Interactions between β-Lactoglobulin and 3,3′-Diindolylmethane in Model System
Abstract
1. Introduction
2. Results and Discussion
2.1. Effects of DIM on Particle Size of β-LG in Solution
2.2. Effects of DIM on Zeta Potential of β-LG
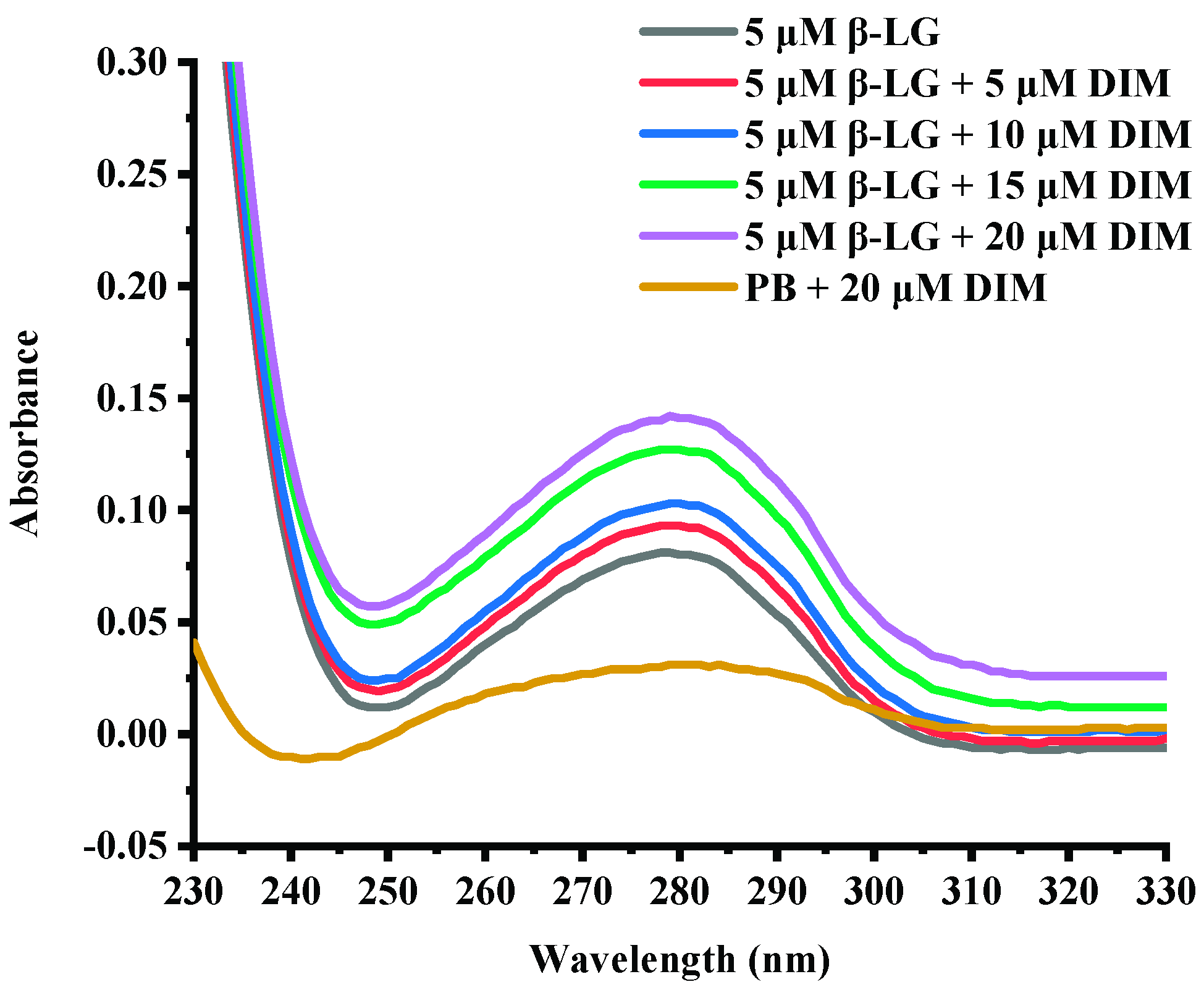
2.3. Effects of DIM on UV-Absorption Spectra of β-LG
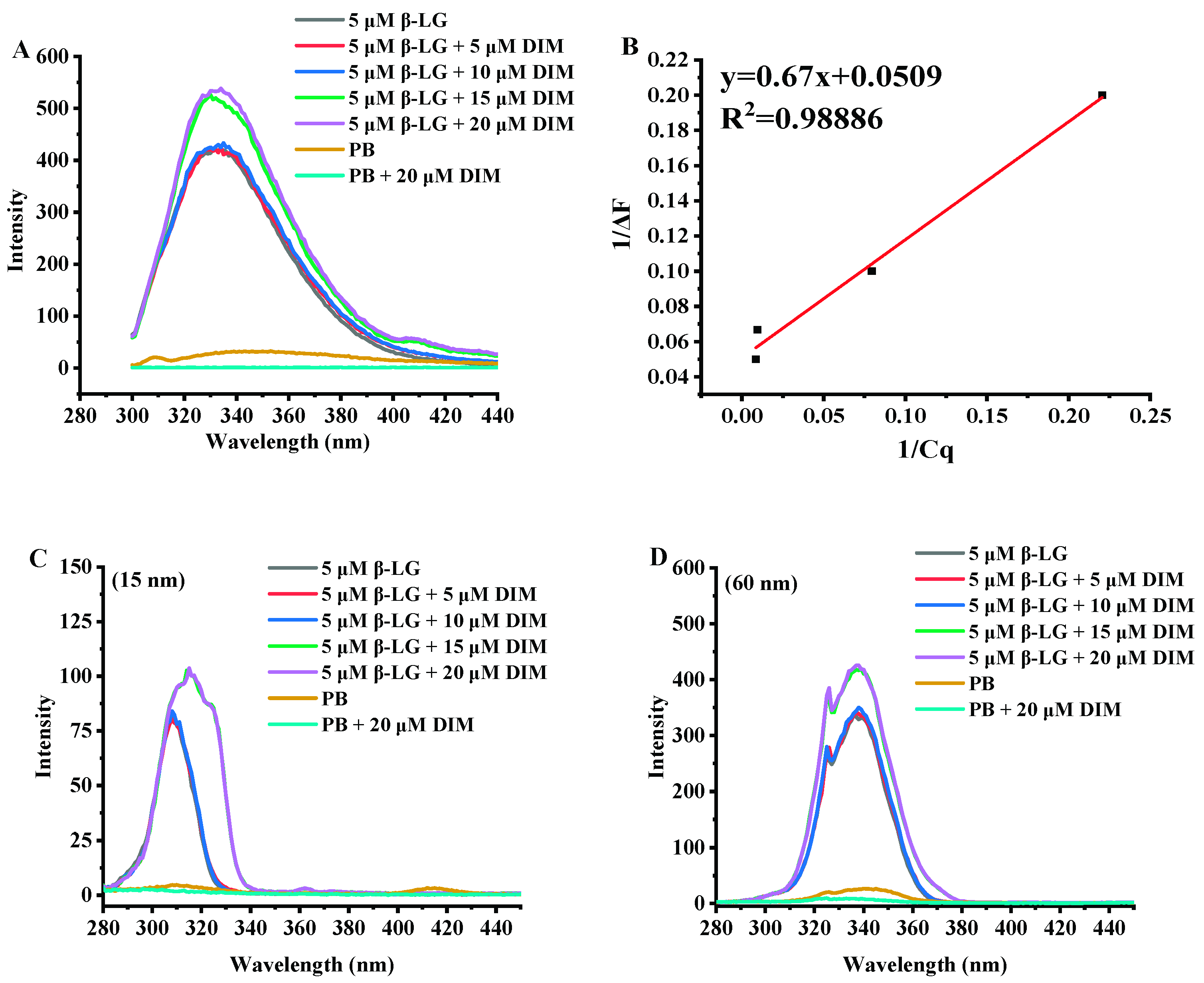
2.4. Effects of DIM on Fluorescence Emission Spectra of β-LG
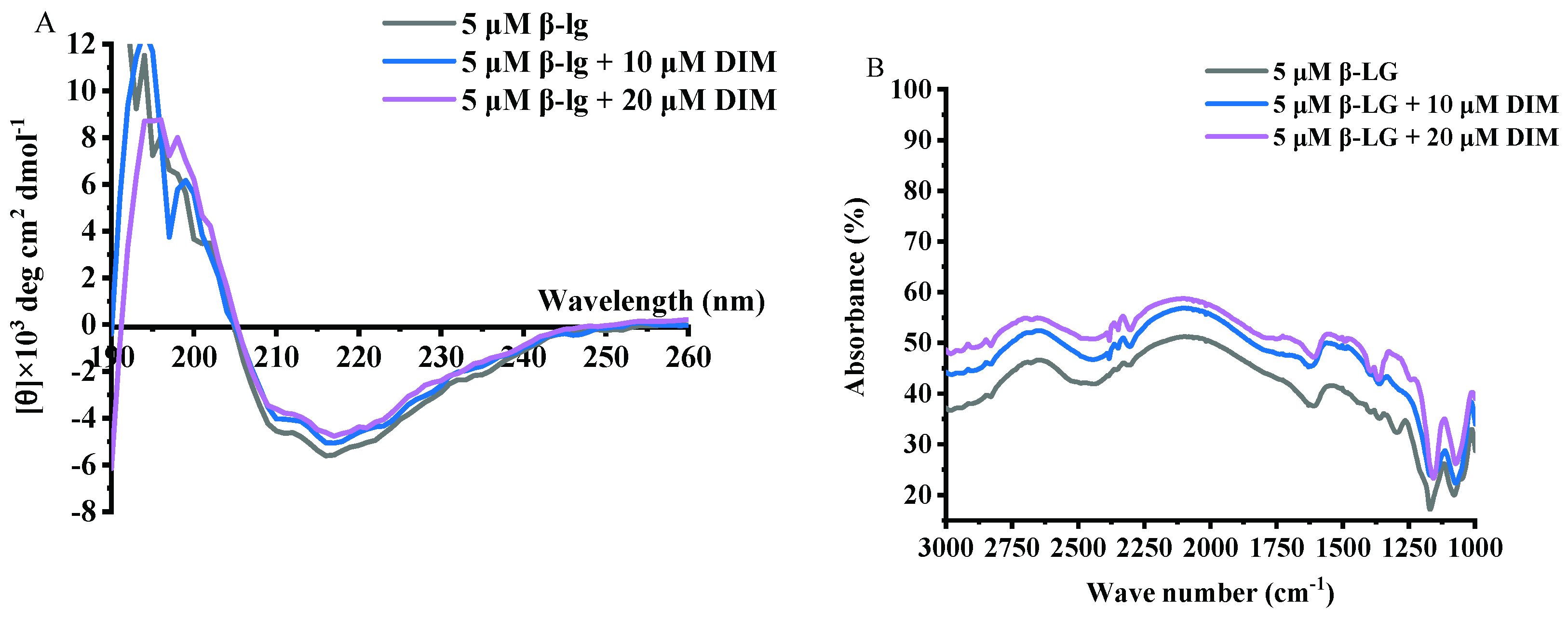
2.5. Effects of DIM on Second Structure of β-LG
2.6. Effects of DIM on Fourier Transform Infrared (FT-IR) Spectra of β-LG
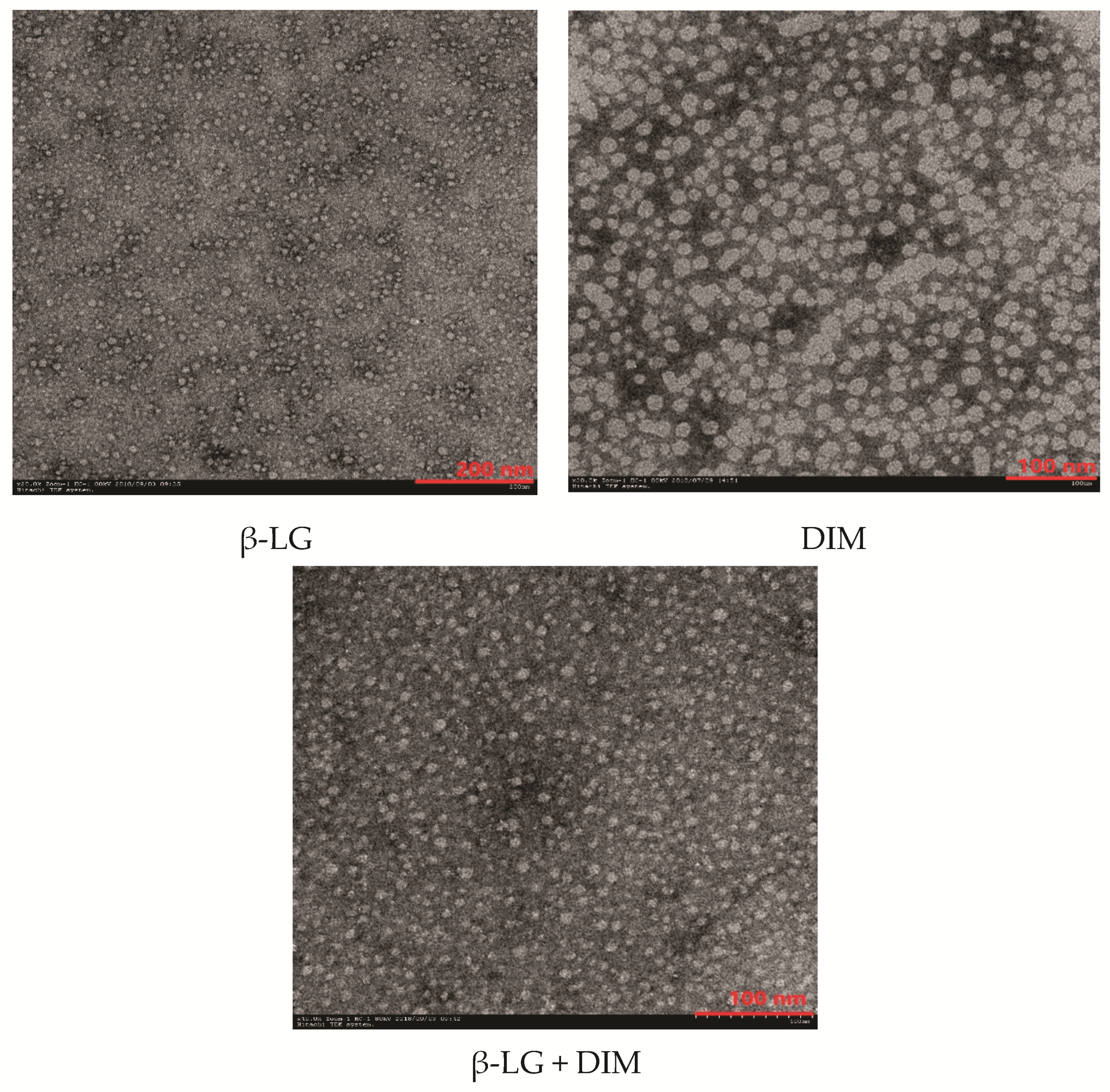
2.7. Effects of DIM on microstructure of β-LG particles
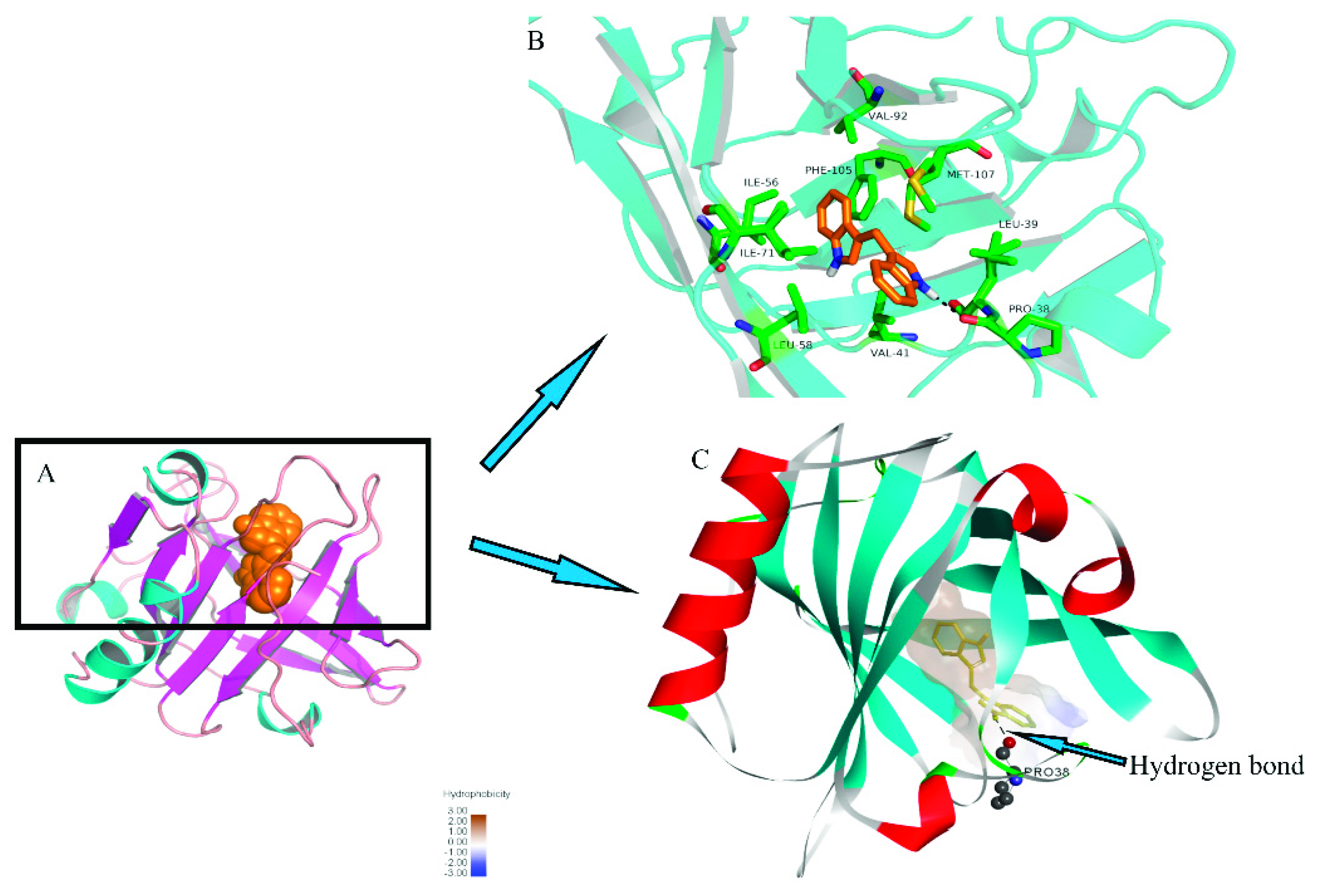
2.8. Molecular Docking between DIM and β-LG
3. Materials and Methods
3.1. Materials
3.2. Preparation of β-LG and DIM Solution
3.3. Particle Size and Zeta Potential Measurement
3.4. Absorption Spectra Measurements
3.5. Fluorescence Spectra Measurements
3.6. Far-Ultraviolet Circular Dichroism (Far-UV CD) Spectroscopy
3.7. Fourier Transform Infrared (FT-IR) Spectroscopy
3.8. Transmission Electron Microscopy (TEM) Analysis
3.9. Molecular Docking
3.10. Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Chen, I.; McDougal, A.; Wang, F.; Safe, S. Aryl hydrocarbon receptor-mediated antiestrogenic and antitumorigenic activity of diindolylmethane. Carcinogenesis 1998, 19, 1631–1639. [Google Scholar] [CrossRef] [PubMed]
- Reed, G.A.; Sunega, J.M.; Sullivan, D.K.; Gray, J.C.; Mayo, M.S.; Crowell, J.A.; Hurwitz, A. Single-dose pharmacokinetics and tolerability of absorption-enhanced 3,3′-diindolylmethane in healthy subjects. Cancer Epidemiol. Biomark. Prev. 2008, 17, 2619–2624. [Google Scholar] [CrossRef] [PubMed]
