Pharmacophoric Site Identification and Inhibitor Design for Autotaxin
Abstract
1. Introduction
2. Results and Discussion
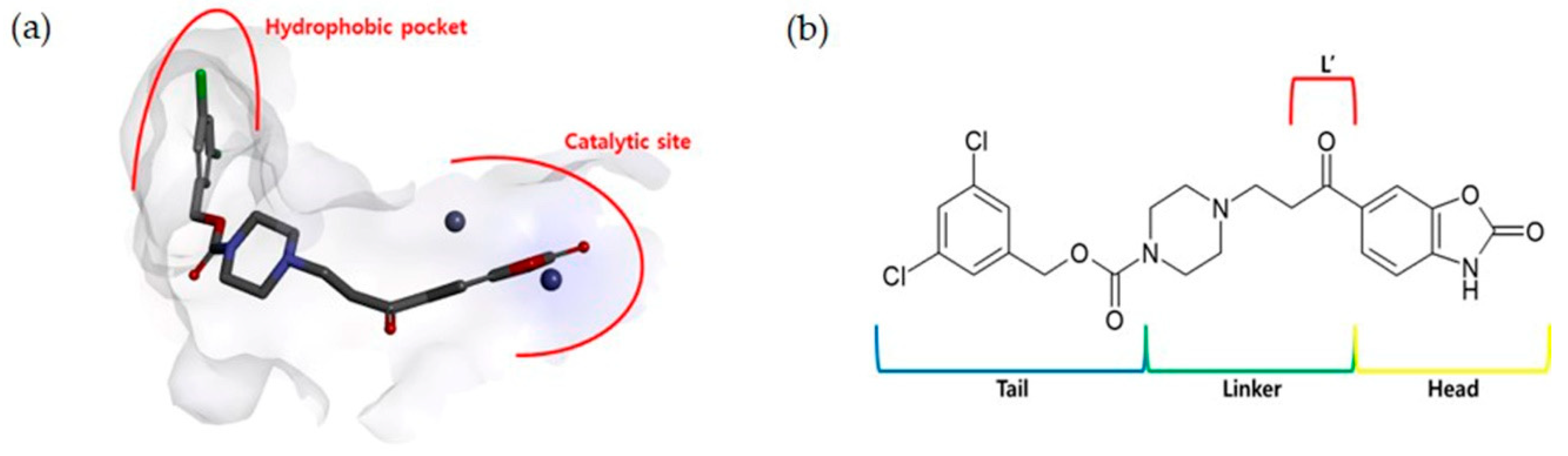
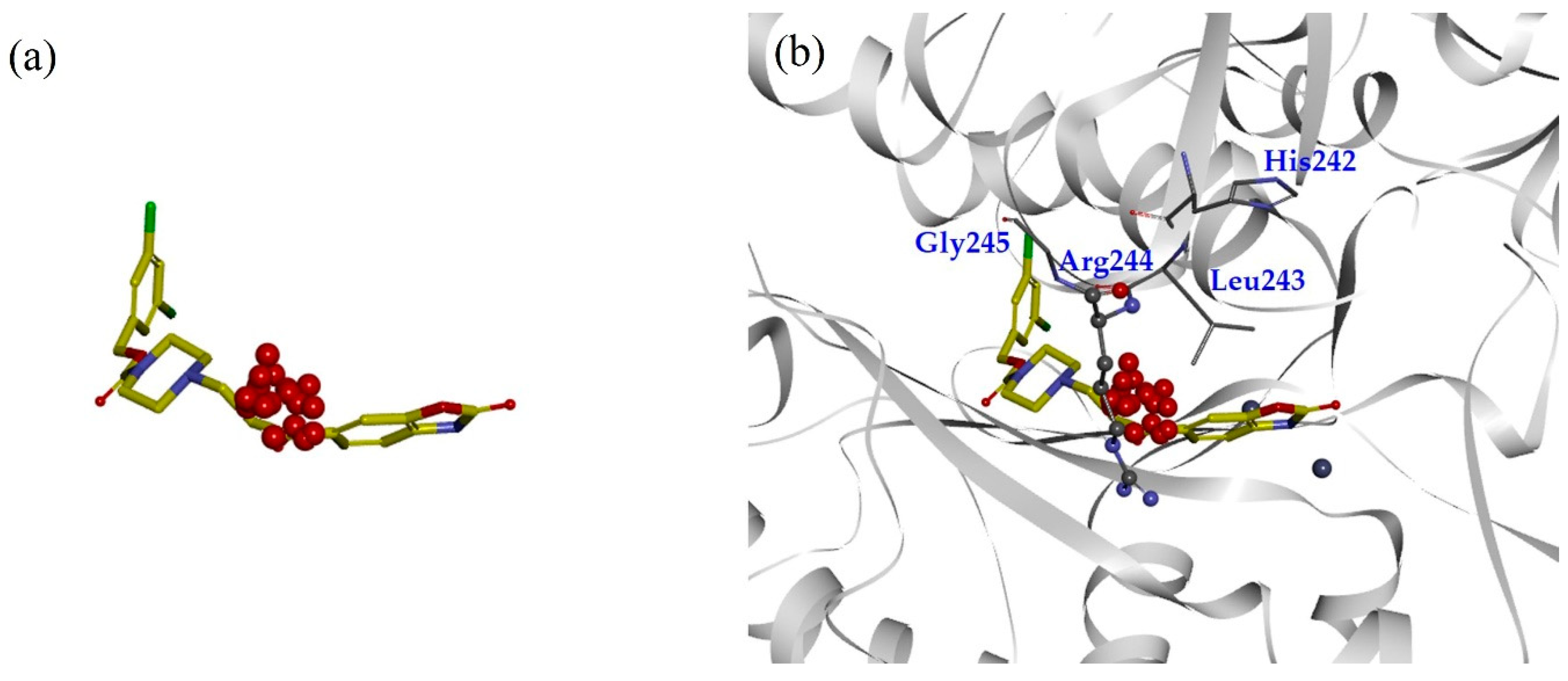
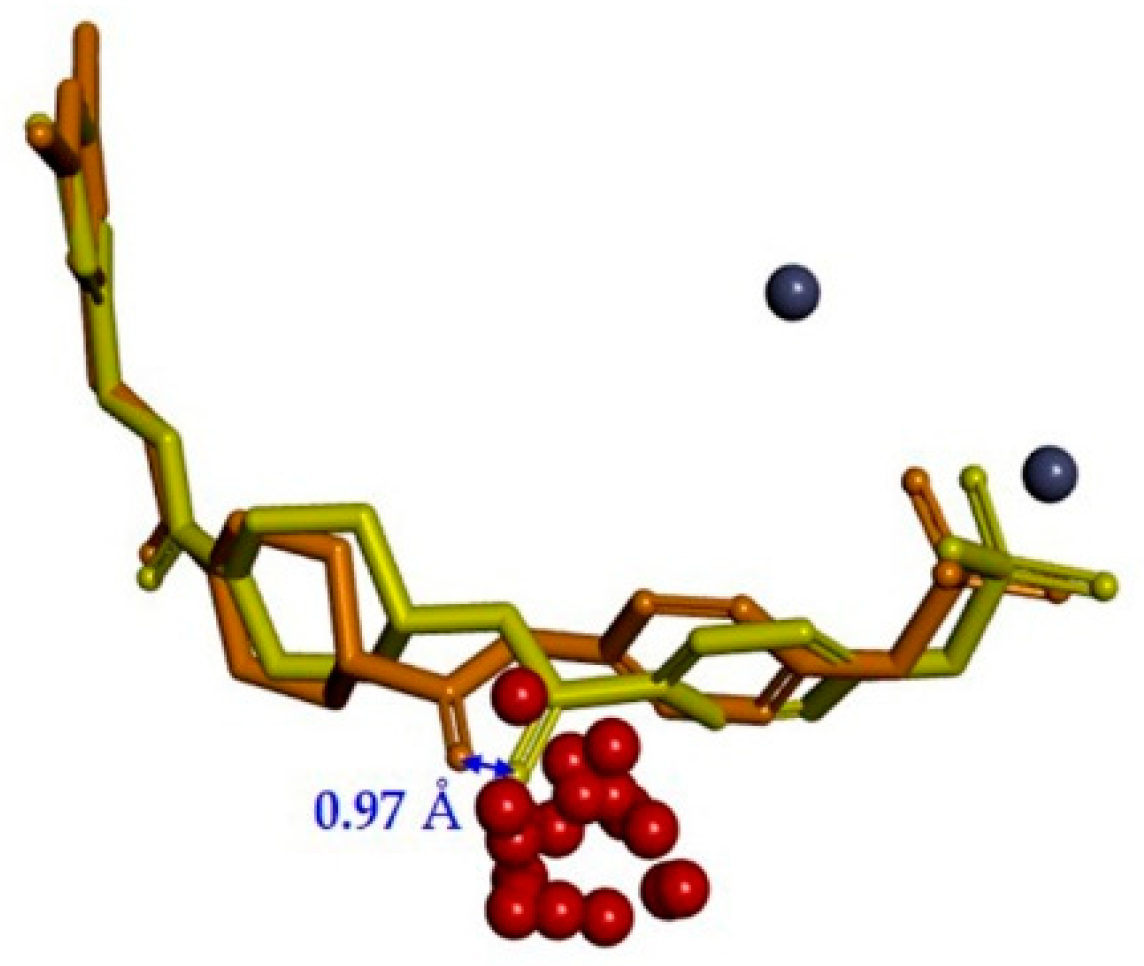
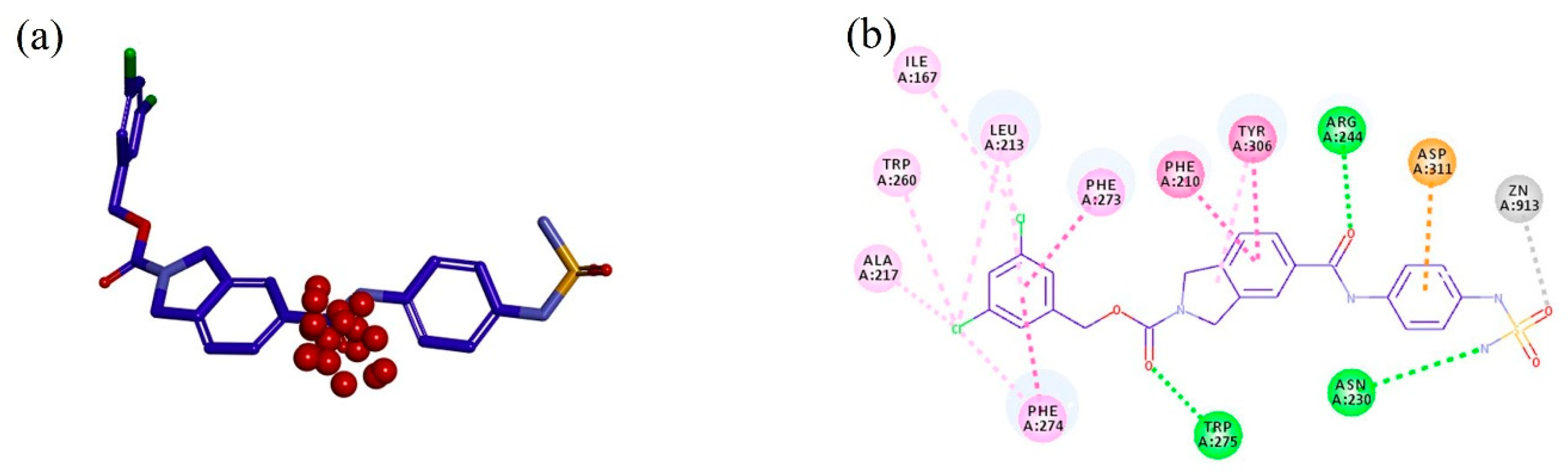
2.1. TWN Analysis and Identification of the Pharmacophoric Sites
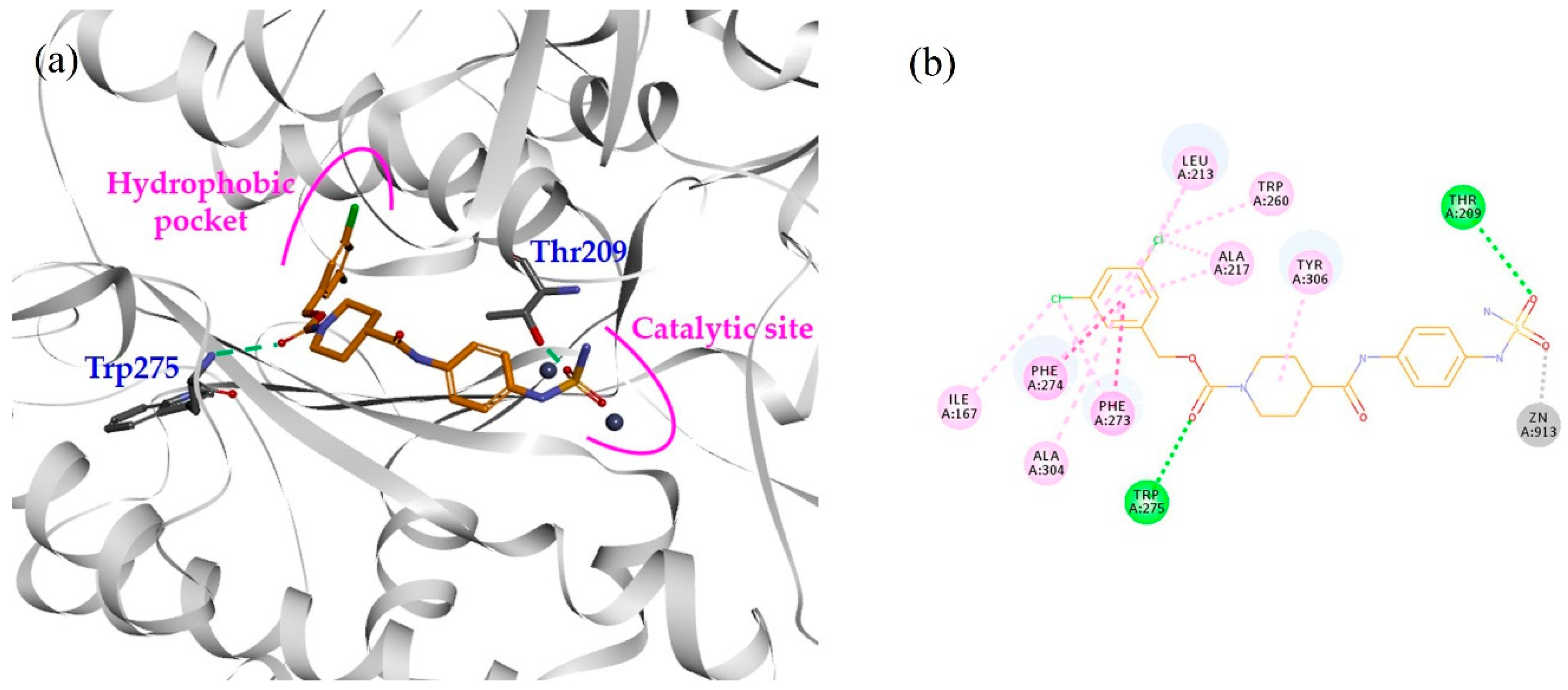
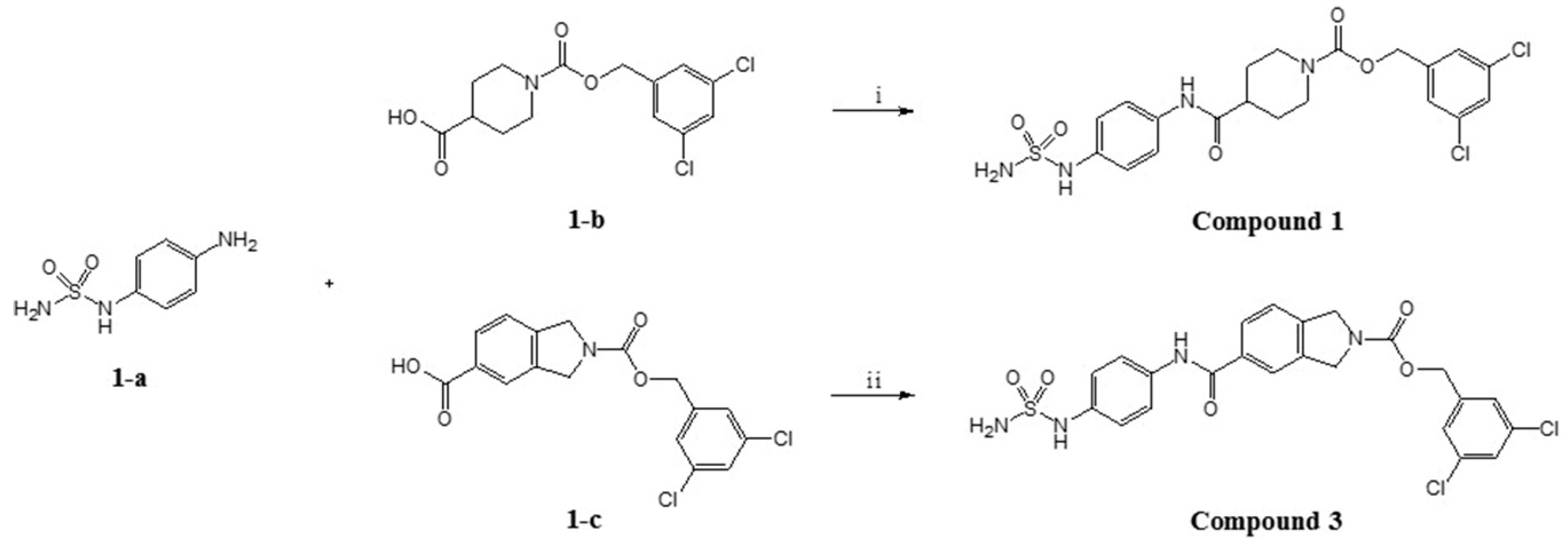
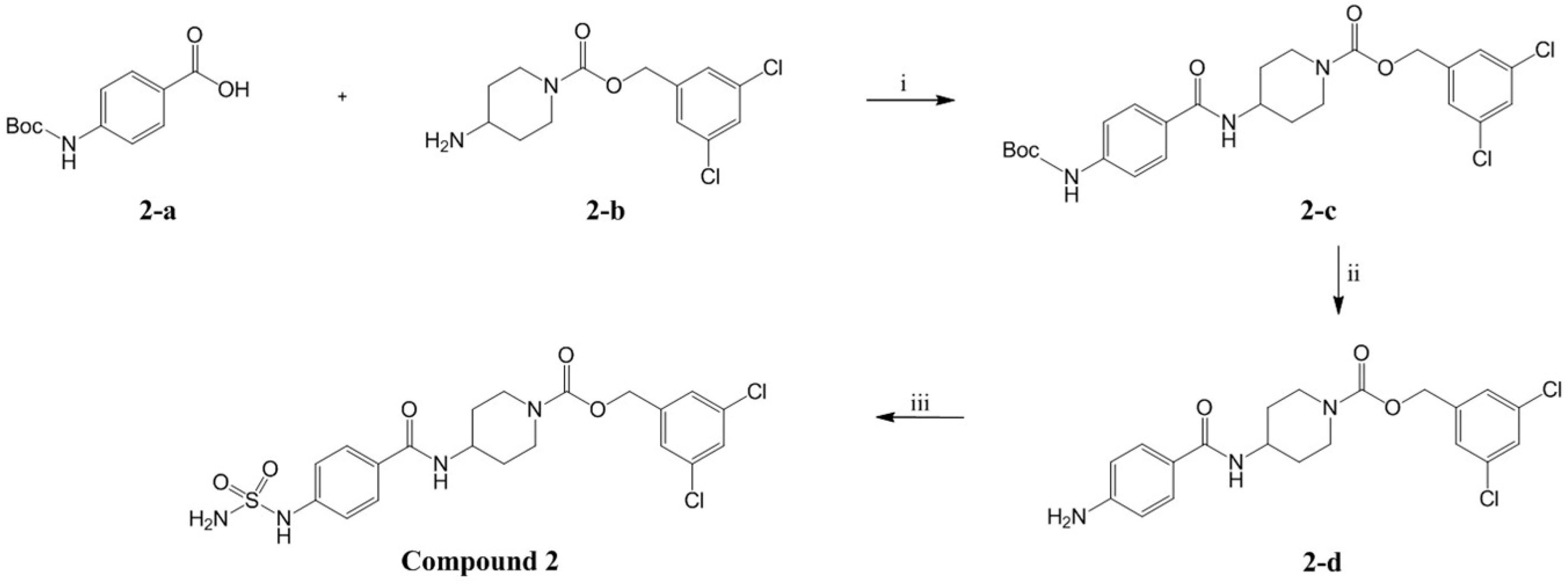
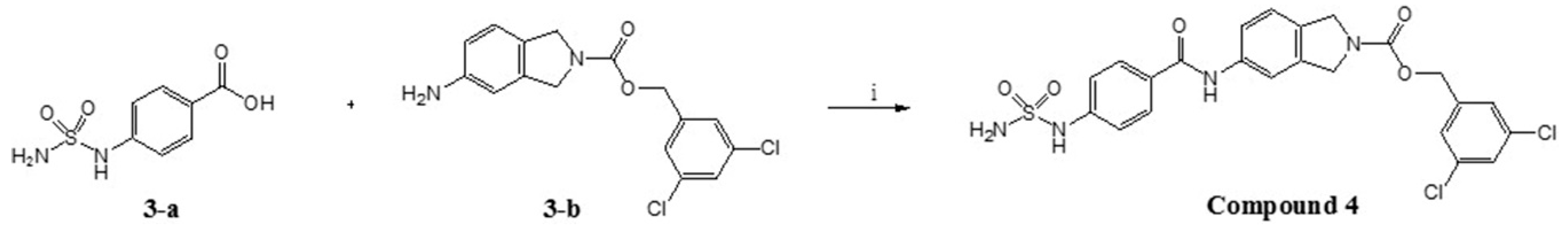
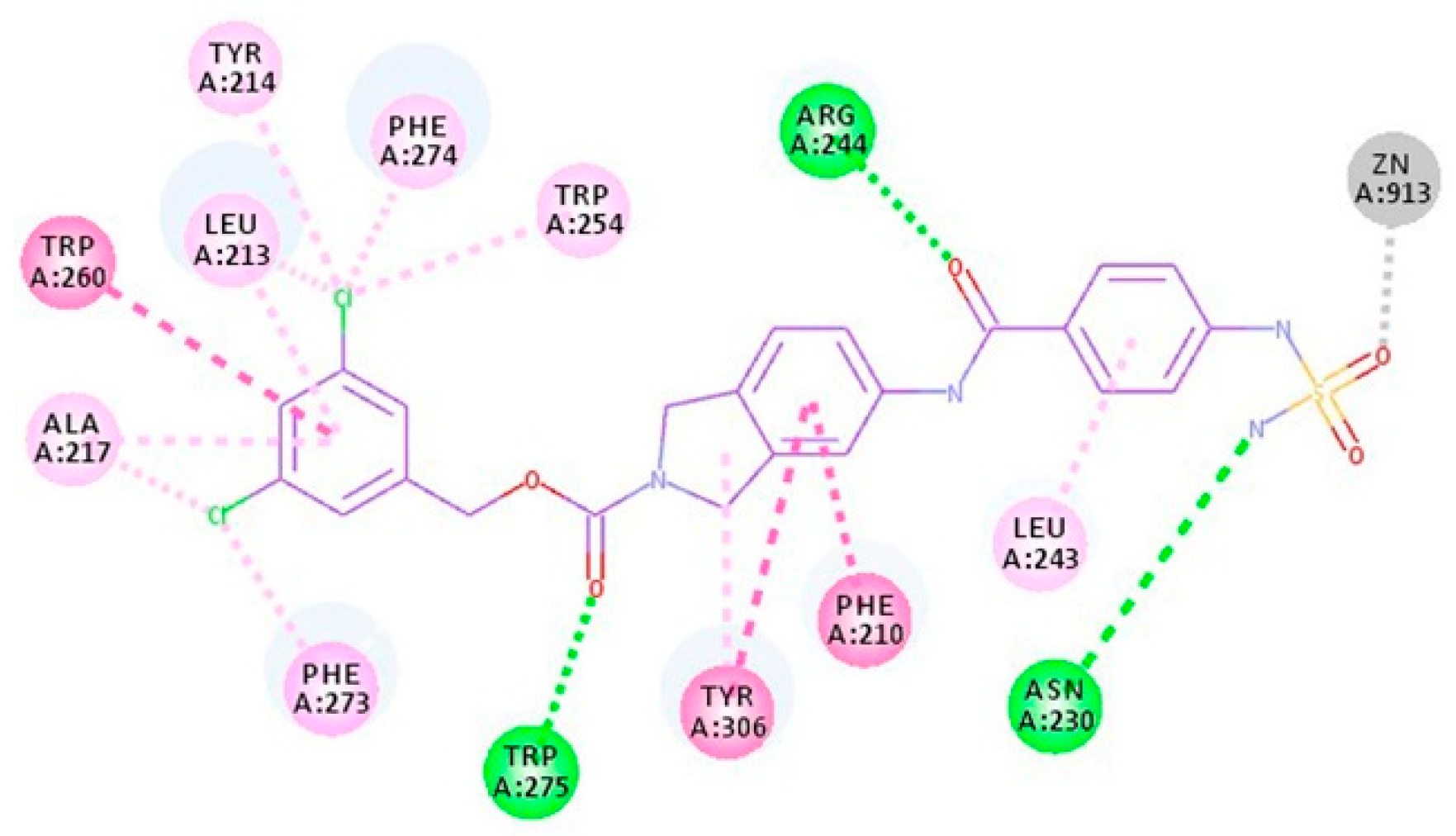
2.2. Design, Synthesis and Activity Evaluation
3. Materials and Methods
3.1. Protein Preparation
3.2. MD Simulation
3.3. TWN Analysis
3.4. Molecular Docking
3.5. Synthesis and Characterization
3.5.1. General Information
3.5.2. (3,5-dichlorophenyl)methyl 4-{[4-(sulfamoylamino)phenyl]carbamoyl}piperidine-1-carboxylate (Compound 1)
3.5.3. (3,5-dichlorophenyl)methyl 4-[4-(sulfamoylamino)benzamido]piperidine-1-carboxylate (Compound 2)
3.5.4. (3,5-dichlorophenyl)methyl 5-{[4-(sulfamoylamino)phenyl]carbamoyl}-2,3-dihydro-1H-isoindole-2-carboxylate (Compound 3)
3.5.5. (3,5-dichlorophenyl)methyl 5-[4-(sulfamoylamino)benzamido]-2,3-dihydro-1H-isoindole-2-carboxylate (Compound 4)
3.6. Biological Experiment
3.6.1. In Vitro bis-pNPP Assay
3.6.2. Ex Vivo Human Plasma Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Tokumura, A.; Majima, E.; Kariya, Y.; Tominaga, K.; Kogure, K.; Yasuda, K.; Fukuzawa, K. Identification of human plasma lysophospholipase D, a lysophosphatidic acid-producing enzyme, as autotaxin, a multifunctional phosphodiesterase. J. Biol. Chem. 2002, 277, 39436–39442. [Google Scholar] [CrossRef] [PubMed]
- Umezu-Goto, M.; Kishi, Y.; Taira, A.; Hama, K.; Dohmae, N.; Takio, K.; Yamori, T.; Mills, G.B.; Inoue, K.; Arai, H. Autotaxin has lysophospholipase D activity leading to tumor cell growth and motility by lysophosphatidic acid production. J. Cell Biol. 2002, 158, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, K.; Herr, D.; Mutoh, T.; Chun, J. Lysophosphatidic acid (LPA) and its receptors. Curr. Opin. Pharmacol. 