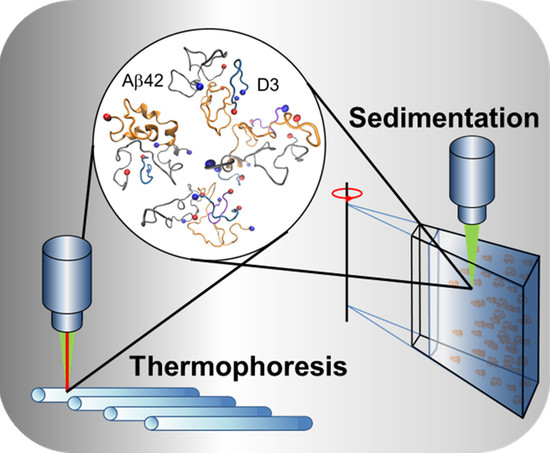
Interference with Amyloid-β Nucleation by Transient Ligand Interaction
Abstract
1. Introduction
2. Results
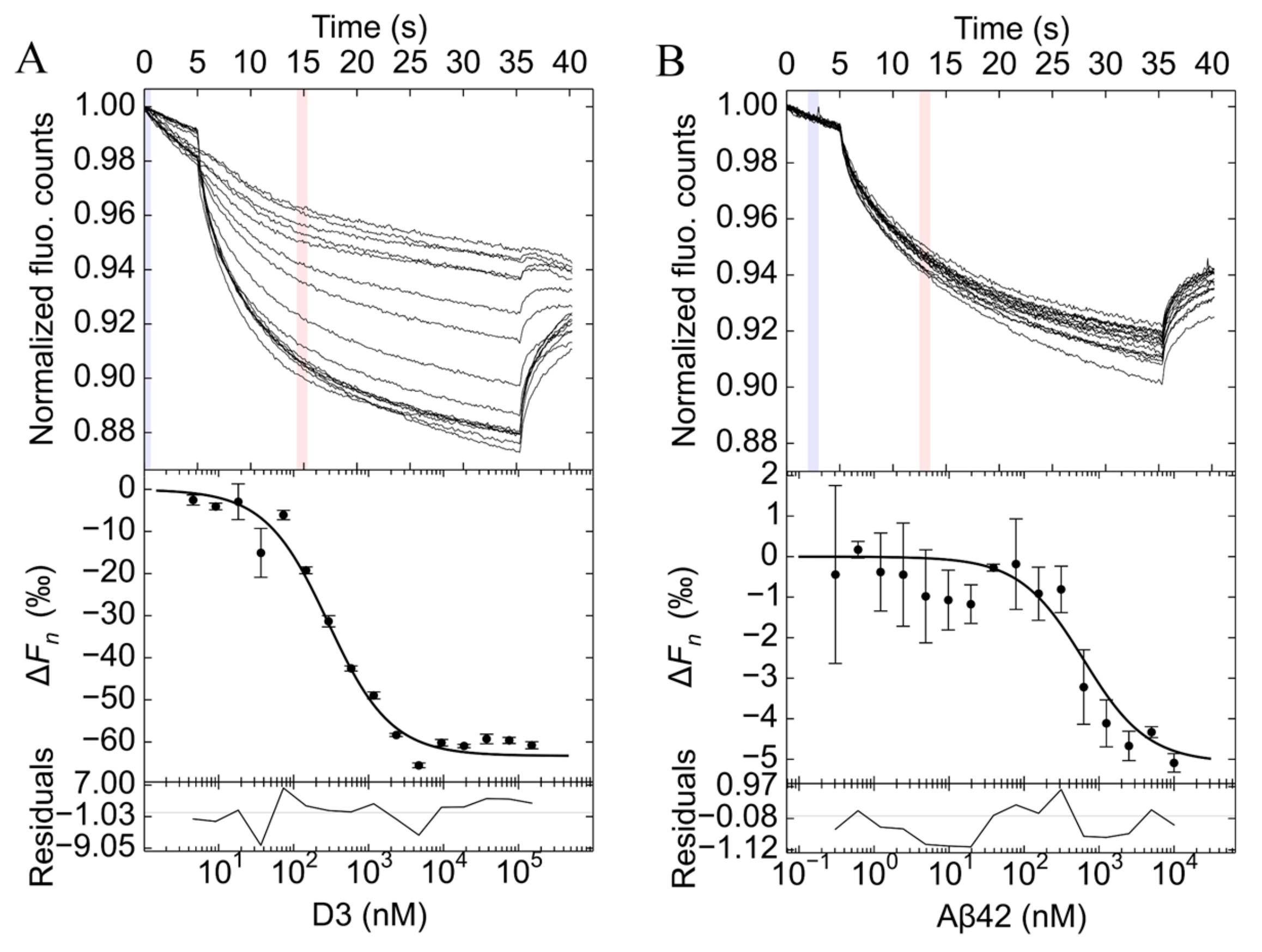
2.1. Characterization of the Dissociation Constant of Aβ42 and D3 Interaction
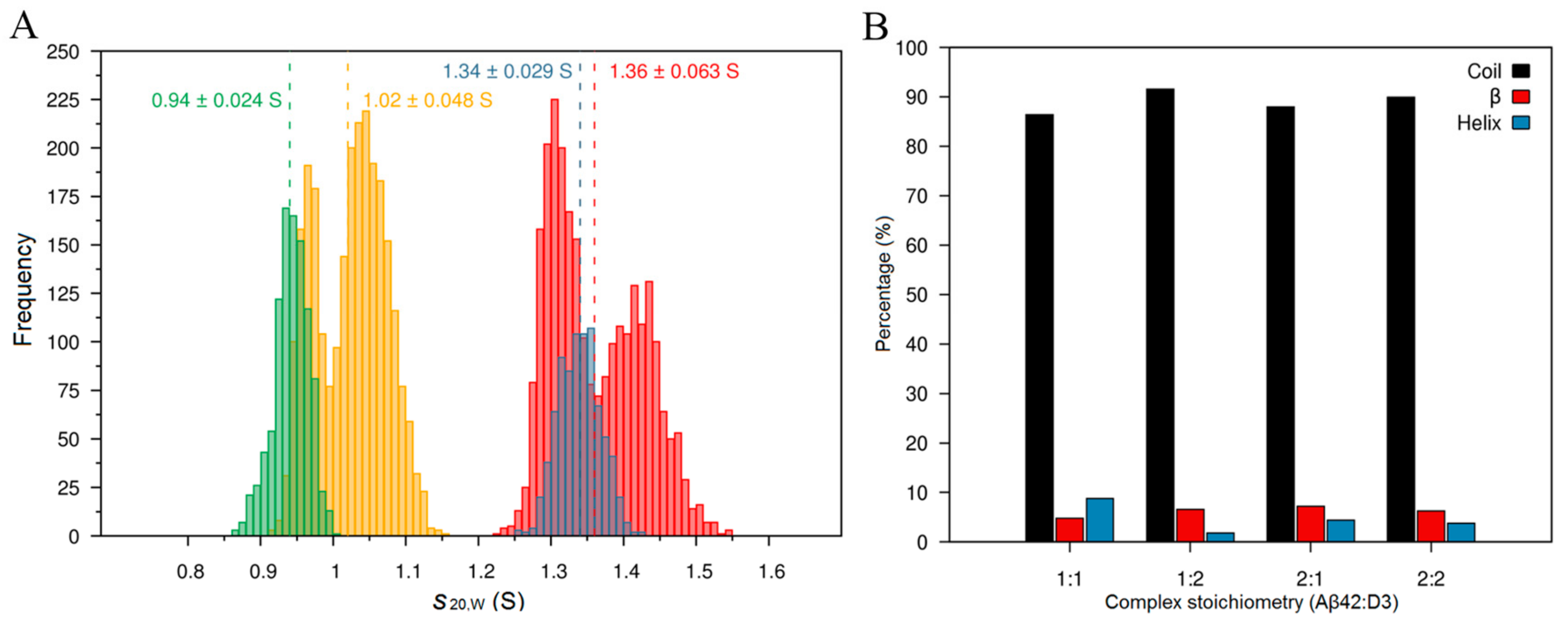
2.2. AUC Analysis of Aβ42 and D3 Mixtures
2.3. Complex Formation Studied by Molecular Dynamics Simulation
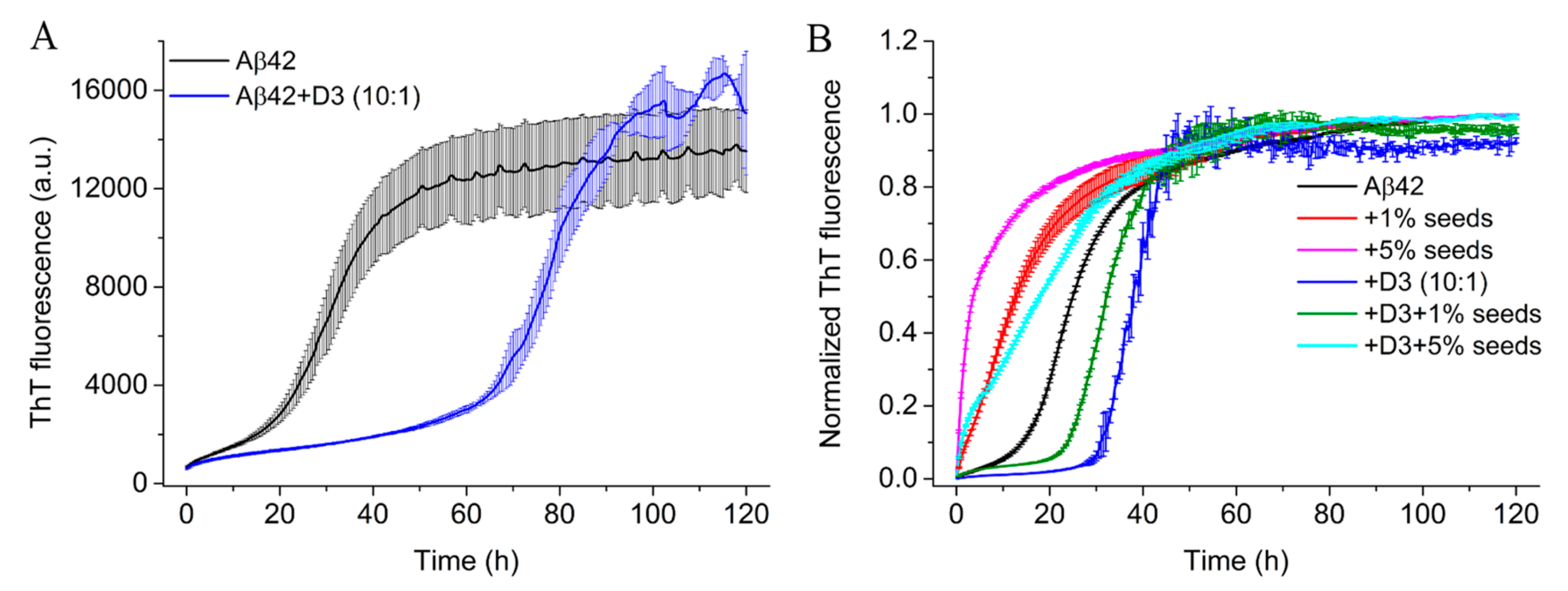
2.4. D3 Retards the Fibrillation of Aβ42 at Substoichiometric Concentrations
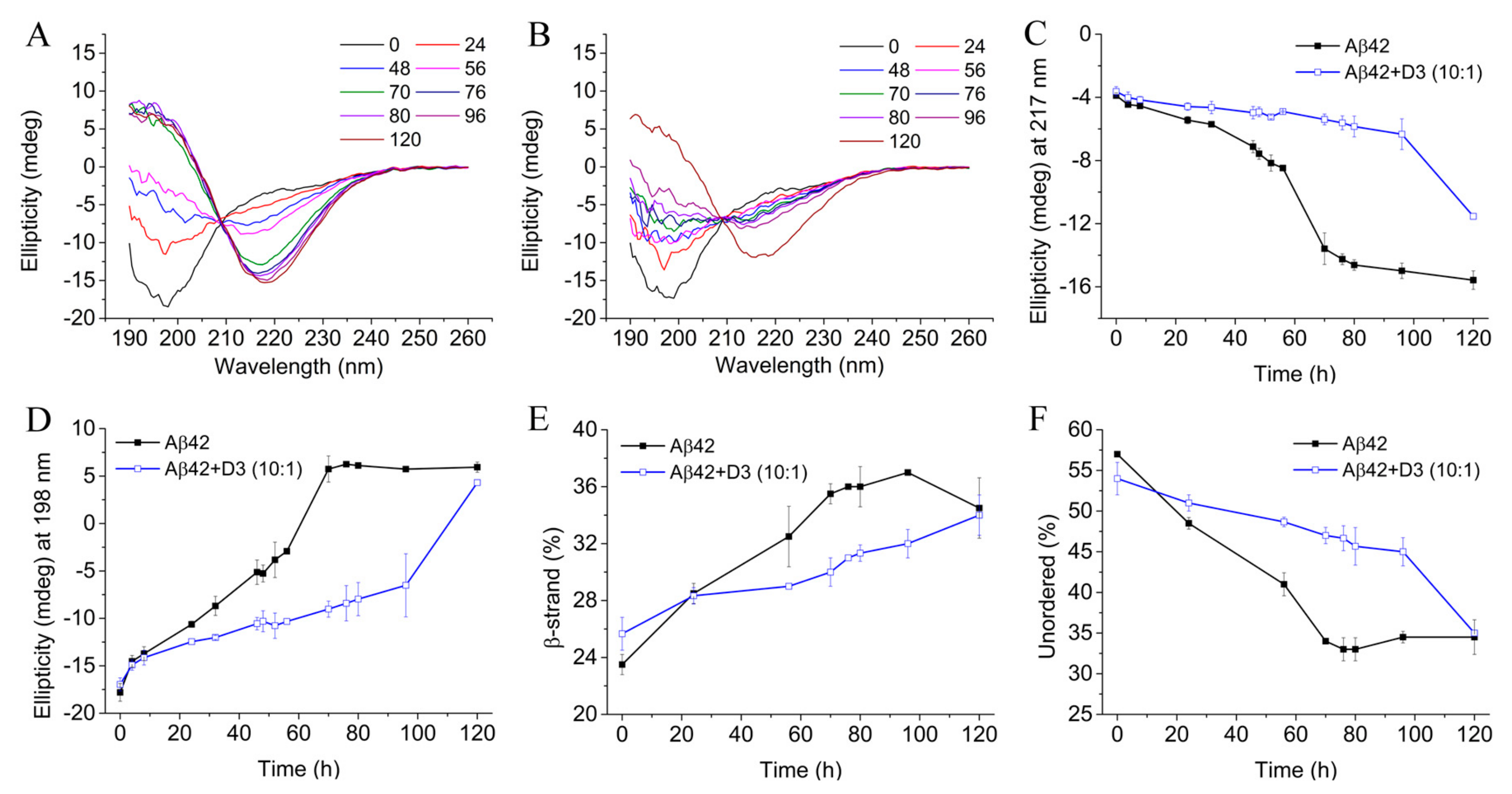
2.5. D3 Slows Down the Secondary Structure Conversion of Aβ42 at Substoichiometric Concentrations
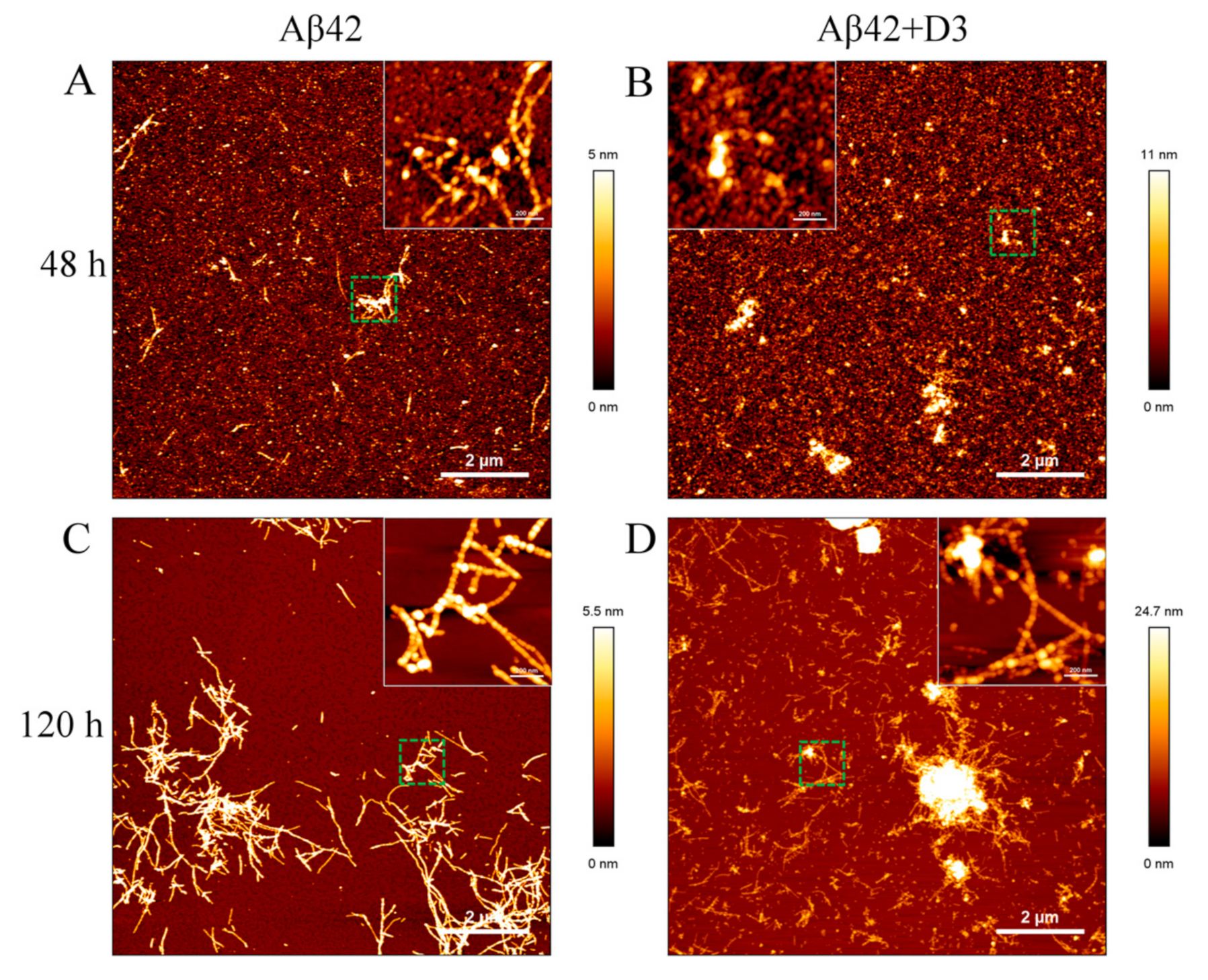
2.6. Morphologies of Aβ42 Samples in the Presence of Substoichiometric D3
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Microscale Thermophoresis
4.3. Analytical Ultracentrifugation
4.4. Molecular Dynamics (MD) Simulation and Data Analysis
4.4.1. Simulation Setup
4.4.2. Prediction of the Sedimentation Coefficient
4.4.3. Secondary Structure
4.5. ThT Assay
4.6. Seeding Experiment
4.7. Circular Dichroism Spectroscopy
4.8. Turbidity Assay
4.9. Atomic Force Microscopy
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Uversky, V.N. Introduction to intrinsically disordered proteins (IDPs). Chem. Rev. 2014, 114, 6557–6560. [Google Scholar] [CrossRef] [PubMed]
- Dyson, H.J.; Wright, P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005, 6, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Oldfield, C.J.; Dunker, A.K. Intrinsically disordered proteins in human diseases: Introducing the D2 concept. Annu. Rev. Biophys. 2008, 37, 215–246. [Google Scholar] [CrossRef] [PubMed]
- Korsak, M.; Kozyreva, T. Beta amyloid hallmarks: From intrinsically disordered proteins to alzheimer’s disease. Adv. Exp. Med. Biol. 2015, 870, 401–421. [Google Scholar] [CrossRef] [PubMed]
- Selkoe, D.J.; Hardy, J. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol. Med. 2016, 8, 595–608. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.F.; Xu, T.H.; Yan, Y.; Zhou, Y.R.; Jiang, Y.; Melcher, K.; Xu, H.E. Amyloid beta: Structure, biology and structure-based therapeutic development. Acta Pharmacol. Sin. 2017, 38, 1205–1235. [Google Scholar] [CrossRef] [PubMed]
