Expression of Bioactive Lunasin Peptide in Transgenic Rice Grains for the Application in Functional Food
Abstract
:1. Practical Application
2. Introduction
3. Materials and Methods
3.1. Reagent
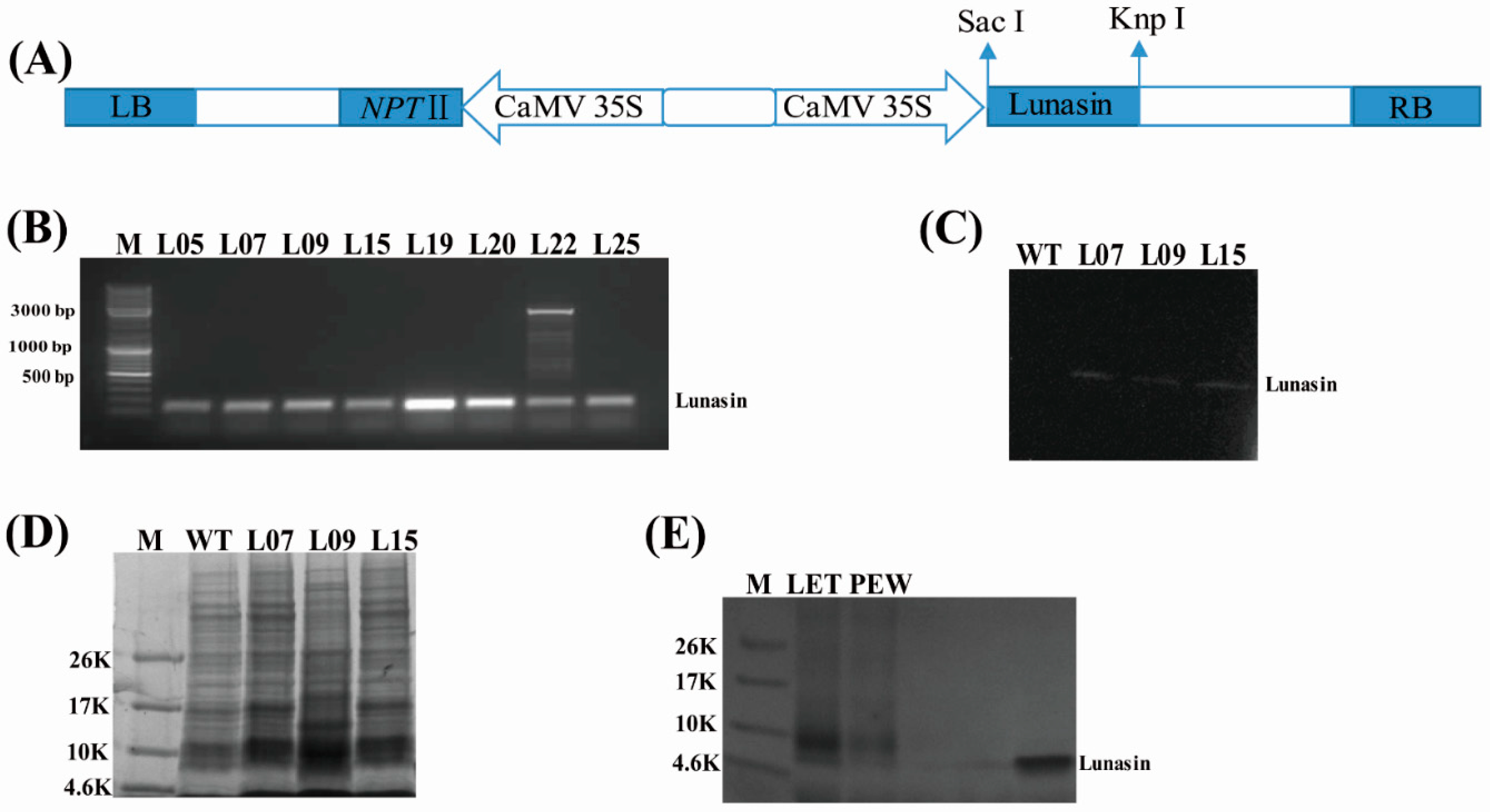
3.2. Construction of Plasmid
3.3. Generation and Selection of Transgenic Lines Expressing Lunasin
3.4. Protein Isolation and Western Blot
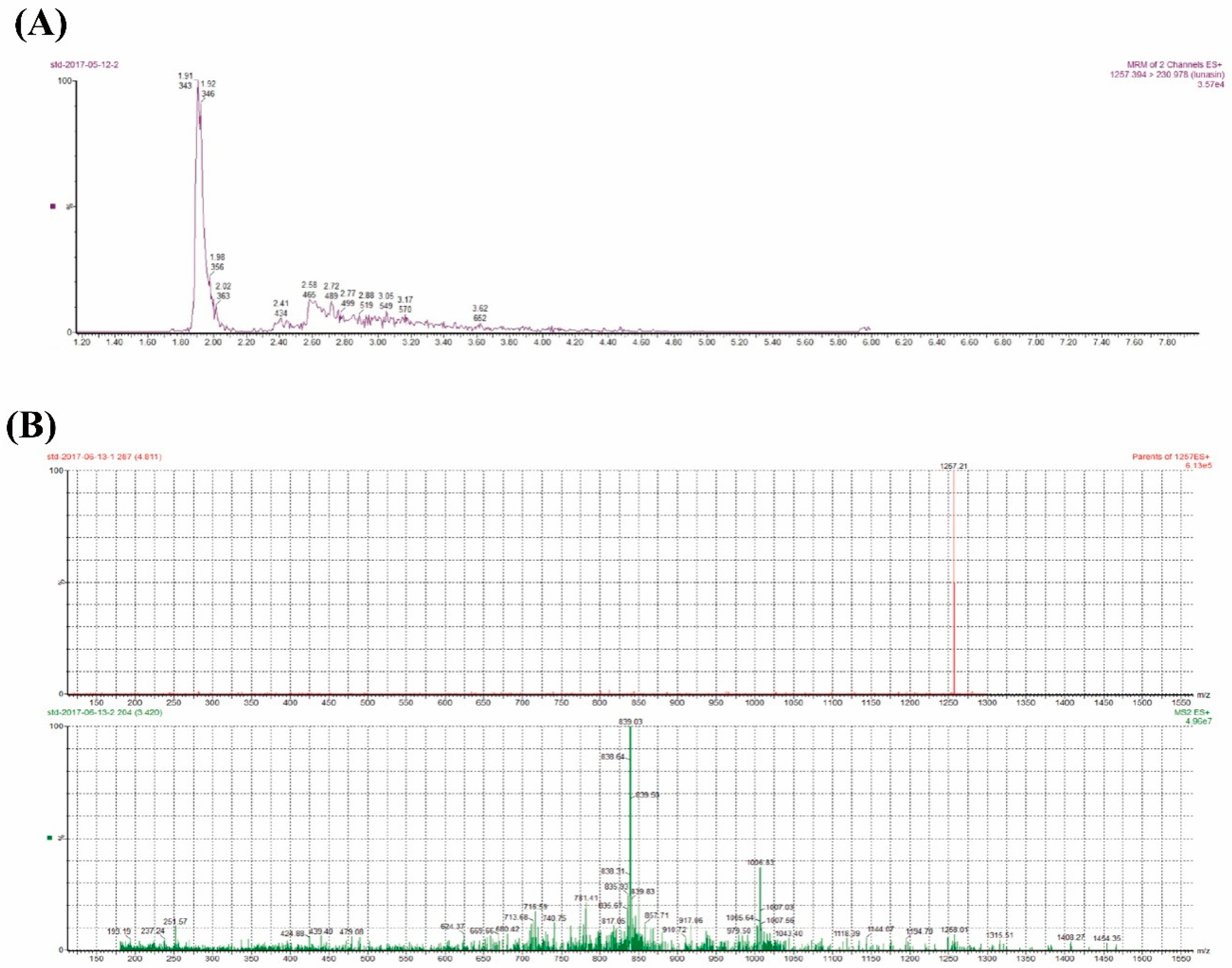