2009, 9, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Moolenaar, W.H.; van Meeteren, L.A.; Giepmans, B.N. The ins and outs of lysophosphatidic acid signaling. Bioessays 2004, 26, 870–881. [Google Scholar] [CrossRef] [PubMed]
- Bourgoin, S.G.; Zhao, C. Autotaxin and lysophospholipids in rheumatoid arthritis. Curr. Opin. Investig. Drugs 2010, 11, 515–526. [Google Scholar] [PubMed]
- Zhao, Y.; Natarajan, V. Lysophosphatidic acid (LPA) and its receptors: Role in airway inflammation and remodeling. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids 2013, 1831, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Tager, A.M.; LaCamera, P.; Shea, B.S.; Campanella, G.S.; Selman, M.; Zhao, Z.; Polosukkhin, V.; Wain, J.; Karimi-Shah, B.A.; Kim, N.D.; et al. The lysophosphatidic acid receptor LPA 1 links pulmonary fibrosis to lung injury by mediating fibroblast recruitment and vascular leak. Nat. Med. 2008, 14, 45. [Google Scholar] [CrossRef]
- Willier, S.; Butt, E.; Grunewald, T.G. Lysophosphatidic acid (LPA) signalling in cell migration and cancer invasion: A focussed review and analysis of LPA receptor gene expression on the basis of more than 1700 cancer microarrays. Biol. Cell 2013, 105, 317–333. [Google Scholar] [CrossRef]
- Rivera-Lopez, C.M.; Tucker, A.L.; Lynch, K.R. Lysophosphatidic acid (LPA) and angiogenesis. Angiogenesis 2008, 11, 301–310. [Google Scholar] [CrossRef]
- Nikolaou, A.; Kokotou, M.G.; Limnios, D.; Psarra, A.; Kokotos, G. Autotaxin inhibitors: A patent review (2012–2016). Expert Opin. Ther. Pat. 2017, 27, 815–829. [Google Scholar] [CrossRef]
- Barbayianni, E.; Magrioti, V.; Moutevelis-Minakakis, P.; Kokotos, G. Autotaxin inhibitors: A patent review. Expert Opin. Ther. Pat. 2013, 23, 1123–1132. [Google Scholar] [CrossRef] [PubMed]