- Zeligs, M.A.; Jacobs, I.C. Compositions and methods of adjusting steroid hormone metabolism through phytochemicals. U.S. Patent 6,086,915, 11 July 2000. [Google Scholar]
- Jellinck, P.H.; Makin, H.L.; Sepkovic, D.W.; Bradlow, H.L. Influence of indole carbinols and growth hormone on the metabolism of 4-androstenedione by rat liver microsomes. J. Steroid Biochem. Mol. Biol. 1993, 46, 791–798. [Google Scholar] [CrossRef]
- Luo, Y.; Wang, T.T.; Teng, Z.; Chen, P.; Sun, J.; Wang, Q. Encapsulation of indole-3-carbinol and 3,3′-diindolylmethane in zein/carboxymethyl chitosan nanoparticles with controlled release property and improved stability. Food Chem. 2013, 139, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Boye, J.; Ma, C.; Ismail, A.; Harwalkar, V.; Kalab, M. Molecular and microstructural studies of thermal denaturation and gelation of beta-lactoglobulins A and B. J. Agric. Food. Chem. 1997, 45, 1608–1618. [Google Scholar] [CrossRef]
- Divsalar, A.; Saboury, A.A.; Ahmad, F.; Moosavimovahedi, A.A. Effects of temperature and chromium (III) ion on the structure of bovine β-lactoglobulin-A. J. Braz. Chem. Soc. 2009, 20, 245–248. [Google Scholar] [CrossRef]
- Yadav, J.S.S.; Yan, S.; Pilli, S.; Kumar, L.; Tyagi, R.D.; Surampalli, R.Y. Cheese whey: A potential resource to transform into bioprotein, functional/nutritional proteins and bioactive peptides. Biotechnol. Adv. 2015, 33, 756–774. [Google Scholar] [CrossRef]
- Liang, L.; Tajmirriahi, H.A.; Subirade, M. Interaction of β-Lactoglobulin with Resveratrol and its Biological Implications. Biomacromolecules 2008, 9, 50–56. [Google Scholar] [CrossRef]
