 2.1692 | 150.0805 | 2.3384 |
| - | 140.0 | 1.6200 | 161.0846 | 2.0207 |
| Compounds | KSV (× 105 L·mol−1) | R2 | KA (× 106 L·mol−1) | n | R2 |
|---|---|---|---|---|---|
| 1 | 4.1104 | 0.9886 | 0.9666 | 1.6920 | 0.9917 |
| 2 | 2.3636 | 0.9843 | 0.7923 | 1.1974 | 0.9933 |
| 3 | 1.3442 | 0.9997 | 0.7604 | 1.1206 | 0.9915 |
| 4 | 0.9569 | 0.9989 | 0.7335 | 1.0917 | 0.9998 |
| 5 | 0.6040 | 0.9920 | 0.5844 | 0.8147 | 0.9955 |
| 6 | 0.5608 | 0.9875 | 0.5057 | 0.7006 | 0.9998 |
| 7 | 2.1656 | 0.9895 | 0.7851 | 1.1733 | 0.9955 |
| 8 | 1.1120 | 0.9904 | 0.7416 | 1.0868 | 0.9986 |
| Luteolin | 0.5614 | 0.9986 | 0.5578 | 0.7662 | 0.9998 |
| Dihydrokaempferol | 0.2479 | 0.9959 | 0.2998 | 0.5758 | 0.9986 |
| Apigenin | 0.4481 | 0.9926 | 0.5142 | 0.4481 | 0.9993 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, H.M.; Kim, J.Y.; Li, Z.P.; Jenis, J.; Ban, Y.J.; Baiseitova, A.; Park, K.H. Effectiveness of Prenyl Group on Flavonoids from Epimedium koreanum Nakai on Bacterial Neuraminidase Inhibition. Molecules 2019, 24, 317. https://doi.org/10.3390/molecules24020317
Choi HM, Kim JY, Li ZP, Jenis J, Ban YJ, Baiseitova A, Park KH. Effectiveness of Prenyl Group on Flavonoids from Epimedium koreanum Nakai on Bacterial Neuraminidase Inhibition. Molecules. 2019; 24(2):317. https://doi.org/10.3390/molecules24020317
Chicago/Turabian StyleChoi, Hong Min, Jeong Yoon Kim, Zuo Peng Li, Janar Jenis, Yeong Jun Ban, Aizhamal Baiseitova, and Ki Hun Park. 2019. "Effectiveness of Prenyl Group on Flavonoids from Epimedium koreanum Nakai on Bacterial Neuraminidase Inhibition" Molecules 24, no. 2: 317. https://doi.org/10.3390/molecules24020317
APA StyleChoi, H. M., Kim, J. Y., Li, Z. P., Jenis, J., Ban, Y. J., Baiseitova, A., & Park, K. H. (2019). Effectiveness of Prenyl Group on Flavonoids from Epimedium koreanum Nakai on Bacterial Neuraminidase Inhibition. Molecules, 24(2), 317. https://doi.org/10.3390/molecules24020317