- Roche, J.; Shen, Y.; Lee, J.H.; Ying, J.; Bax, A. Monomeric Abeta(1-40) and Abeta(1-42) Peptides in Solution Adopt Very Similar Ramachandran Map Distributions That Closely Resemble Random Coil. Biochemistry 2016, 55, 762–775. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Nam, E.; Lee, H.J.; Savelieff, M.G.; Lim, M.H. Towards an understanding of amyloid-beta oligomers: Characterization, toxicity mechanisms, and inhibitors. Chem. Soc. Rev. 2017, 46, 310–323. [Google Scholar] [CrossRef] [PubMed]
- Gremer, L.; Scholzel, D.; Schenk, C.; Reinartz, E.; Labahn, J.; Ravelli, R.B.G.; Tusche, M.; Lopez-Iglesias, C.; Hoyer, W.; Heise, H.; et al. Fibril structure of amyloid-beta(1-42) by cryo-electron microscopy. Science 2017, 358, 116–119. [Google Scholar] [CrossRef] [PubMed]
- Wiesehan, K.; Buder, K.; Linke, R.P.; Patt, S.; Stoldt, M.; Unger, E.; Schmitt, B.; Bucci, E.; Willbold, D. Selection of D-amino-acid peptides that bind to Alzheimer’s disease amyloid peptide abeta1-42 by mirror image phage display. Chembiochem 2003, 4, 748–753. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, T.N.; Mayr, L.M.; Minor, D.L., Jr.; Milhollen, M.A.; Burgess, M.W.; Kim, P.S. Identification of D-peptide ligands through mirror-image phage display. Science 1996, 271, 1854–1857. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.; Funke, S.A.; Willbold, D. Mirror image phage display—Generating stable therapeutically and diagnostically active peptides with biotechnological means. J. Biotechnol. 2012, 161, 121–125. [Google Scholar] [CrossRef] [PubMed]
- Ziehm, T.; Brener, O.; van Groen, T.; Kadish, I.; Frenzel, D.; Tusche, M.; Kutzsche, J.; Reiss, K.; Gremer, L.; Nagel-Steger, L.; et al. Increase of positive net charge and conformational rigidity enhances the efficacy of d-enantiomeric peptides designed to eliminate cytotoxic abeta species. ACS Chem. Neurosci. 2016, 7, 1088–1096. [Google Scholar] [CrossRef] [PubMed]
- Elfgen, A.; Santiago-Schubel, B.; Gremer, L.; Kutzsche, J.; Willbold, D. Surprisingly high stability of the Abeta oligomer eliminating all-d-enantiomeric peptide D3 in media simulating the route of orally administered drugs. Eur. J. Pharm. Sci. 2017, 107, 203–207. [Google Scholar] [CrossRef]
- Funke, S.A.; Willbold, D. Mirror image phage display—A method to generate D-peptide ligands for use in diagnostic or therapeutical applications. Mol. Biosyst. 2009, 5, 783–786. [Google Scholar] [CrossRef]