- Li, M.; Ma, Y.; Ngadi, M.O. Binding of curcumin to β-lactoglobulin and its effect on antioxidant characteristics of curcumin. Food Chem. 2013, 141, 1504–1511. [Google Scholar] [CrossRef]
- Futterman, S.; Heller, J. The enhancement of fluorescence and the decreased susceptibility to enzymatic oxidation of retinol complexed with bovine serum albumin, -lactoglobulin, and the retinol-binding protein of human plasma. J. Biol. Chem. 1972, 247, 5168–5172. [Google Scholar]
- Le, M.S.; Giblin, L.; Croguennec, T.; Bouhallab, S.; Brodkorb, A. β-Lactoglobulin as a molecular carrier of linoleate: Characterization and effects on intestinal epithelial cells in vitro. J. Agric. Food. Chem. 2012, 60, 9476–9483. [Google Scholar]
- Yi, J.; Fan, Y.; Yokoyama, W.; Zhang, Y.; Zhao, L. Characterization of milk proteins-lutein complexes and the impact on lutein chemical stability. Food Chem. 2016, 200, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Mirpoor, S.F.; Hosseini, S.M.H.; Yousefi, G.H. Mixed biopolymer nanocomplexes conferred physicochemical stability and sustained release behavior to introduced curcumin. Food Hydrocoll. 2017, 71, 216–224. [Google Scholar] [CrossRef]
- Zhu, J.X.; Sun, X.W.; Wang, S.H.; Xu, Y.; Wang, D.F. Formation of nanocomplexes comprising whey proteins and fucoxanthin: Characterization, spectroscopic analysis, and molecular docking. Food Hydrocoll. 2017, 63, 391–403. [Google Scholar] [CrossRef]
- Shen, X.; Zhao, C.; Lu, J.; Guo, M. Physicochemical properties of whey-protein-stabilized astaxanthin nanodispersion and its transport via a Caco-2 monolayer. J. Agric. Food. Chem. 2018, 66, 1472–1478. [Google Scholar] [CrossRef] [PubMed]