- Castagna, D.; Budd, D.C.; Macdonald, S.J.; Jamieson, C.; Watson, A.J. Development of autotaxin inhibitors: An overview of the patent and primary literature: Miniperspective. J. Med. Chem. 2016, 59, 5604–5621. [Google Scholar] [CrossRef] [PubMed]
- Desroy, N.; Housseman, C.; Bock, X.; Joncour, A.; Bienvenu, N.; Cherel, L.; Laveguere, V.; Rondet, E.; Peixoto, C.; Grassot, J.; et al. Discovery of 2-[[2-Ethyl-6-[4-[2-(3-hydroxyazetidin-1-yl)-2-oxoethyl] piperazin-1-yl]-8-methylimidazo [1–a] pyridin-3-yl] methylamino]-4-(4-fluorophenyl) thiazole-5-carbonitrile (GLPG1690), a first-in-class autotaxin inhibitor undergoing clinical evaluation for the treatment of idiopathic pulmonary fibrosis. J. Med. Chem. 2017, 60, 3580–3590. [Google Scholar] [PubMed]
- Frey, M. Water structure associated with proteins and its role in crystallization. Acta Crystallogr. D 1994, 50, 663–666. [Google Scholar] [CrossRef] [PubMed]
- Bellissent-Funel, M.-C.; Hassanali, A.; Havenith, M.; Henchman, R.; Pohl, P.; Sterpone, F.; Van Der Spoel, D.; Xu, Y.; Garcia, A.E. Water determines the structure and dynamics of proteins. Chem. Rev. 2016, 116, 7673–7697. [Google Scholar] [CrossRef] [PubMed]
- Volkhard, H. Protein dynamics tightly connected to the dynamics of surrounding and internal water molecules. ChemPhysChem 2007, 8, 23–33. [Google Scholar]
- Baron, R.; Setny, P.; McCammon, J.A. Water in cavity−ligand recognition. J. Am. Chem. Soc. 2010, 132, 12091–12097. [Google Scholar] [CrossRef] [PubMed]
- Baron, R.; Setny, P.; Paesani, F. Water structure, dynamics, and spectral signatures: Changes upon model cavity-ligand recognition. J. Phys. Chem. B 2012, 116, 13774–13780. [Google Scholar] [CrossRef]
- Ladbury, J.E. Just add water! The effect of water on the specificity of protein-ligand binding sites and its potential application to drug design. Chem. Biol. 1996, 3, 973–980. [Google Scholar] [CrossRef]
- Poornima, C.; Dean, P. Hydration in drug design. 1. Multiple hydrogen-bonding features of water molecules in mediating protein-ligand interactions. J. Comput. Aided Mol. Des. 1995, 9, 500–512. [Google Scholar] [CrossRef]