- Borgia, A.; Borgia, M.B.; Bugge, K.; Kissling, V.M.; Heidarsson, P.O.; Fernandes, C.B.; Sottini, A.; Soranno, A.; Buholzer, K.J.; Nettels, D.; et al. Extreme disorder in an ultrahigh-affinity protein complex. Nature 2018, 555, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Jerabek-Willemsen, M.; Wienken, C.J.; Braun, D.; Baaske, P.; Duhr, S. Molecular interaction studies using microscale thermophoresis. Assay Drug Dev. Technol. 2011, 9, 342–353. [Google Scholar] [CrossRef] [PubMed]
- Seidel, S.A.; Dijkman, P.M.; Lea, W.A.; van den Bogaart, G.; Jerabek-Willemsen, M.; Lazic, A.; Joseph, J.S.; Srinivasan, P.; Baaske, P.; Simeonov, A.; et al. Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods 2013, 59, 301–315. [Google Scholar] [CrossRef] [PubMed]
- Asmari, M.; Ratih, R.; Alhazmi, H.A.; El Deeb, S. Thermophoresis for characterizing biomolecular interaction. Methods 2018, 145, 107–119. [Google Scholar] [CrossRef]
- Chaturvedi, S.K.; Ma, J.; Zhao, H.; Schuck, P. Use of fluorescence-detected sedimentation velocity to study high-affinity protein interactions. Nat. Protoc. 2017, 12, 1777–1791. [Google Scholar] [CrossRef]
- Zhao, H.; Mayer, M.L.; Schuck, P. Analysis of protein interactions with picomolar binding affinity by fluorescence-detected sedimentation velocity. Anal. Chem. 2014, 86, 3181–3187. [Google Scholar] [CrossRef]
- Kingsbury, J.S.; Laue, T.M. Fluorescence-detected sedimentation in dilute and highly concentrated solutions. Methods Enzymol. 2011, 492, 283–304. [Google Scholar] [CrossRef]
- Schuck, P. Diffusion of the reaction boundary of rapidly interacting macromolecules in sedimentation velocity. Biophys. J. 2010, 98, 2741–2751. [Google Scholar] [CrossRef][Green Version]
- Schuck, P. Sedimentation patterns of rapidly reversible protein interactions. Biophys. J. 2010, 98, 2005–2013. [Google Scholar] [CrossRef]
- Wolff, M.; Zhang-Haagen, B.; Decker, C.; Barz, B.; Schneider, M.; Biehl, R.; Radulescu, A.; Strodel, B.; Willbold, D.; Nagel-Steger, L. Abeta42 pentamers/hexamers are the smallest detectable oligomers in solution. Sci. Rep. 2017, 7, 2493. [Google Scholar] [CrossRef]
- Knowles, T.P.; Vendruscolo, M.; Dobson, C.M. The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 2014, 15, 384–396. [Google Scholar] [CrossRef]
- Gurry, T.; Stultz, C.M. Mechanism of amyloid-beta fibril elongation. Biochemistry 2014, 53, 6981–6991. [Google Scholar] [CrossRef]