- Mehrad, B.; Ravanfar, R.; Licker, J.; Regenstein, J.M.; Abbaspourrad, A. Enhancing the physicochemical stability of β-carotene solid lipid nanoparticle (SLNP) using whey protein isolate. Food Res. Int. 2018, 105, 962–969. [Google Scholar] [CrossRef] [PubMed]
- Evoli, S.; Guzzi, R.; Rizzuti, B. Molecular simulations of β-lactoglobulin complexed with fatty acids reveal the structural basis of ligand affinity to internal and possible external binding sites. Proteins Struct. Funct. Bioinform. 2015, 82, 2609–2619. [Google Scholar] [CrossRef] [PubMed]
- Visentini, F.F.; Sponton, O.E.; Perez, A.A.; Santiago, L.G. Formation and colloidal stability of ovalbumin-retinol nanocomplexes. Food Hydrocoll. 2017, 67, 130–138. [Google Scholar] [CrossRef]
- Lange, D.C.; Kothari, R.; Patel, R.C.; Patel, S.C. Retinol and retinoic acid bind to a surface cleft in bovine beta-lactoglobulin: A Method of binding site determination using fluorescence resonance energy transfer. Biophys. Chem. 1998, 74, 45–51. [Google Scholar] [CrossRef]
- Dodin, G.; Andrieux, M.; Kabbani, H.A. Binding of ellipticine to β-lactoglobulin: A physico-chemical study of the specific interaction of an antitumor drug with a transport protein. Eur. J. Biochem. 1990, 193, 697–700. [Google Scholar] [CrossRef] [PubMed]
- Zi, T.; Xu, R.; Qin, W. ChemInform Abstract: Beta-lactoglobulin-Based Encapsulating Systems as Emerging Bioavailability Enhancers for Nutraceuticals: A Review. RSC Adv. 2015, 5, 35138–35154. [Google Scholar]
- Chung, C.; Rojanasasithara, T.; Mutilangi, W.; McClements, D.J. Enhanced stability of anthocyanin-based color in model beverage systems through whey protein isolate complexation. Food Res. Int. 2015, 76, 761–768. [Google Scholar] [CrossRef] [PubMed]