- Hummer, G. Molecular binding: Under water’s influence. Nat. Chem. 2010, 2, 906. [Google Scholar] [CrossRef]
- Quiocho, F.; Wilson, D.K.; Vyas, N. Substrate specificity and affinity of a protein modulated by bound water molecules. Nature 1989, 340, 404. [Google Scholar] [CrossRef]
- Connelly, P.; Aldape, R.; Bruzzese, F.; Chambers, S.; Fitzgibbon, M.; Fleming, M.; Itoh, S.; Livingston, D.; Navia, M.; Thomson, J.; et al. Enthalpy of hydrogen bond formation in a protein-ligand binding reaction. Proc. Natl. Acad. Sci. USA 1994, 91, 1964–1968. [Google Scholar] [CrossRef]
- Otting, G.; Liepinsh, E.; Wuthrich, K. Protein hydration in aqueous solution. Science 1991, 254, 974–980. [Google Scholar] [CrossRef]
- Barillari, C.; Taylor, J.; Viner, R.; Essex, E. Classification of water molecules in protein binding sites. J. Am. Chem. Soc. 2007, 129, 2577–2587. [Google Scholar] [CrossRef]
- Uehara, S.; Tanaka, S. AutoDock-GIST: Incorporating thermodynamics of active-site water into scoring function for accurate protein-ligand docking. Molecules 2016, 21, 1604. [Google Scholar] [CrossRef]
- Li, Z.; Lazaridis, T. Thermodynamics of buried water clusters at a protein-ligand binding interface. J. Phys. Chem. B 2006, 110, 1464–1475. [Google Scholar] [CrossRef]
- Chen, J.M.; Xu, S.L.; Wawrzak, Z.; Basarab, G.S.; Jordan, D.B. Structure-based design of potent inhibitors of scytalone dehydratase: Displacement of a water molecule from the active site. Biochemistry 1998, 37, 17735–17744. [Google Scholar] [CrossRef]
- Tiwary, P.; Mondal, J.; Morrone, J.A.; Berne, B.J. Role of water and steric constraints in the kinetics of cavity-ligand unbinding. Proc. Natl. Acad. Sci. USA 2015, 112, 12015–12019. [Google Scholar] [CrossRef]
- Kouza, M.; Banerji, A.; Kolinski, A.; Buhimschi, I.; Kloczkowski, A. Role of resultant dipole moment in mechanical dissociation of biological complexes. Molecules 2018, 23, 1995. [Google Scholar] [CrossRef]