- Arosio, P.; Knowles, T.P.; Linse, S. On the lag phase in amyloid fibril formation. Phys. Chem. Chem. Phys. PCCP 2015, 17, 7606–7618. [Google Scholar] [CrossRef]
- Linse, S. Monomer-dependent secondary nucleation in amyloid formation. Biophys. Rev. 2017, 9, 329–338. [Google Scholar] [CrossRef]
- Tornquist, M.; Michaels, T.C.T.; Sanagavarapu, K.; Yang, X.; Meisl, G.; Cohen, S.I.A.; Knowles, T.P.J.; Linse, S. Secondary nucleation in amyloid formation. Chem. Commun. 2018, 54, 8667–8684. [Google Scholar] [CrossRef]
- Habchi, J.; Arosio, P.; Perni, M.; Costa, A.R.; Yagi-Utsumi, M.; Joshi, P.; Chia, S.; Cohen, S.I.; Muller, M.B.; Linse, S.; et al. An anticancer drug suppresses the primary nucleation reaction that initiates the production of the toxic Abeta42 aggregates linked with Alzheimer’s disease. Sci. Adv. 2016, 2, e1501244. [Google Scholar] [CrossRef] [PubMed]
- Arosio, P.; Vendruscolo, M.; Dobson, C.M.; Knowles, T.P. Chemical kinetics for drug discovery to combat protein aggregation diseases. Trends Pharmacol. Sci. 2014, 35, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Zeineddine, R.; Yerbury, J.J. The role of macropinocytosis in the propagation of protein aggregation associated with neurodegenerative diseases. Front. Physiol. 2015, 6, 277. [Google Scholar] [CrossRef] [PubMed]
- Meisl, G.; Kirkegaard, J.B.; Arosio, P.; Michaels, T.C.; Vendruscolo, M.; Dobson, C.M.; Linse, S.; Knowles, T.P. Molecular mechanisms of protein aggregation from global fitting of kinetic models. Nat. Protoc. 2016, 11, 252–272. [Google Scholar] [CrossRef] [PubMed]
- Arimon, M.; Diez-Perez, I.; Kogan, M.J.; Durany, N.; Giralt, E.; Sanz, F.; Fernandez-Busquets, X. Fine structure study of Abeta1-42 fibrillogenesis with atomic force microscopy. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2005, 19, 1344–1346. [Google Scholar] [CrossRef]
- Aileen Funke, S.; van Groen, T.; Kadish, I.; Bartnik, D.; Nagel-Steger, L.; Brener, O.; Sehl, T.; Batra-Safferling, R.; Moriscot, C.; Schoehn, G.; et al. Oral treatment with the d-enantiomeric peptide D3 improves the pathology and behavior of Alzheimer’s Disease transgenic mice. ACS Chem. Neurosci. 2010, 1, 639–648. [Google Scholar] [CrossRef]