- Hasni, I.; Bourassa, P.; Tajmir-Riahi, H.A. Binding of cationic lipids to milk β-lactoglobulin. J. Phys. Chem. B 2011, 115, 6683–6690. [Google Scholar] [CrossRef] [PubMed]
- André, S.; Arne, S. The menagerie of human lipocAlins: A natural protein scaffold for molecular recognition of physiological compounds. Acc Chem. Res. 2015, 48, 976–985. [Google Scholar]
- Hufnagl, K.; Ghosh, D.; Wagner, S.; Fiocchi, A.; Dahdah, L.; Bianchini, R.; Braun, N.; Steinborn, R.; Hofer, M.; Blaschitz, M. Retinoic acid prevents immunogenicity of milk lipocalin Bos d 5 through binding to its immunodominant T-cell epitope. Sci. Rep. 2018, 8, 1598. [Google Scholar] [CrossRef]
- Chatterton, D.; Smithers, G.; Roupas, P.; Brodkorb, A. Bioactivity of β-lactoglobulin and α-lactalbumin-Technological implications for processing. Int. Dairy J. 2006, 16, 1229–1240. [Google Scholar] [CrossRef]
- Walsh, H. Functional Properties of Whey Protein and Its Application in Nanocomposite Materials and Functional Foods. Ph.D. Thesis, The University of Vermont, Burlington, VT, USA, 2014; pp. 5904–5909. [Google Scholar]
- Ho-Kyung, H.; Wook, K.J.; Mee-Ryung, L.; Woojin, J.; Won-Jae, L. Cellular uptake and cytotoxicity of β-lactoglobulin nanoparticles: The effects of particle size and surface charge. Asian Aust. J. Anim. Sci. 2015, 28, 420–427. [Google Scholar]
- Zhang, Y.; Zhong, Q.X. Binding between bixin and whey protein at pH 7.4 studied by spectroscopy and isothermal titration calorimetry. J. Agr. Food Chem. 2012, 60, 1880–1886. [Google Scholar] [CrossRef]
- Aitken, A.; Learmonth, M. Protein Determination by UV Absorption; Humana Press: Totowa, NJ, USA, 1996; pp. 3–6. [Google Scholar]
- Zhang, Y.; Zhong, Q.X. Probing the binding between norbixin and dairy proteins by spectroscopy methods. Food Chem. 2013, 139, 611–616. [Google Scholar] [CrossRef]