- Thirumalai, D.; Reddy, G.; Straub, J.E. Role of water in protein aggregation and amyloid polymorphism. Acc. Chem. Res. 2012, 45, 83–92. [Google Scholar] [CrossRef]
- Kouza, M.; Co, N.T.; Li, M.S.; Kmiecik, S.; Kolinski, A.; Kloczkowski, A.; Buhimschi, I.A. Kinetics and mechanical stability of the fibril state control fibril formation time of polypeptide chains: A computational study. J. Chem. Phys. 2018, 148, 215106. [Google Scholar] [CrossRef]
- Balupuri, A.; Choi, K.-E.; Kang, N.S. Computational insights into the role of α-strand/sheet in aggregation of α-synuclein. Sci. Rep. 2019, 9, 59. [Google Scholar] [CrossRef]
- Bucher, D.; Stouten, P.; Triballeau, N. Shedding light on important waters for drug design: Simulations versus grid-based methods. J. Chem. Inf. Model. 2018, 58, 692–699. [Google Scholar] [CrossRef]
- Horbert, R.; Pinchuk, B.; Johannes, E.; Schlosser, J.; Schmidt, D.; Cappel, D.; Totzke, F.; Schaächtele, C.; Peifer, C. Optimization of potent DFG-in inhibitors of platelet derived growth factor receptorβ (PDGF-Rβ) guided by water thermodynamics. J. Med. Chem. 2014, 58, 170–182. [Google Scholar] [CrossRef]
- Marrone, T.J.; Briggs, J.M.; McCammon, J.A. Structure-based drug design: Computational advances. Annu. Rev. Pharmacol. 1997, 37, 71–90. [Google Scholar] [CrossRef]
- Roberts, B.C.; Mancera, R.L. Ligand-protein docking with water molecules. J. Chem. Inf. Model. 2008, 48, 397–408. [Google Scholar] [CrossRef]
- De Graff, C.; Oostenbrink, C.; Keizers, P.H.J.; van der Wijst, T.; Jongejan, A.; Vermeulen, N.P.E. Catalytic site prediction and virtual screening of cytochrome P450 2D6 substrates by consideration of water and rescoring in automated docking. J. Med. Chem. 2006, 49, 2417–2430. [Google Scholar] [CrossRef]
- Jang, W.D.; Kim, J.-T.; Kang, N.S. The analysis of water network for kinase selectivity based on the MD simulations. J. Mol. Liq. 2014, 191, 37–41. [Google Scholar] [CrossRef]