- van Groen, T.; Schemmert, S.; Brener, O.; Gremer, L.; Ziehm, T.; Tusche, M.; Nagel-Steger, L.; Kadish, I.; Schartmann, E.; Elfgen, A.; et al. The Abeta oligomer eliminating D-enantiomeric peptide RD2 improves cognition without changing plaque pathology. Sci. Rep. 2017, 7, 16275. [Google Scholar] [CrossRef]
- Zhang, Z.; Witham, S.; Alexov, E. On the role of electrostatics in protein-protein interactions. Phys. Biol. 2011, 8, 035001. [Google Scholar] [CrossRef]
- Liu, J.; Wang, W.; Zhang, Q.; Zhang, S.; Yuan, Z. Study on the efficiency and interaction mechanism of a decapeptide inhibitor of beta-amyloid aggregation. Biomacromolecules 2014, 15, 931–939. [Google Scholar] [CrossRef]
- Dogan, J.; Gianni, S.; Jemth, P. The binding mechanisms of intrinsically disordered proteins. Phys. Chem. Chem. Phys. PCCP 2014, 16, 6323–6331. [Google Scholar] [CrossRef]
- Ganguly, D.; Otieno, S.; Waddell, B.; Iconaru, L.; Kriwacki, R.W.; Chen, J. Electrostatically accelerated coupled binding and folding of intrinsically disordered proteins. J. Mol. Biol. 2012, 422, 674–684. [Google Scholar] [CrossRef] [PubMed]
- Cox, D.; Selig, E.; Griffin, M.D.; Carver, J.A.; Ecroyd, H. Small heat-shock proteins prevent alpha-synuclein aggregation via transient interactions and their efficacy is affected by the rate of aggregation. J. Biol. Chem. 2016, 291, 22618–22629. [Google Scholar] [CrossRef] [PubMed]
- Assarsson, A.; Hellstrand, E.; Cabaleiro-Lago, C.; Linse, S. Charge dependent retardation of amyloid beta aggregation by hydrophilic proteins. ACS Chem. Neurosci. 2014, 5, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Walti, M.A.; Steiner, J.; Meng, F.; Chung, H.S.; Louis, J.M.; Ghirlando, R.; Tugarinov, V.; Nath, A.; Clore, G.M. Probing the mechanism of inhibition of amyloid-beta(1-42)-induced neurotoxicity by the chaperonin GroEL. Proc. Natl. Acad. Sci. USA 2018, 115, E11924–E11932. [Google Scholar] [CrossRef] [PubMed]
- Nick, M.; Wu, Y.; Schmidt, N.W.; Prusiner, S.B.; Stohr, J.; DeGrado, W.F. A long-lived Abeta oligomer resistant to fibrillization. Biopolymers 2018, 109, e23096. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Aucoin, D.; Davis, J.; Van Nostrand, W.E.; Smith, S.O. Mechanism of nucleated conformational conversion of abeta42. Biochemistry 2015, 54, 4197–4207. [Google Scholar] [CrossRef] [PubMed]
- Hasecke, F.; Miti, T.; Perez, C.; Barton, J.; Scholzel, D.; Gremer, L.; Gruning, C.S.R.; Matthews, G.; Meisl, G.; Knowles, T.P.J.; et al. Origin of metastable oligomers and their effects on amyloid fibril self-assembly. Chem. Sci. 2018, 9, 5937–5948. [Google Scholar] [CrossRef] [PubMed]