- Ma, S.; Wang, C.; Guo, M. Changes in structure and antioxidant activity of β-lactoglobulin by ultrasound and enzymatic treatment. Ultrason. Sonochem. 2018, 43, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Jian, W.; Wang, L.; Wu, L.; Sun, Y.M. Physicochemical properties of bovine serum albumin-glucose and bovine serum albumin-mannose conjugates prepared by pulsed electric fields treatment. Molecules 2018, 23, 570. [Google Scholar] [CrossRef] [PubMed]
- Kanakis, C.; Hasni, I.; Bourassa, P.; Tarantilis, P.; Polissiou, M.; Tajmir-Riahi, H.-A. Milk β-lactoglobulin complexes with tea polyphenols. Food Chem. 2011, 127, 1046–1055. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.Y.; Xu, S.Q.; Zhu, X.S.; Gong, A.Q. Study on the interaction between methyl violet and bovine serum albumin by spectral analyses. Spectrochim. Acta A 2009, 74, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Albani, J.R.; Vogelaer, J.; Bretesche, L.; Kmiecik, D. Tryptophan 19 residue is the origin of bovine β-lactoglobulin fluorescence. J. Pharm. Biomed. Anal. 2014, 91, 144–150. [Google Scholar] [CrossRef] [PubMed]
- Lucia, D.L.H.; Flaviam, N. Structural modifications of β-lactoglobulin subjected to gamma radiation. Int. Dairy J. 2008, 18, 1126–1132. [Google Scholar]
- Li, W.; Zhao, H.; He, Z.; Zeng, M.; Qin, F.; Chen, J. Modification of soy protein hydrolysates by Maillard reaction: Effects of carbohydrate chain length on structural and interfacial properties. Colloids Surf. B Biointerfaces 2016, 138, 70–77. [Google Scholar] [CrossRef]