- Jang, W.D.; Lee, M.H.; Kang, N.S. Quantitative assessment of kinase selectivity based the water-ring network in protein binding sites using molecular dynamics simulations. J. Mol. Liq. 2016, 221, 316–322. [Google Scholar] [CrossRef]
- Lee, M.; Balupuri, A.; Jung, Y.-R.; Choi, S.; Lee, A.; Cho, Y.; Kang, N.S. Design of a novel and selective IRAK4 inhibitor using topological water network analysis and molecular modeling approaches. Molecules 2018, 23, 3136. [Google Scholar] [CrossRef]
- Choi, K.-E.; Chae, E.; Balupuri, A.; Yoon, H.R.; Kang, N.S. Topological water network analysis around amino acids. Molecules 2019, 24, 2653. [Google Scholar] [CrossRef]
- Balupuri, A.; Lee, D.Y.; Lee, M.H.; Chae, S.; Jung, E.; Kim, Y.; Ryu, J.; Kang, N.S. Design, synthesis, docking and biological evaluation of 4-phenyl-thiazole derivatives as autotaxin (ATX) inhibitors. Bioorg. Med. Chem. Lett. 2017, 27, 4156–4164. [Google Scholar] [CrossRef]
- Balupuri, A.; Lee, M.H.; Chae, S.; Jung, E.; Yoon, W.; Kim, Y.; Son, S.J.; Ryu, J.; Kang, D.H.; Yang, Y.J.; et al. Discovery and optimization of ATX inhibitors via modeling, synthesis and biological evaluation. Eur. J. Med. Chem. 2018, 148, 397–409. [Google Scholar] [CrossRef]
- Gierse, J.; Thorarensen, A.; Beltey, K.; Bradshaw-Pierce, E.; Cortes-Burgos, L.; Hall, T.; Johnston, A.; Murphy, M.; Nemirovskiy, O.; Ogawa, S.; et al. A novel autotaxin inhibitor reduces lysophosphatidic acid levels in plasma and the site of inflammation. J. Pharmacol. Exp. Ther. 2010, 334, 310–317. [Google Scholar] [CrossRef]
- Van Meeteren, L.A.; Ruurs, P.; Christodoulou, E.; Goding, J.W.; Takakusa, H.; Kikuchi, K.; Perrakis, A.; Nagano, T.; Moolenaar, W.H. Inhibition of autotaxin by lysophosphatidic acid and sphingosine 1-phosphate. J. Biol. Chem. 2005, 280, 21155–21161. [Google Scholar] [CrossRef]
- Nishimasu, H.; Okudaira, S.; Hama, K.; Mihara, E.; Dohmae, N.; Inoue, A.; Ishitani, R.; Takagi, J.; Aoki, J.; Nureki, O. Crystal structure of autotaxin and insight into GPCR activation by lipid mediators. Nat. Struct. Mol. Biol. 2011, 18, 205. [Google Scholar] [CrossRef]