- Paravastu, A.K.; Leapman, R.D.; Yau, W.M.; Tycko, R. Molecular structural basis for polymorphism in Alzheimer’s beta-amyloid fibrils. Proc. Natl. Acad. Sci. USA 2008, 105, 18349–18354. [Google Scholar] [CrossRef]
- Olubiyi, O.O.; Strodel, B. Structures of the amyloid beta-peptides Abeta1-40 and Abeta1-42 as influenced by pH and a D-peptide. J. Phys. Chem. B 2012, 116, 3280–3291. [Google Scholar] [CrossRef]
- Liao, Q.; Owen, M.C.; Bali, S.; Barz, B.; Strodel, B. Abeta under stress: The effects of acidosis, Cu(2+)-binding, and oxidation on amyloid beta-peptide dimers. Chem. Commun. 2018, 54, 7766–7769. [Google Scholar] [CrossRef]
- Liu, H.; Morris, C.; Lantz, R.; Kent, T.W.; Elbassal, E.A.; Wojcikiewicz, E.P.; Du, D. Residue-specific dynamics and local environmental changes in abeta40 oligomer and fibril formation. Angew. Chem. Int. Ed. Engl. 2018, 57, 8017–8021. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.; Davis, J.; Aucoin, D.; Sato, T.; Ahuja, S.; Aimoto, S.; Elliott, J.I.; Van Nostrand, W.E.; Smith, S.O. Structural conversion of neurotoxic amyloid-beta(1-42) oligomers to fibrils. Nat. Struct. Mol. Biol. 2010, 17, 561–567. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Liu, C.; Guo, Z. Structural insights into Abeta42 oligomers using site-directed spin labeling. J. Biol. Chem. 2013, 288, 18673–18683. [Google Scholar] [CrossRef] [PubMed]
- Scheuermann, T.H.; Padrick, S.B.; Gardner, K.H.; Brautigam, C.A. On the acquisition and analysis of microscale thermophoresis data. Anal. Biochem. 2016, 496, 79–93. [Google Scholar] [CrossRef] [PubMed]
- Brautigam, C.A. Calculations and publication-quality illustrations for analytical ultracentrifugation data. Methods Enzymol. 2015, 562, 109–133. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.L.; Lary, J.W.; Moody, T.P.; Laue, T.M. Analytical ultracentrifugation: Sedimentation velocity and sedimentation equilibrium. Methods Cell Biol. 2008, 84, 143–179. [Google Scholar] [CrossRef]
- Choi, T.S.; Lee, H.J.; Han, J.Y.; Lim, M.H.; Kim, H.I. Molecular Insights into Human Serum Albumin as a receptor of amyloid-beta in the extracellular region. J. Am. Chem. Soc. 2017, 139, 15437–15445. [Google Scholar] [CrossRef]