- Jia, Z.; Zheng, M.; Tao, F.; Chen, W.; Huang, G.; Jiang, J. Effect of covalent modification by (−)-epigallocatechin-3-gallate on physicochemical and functional properties of whey protein isolate. LWT Food Sci. Technol. 2016, 66, 305–310. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, Z.; Du, J.; Yao, X.; Lei, R.; Zheng, X.; Liu, J.; Hu, H.; Li, H. Interaction of curcumin with intravenous immunoglobulin: A fluorescence quenching and Fourier transformation infrared spectroscopy study. Immunobiology 2008, 213, 651–661. [Google Scholar] [CrossRef]
- Zhang, X.; Xiaomeng, S.; Gao, F.; Wang, J.; Wang, C. Systematical characterization of physiochemical and rheological properties of thermal induced polymerized whey protein: Properties of thermal induced polymerized whey protein. J. Sci. Food Agr. 2018, 99, 923–932. [Google Scholar] [CrossRef]
- Bhattacharyya, J.; Bhattacharyya, M.; Chakrabarty, A.S.; Chaudhuri, U.; Poddar, R.K. Interaction of chlorpromazine with myoglobin and hemoglobin. A comparative study. Biochem. Pharmacol. 1994, 47, 2049–2053. [Google Scholar] [CrossRef]
- Wang, C.N.; Zhang, X.F.; Wang, H.; Wang, J.Q.; Guo, M.R. Effects of amidated low methoxyl pectin on physiochemical and structural properties of polymerized whey proteins. Cyta-J. Food 2018, 16, 923–930. [Google Scholar] [CrossRef]
- University of London. DICHROWEB. Available online: http://dichroweb.cryst.bbk. ac.uk/html/home.shtml (accessed on 8 April 2019).
- Whitmore, L.; Wallace, B.A. Protein secondary structure analyses from circular dichroism spectroscopy: Methods and reference databases. Biopolymers 2008, 89, 392–400. [Google Scholar] [CrossRef] [PubMed]
- U.S. National Institutes of Health (NIH). PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov (accessed on 8 April 2019).