- Gijsbers, R.; Aoki, J.; Arai, H.; Bollen, M. The hydrolysis of lysophospholipids and nucleotides by autotaxin (NPP2) involves a single catalytic site. FEBS Lett. 2003, 538, 60–64. [Google Scholar] [CrossRef]
- Moolenaar, W.H.; Perrakis, A. Insights into autotaxin: How to produce and present a lipid mediator. Nat. Rev. Mol. Cell Biol. 2011, 12, 674. [Google Scholar] [CrossRef]
- Kawaguchi, M.; Okabe, T.; Okudaira, S.; Nishimasu, H.; Ishitani, R.; Kojima, H.; Nureki, O.; Aoki, J.; Nagano, T. Screening and X-ray crystal structure-based optimization of autotaxin (ENPP2) inhibitors, using a newly developed fluorescence probe. ACS Chem. Biol. 2013, 8, 1713–1721. [Google Scholar] [CrossRef]
- Berman, H.M.; Battistuz, T.; Bhat, T.N.; Bluhm, W.F.; Bourne, P.E.; Burkhardt, K.; Feng, Z.; Gilliand, G.L.; Iype, L.; Jin, S.; et al. The protein data bank. Acta Crystallogr. D Biol. Crystallogr. 2002, 58, 899–907. [Google Scholar] [CrossRef]
- Pronk, S.; Páll, S.; Schulz, R.; Larsson, P.; Bjelkmar, P.; Apostolov, R.; Shirts, M.R.; Smith, J.C.; Kasson, P.M.; Spoel, D.; et al. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013, 29, 845–854. [Google Scholar] [CrossRef]
- Schuler, L.D.; Daura, X.; Van Gunsteren, W.F. An improved GROMOS96 force field for aliphatic hydrocarbons in the condensed phase. J. Comput. Chem. 2001, 22, 1205–1218. [Google Scholar] [CrossRef]
- Berendsen, H.J.; Postma, J.V.; van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N⋅log (N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.; Fraaije, J.G. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Stein, A.J.; Bain, G.; Prodanovich, P.; Santini, A.M.; Darlington, J.; Stelzer, N.M.; Sidhu, R.S.; Schaub, J.; Goulet, L.; Lonerga, D.; et al. Structural basis for inhibition of human autotaxin by four potent compounds with distinct modes of binding. Mol. Pharmacol. 2015, 88, 982–992. [Google Scholar] [CrossRef]