- Carrotta, R.; Canale, C.; Diaspro, A.; Trapani, A.; Biagio, P.L.; Bulone, D. Inhibiting effect of alpha(s1)-casein on Abeta(1-40) fibrillogenesis. Biochim. Biophys. Acta 2012, 1820, 124–132. [Google Scholar] [CrossRef]
- Algamal, M.; Ahmed, R.; Jafari, N.; Ahsan, B.; Ortega, J.; Melacini, G. Atomic-resolution map of the interactions between an amyloid inhibitor protein and amyloid beta (Abeta) peptides in the monomer and protofibril states. J. Biol. Chem. 2017, 292, 17158–17168. [Google Scholar] [CrossRef]
- Schuck, P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 2000, 78, 1606–1619. [Google Scholar] [CrossRef]
- Durchschlag, H.; Zipper, P. Calculation of the partial volume of organic compounds and polymers. In Progress in Colloid & Polymer Science; Springer: Berlin/Heidelberg, Germany, 1994; Volume 94, pp. 20–39. [Google Scholar]
- Durchschlag, H. Determination of the partial specific volume of conjugated proteins. Colloid Polym. Sci. 1989, 267, 1139–1150. [Google Scholar] [CrossRef]
- Hess, B.; Kutzner, C.; van der Spoel, D.; Lindahl, E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008, 4, 435–447. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Maxwell, D.S.; Tirado-Rives, J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 1996, 118, 11225–11236. [Google Scholar] [CrossRef]
- Kaminski, G.A.; Friesner, R.A.; Tirado-Rives, J.; Jorgensen, W.L. Evaluation and reparametrization of the opls-aa force field for proteins via comparison with accurate quantum chemical calculations on peptides. J. Phys. Chem. B 2001, 105, 6474–6487. [Google Scholar] [CrossRef]
- Qiu, D.; Shenkin, P.S.; Hollinger, F.P.; Still, W.C. The GB/SA continuum model for solvation. a fast analytical method for the calculation of approximate born radii. J. Phys. Chem. A 1997, 101, 3005–3014. [Google Scholar] [CrossRef]
- Anton, F.K.; Berk, H.; Berendsen, H.J. Improving efficiency of large time-scale molecular dynamics simulations of hydrogen-rich systems. J. Comput. Chem. 1999, 20, 786–798. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef]
- Xavier, D.; Karl, G.; Bernhard, J.; Dieter, S.; Van Gunsteren, W.F.; Mark, A.E. Peptide folding: When simulation meets experiment. Angew. Chem. Int. Ed. 1999, 38, 236–240. [Google Scholar] [CrossRef]
- Abraham, M.J.; Spoel, D.V.D.; Lindahl, E.; Hess, B.; Team, T.G.D. GROMACS User Manual version 2018. Available online: www.gromacs.org (accessed on 12 January 2019).