- RCSB Protein Data Bank. Available online: http://www.rcsb.org (accessed on 8 April 2019).
Sample Availability: Samples of the compounds are not available from the authors. |
| PDI | Dh | Zeta Potential | |
|---|---|---|---|
| 50 μM β-LG | 0.27 ± 0.01 a | 4.86 ± 0.26 a | −8.63 ± 0.50 a |
| 50 μM β-LG + 50 μM DIM | 0.32 ± 0.01 b | 5.31 ± 0.56 ab | −17.93 ± 0.70 b |
| 50 μM β-LG + 100 μM DIM | 0.32 ± 0.01 b | 5.82 ± 0.39 b | −17.13 ± 0.87 b |
| 50 μM β-LG + 150 μM DIM | 0.33 ± 0.05 b | 5.81 ± 0.77 b | −18.03 ± 0.85 b |
| 50 μM β-LG + 200 μM DIM | 0.34 ± 0.02 b | 5.72 ± 0.31 b | −17.60 ± 0.56 b |
| PB + 200 μM DIM | - | - | −10.63 ± 0.46 c |
| Helix | Sheet | Turn | Unordered | |
|---|---|---|---|---|
| β-LG | 16.15 | 34.85 | 22.55 | 26.50 |
| β-LG/DIM (10 μM) | 8.75 | 38.3 | 19.35 | 33.40 |
| β-LG/DIM (20 μM) | 5.35 | 47.9 | 16.95 | 29.75 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Zhou, X.; Wang, H.; Sun, X.; Guo, M. Interactions between β-Lactoglobulin and 3,3′-Diindolylmethane in Model System. Molecules 2019, 24, 2151. https://doi.org/10.3390/molecules24112151
Wang C, Zhou X, Wang H, Sun X, Guo M. Interactions between β-Lactoglobulin and 3,3′-Diindolylmethane in Model System. Molecules. 2019; 24(11):2151. https://doi.org/10.3390/molecules24112151
Chicago/Turabian StyleWang, Cuina, Xinhui Zhou, Hao Wang, Xiaomeng Sun, and Mingruo Guo. 2019. "Interactions between β-Lactoglobulin and 3,3′-Diindolylmethane in Model System" Molecules 24, no. 11: 2151. https://doi.org/10.3390/molecules24112151
APA StyleWang, C., Zhou, X., Wang, H., Sun, X., & Guo, M. (2019). Interactions between β-Lactoglobulin and 3,3′-Diindolylmethane in Model System. Molecules, 24(11), 2151. https://doi.org/10.3390/molecules24112151