- Momany, F.A.; Rone, R. Validation of the general purpose QUANTA® 3.2/CHARMm® force field. J. Comput. Chem. 1992, 13, 888–900. [Google Scholar] [CrossRef]
- Brooks, B.R.; Bruccoleri, R.E.; Olafson, B.D.; States, D.J.; Swaminathan, S.A.; Karplus, M. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983, 4, 187–217. [Google Scholar] [CrossRef]
- Wu, G.; Robertson, D.H.; Brooks III, C.L.; Vieth, M. Detailed analysis of grid-based molecular docking: A case study of CDOCKER—A CHARMm-based MD docking algorithm. J. Comput. Chem. 2003, 24, 1549–1562. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |
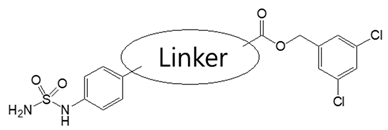 | |||||
|---|---|---|---|---|---|
| Compound | Linker | Bis-pNPP IC50 (nM) | MW | ClogP a | PSA b |
| 1 | 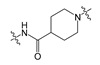 | 250.00 | 501.38 | 3.59 | 139.21 |
| 2 | 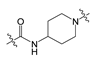 | 44.50 | 501.38 | 3.63 | 139.21 |
| 3 | 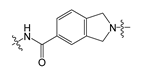 | 3.86 | 535.40 | 4.36 | 139.21 |
| 4 | 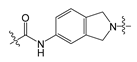 | 12.70 | 535.40 | 4.70 | 139.21 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, M.H.; Lee, D.-Y.; Balupuri, A.; Jeong, J.-W.; Kang, N.S. Pharmacophoric Site Identification and Inhibitor Design for Autotaxin. Molecules 2019, 24, 2808. https://doi.org/10.3390/molecules24152808
Lee MH, Lee D-Y, Balupuri A, Jeong J-W, Kang NS. Pharmacophoric Site Identification and Inhibitor Design for Autotaxin. Molecules. 2019; 24(15):2808. https://doi.org/10.3390/molecules24152808
Chicago/Turabian StyleLee, Myeong Hwi, Dae-Yon Lee, Anand Balupuri, Jong-Woo Jeong, and Nam Sook Kang. 2019. "Pharmacophoric Site Identification and Inhibitor Design for Autotaxin" Molecules 24, no. 15: 2808. https://doi.org/10.3390/molecules24152808
APA StyleLee, M. H., Lee, D.-Y., Balupuri, A., Jeong, J.-W., & Kang, N. S. (2019). Pharmacophoric Site Identification and Inhibitor Design for Autotaxin. Molecules, 24(15), 2808. https://doi.org/10.3390/molecules24152808