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Allen, M.P.; Tildesley, D.J. Computer Simulation of Liquids; Clarendon Press: Oxford, UK, 1987. [Google Scholar]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L. A smooth particle mesh ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh ewald: An N⋅log(N) method for ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Ortega, A.; Amoros, D.; De La Torre, J.G. Prediction of hydrodynamic and other solution properties of rigid proteins from atomic- and residue-level models. Biophys. J. 2011, 101, 892–898. [Google Scholar] [CrossRef] [PubMed]
- Voss, N.R.; Gerstein, M.; Steitz, T.A.; Moore, P.B. The geometry of the ribosomal polypeptide exit tunnel. J. Mol. Biol. 2006, 360, 893–906. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Gr. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Kabsch, W.; Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef]
- Luo, J.; Yu, C.H.; Yu, H.; Borstnar, R.; Kamerlin, S.C.; Graslund, A.; Abrahams, J.P.; Warmlander, S.K. Cellular polyamines promote amyloid-beta (Abeta) peptide fibrillation and modulate the aggregation pathways. ACS Chem. Neurosci. 2013, 4, 454–462. [Google Scholar] [CrossRef]
- Hellstrand, E.; Boland, B.; Walsh, D.M.; Linse, S. Amyloid beta-protein aggregation produces highly reproducible kinetic data and occurs by a two-phase process. ACS Chem. Neurosci. 2010, 1, 13–18. [Google Scholar] [CrossRef]
- Ehrnhoefer, D.E.; Bieschke, J.; Boeddrich, A.; Herbst, M.; Masino, L.; Lurz, R.; Engemann, S.; Pastore, A.; Wanker, E.E. EGCG redirects amyloidogenic polypeptides into unstructured, off-pathway oligomers. Nat. Struct. Mol. Biol. 2008, 15, 558–566. [Google Scholar] [CrossRef]
- Whitmore, L.; Wallace, B.A. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004, 32, W668–W673. [Google Scholar] [CrossRef] [PubMed]
- Lobley, A.; Whitmore, L.; Wallace, B.A. DICHROWEB: An interactive website for the analysis of protein secondary structure from circular dichroism spectra. Bioinformatics 2002, 18, 211–212. [Google Scholar] [CrossRef] [PubMed]
- Compton, L.A.; Johnson, W.C., Jr. Analysis of protein circular dichroism spectra for secondary structure using a simple matrix multiplication. Anal. Biochem. 1986, 155, 155–167. [Google Scholar] [CrossRef]
- Sreerama, N.; Woody, R.W. Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000, 287, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Wood, S.J.; Maleeff, B.; Hart, T.; Wetzel, R. Physical, morphological and functional differences between ph 5.8 and 7.4 aggregates of the Alzheimer’s amyloid peptide Abeta. J. Mol. Biol. 1996, 256, 870–877. [Google Scholar] [CrossRef]
- Novo, M.; Freire, S.; Al-Soufi, W. Critical aggregation concentration for the formation of early Amyloid-beta (1-42) oligomers. Sci. Rep. 2018, 8, 1783. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are available from the authors. |

| Sample | k | ||
|---|---|---|---|
| Aβ42 | 30.0 ± 0.7 | 0.15 ± 0.01 | 17.3 ± 1.6 |
| Aβ42 + D3 | 79.3 ± 2.3 | 0.18 ± 0.07 | 67.6 ± 3.8 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, T.; Loschwitz, J.; Strodel, B.; Nagel-Steger, L.; Willbold, D. Interference with Amyloid-β Nucleation by Transient Ligand Interaction. Molecules 2019, 24, 2129. https://doi.org/10.3390/molecules24112129
Zhang T, Loschwitz J, Strodel B, Nagel-Steger L, Willbold D. Interference with Amyloid-β Nucleation by Transient Ligand Interaction. Molecules. 2019; 24(11):2129. https://doi.org/10.3390/molecules24112129
Chicago/Turabian StyleZhang, Tao, Jennifer Loschwitz, Birgit Strodel, Luitgard Nagel-Steger, and Dieter Willbold. 2019. "Interference with Amyloid-β Nucleation by Transient Ligand Interaction" Molecules 24, no. 11: 2129. https://doi.org/10.3390/molecules24112129
APA StyleZhang, T., Loschwitz, J., Strodel, B., Nagel-Steger, L., & Willbold, D. (2019). Interference with Amyloid-β Nucleation by Transient Ligand Interaction. Molecules, 24(11), 2129. https://doi.org/10.3390/molecules24112129