3.5. UPLC-MS/MS Analysis
3.6. Protein Purification
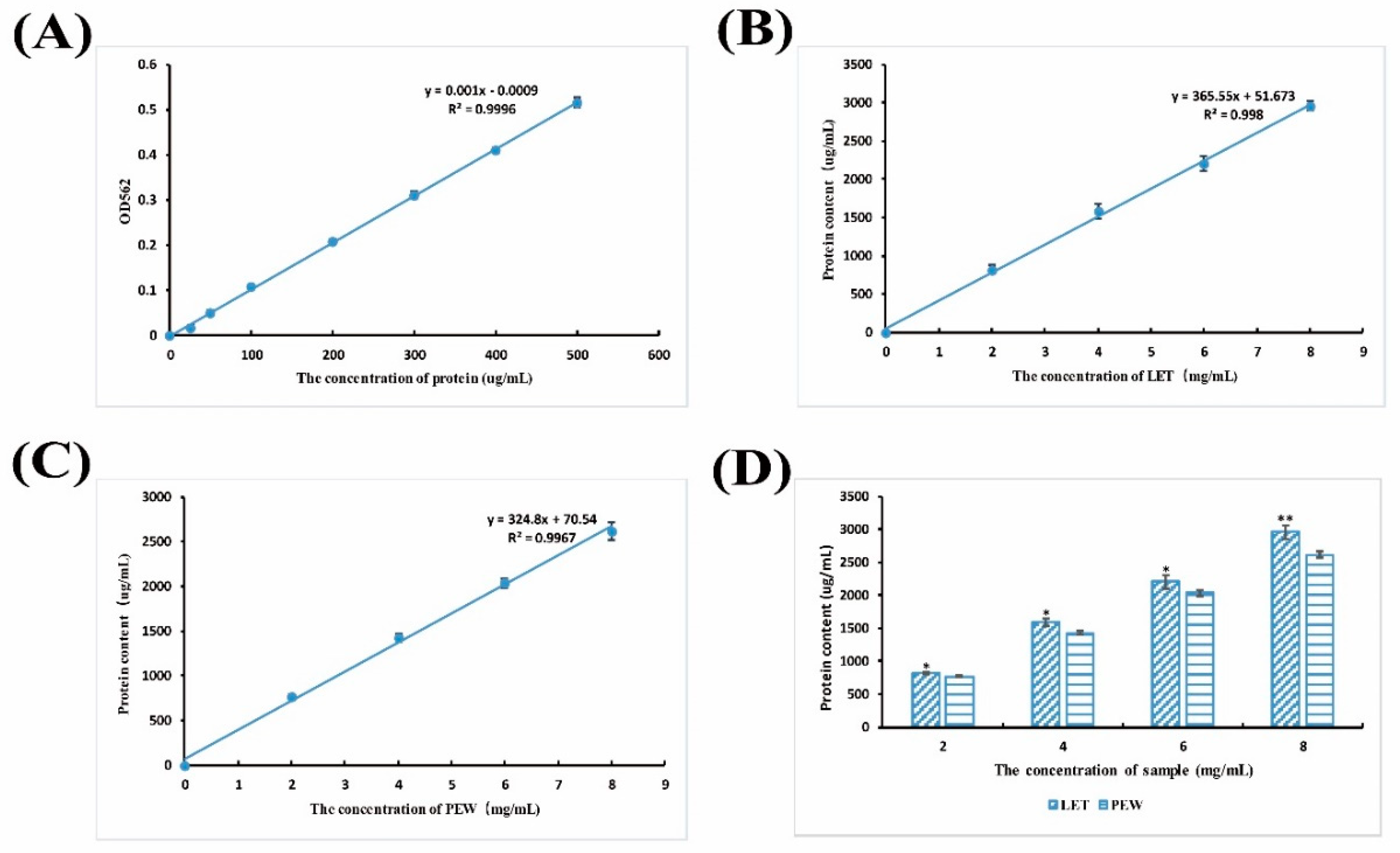
3.7. Protein Content Detection
3.8. Flavone, Total Polyphenol, and Phenolic Acid Analysis
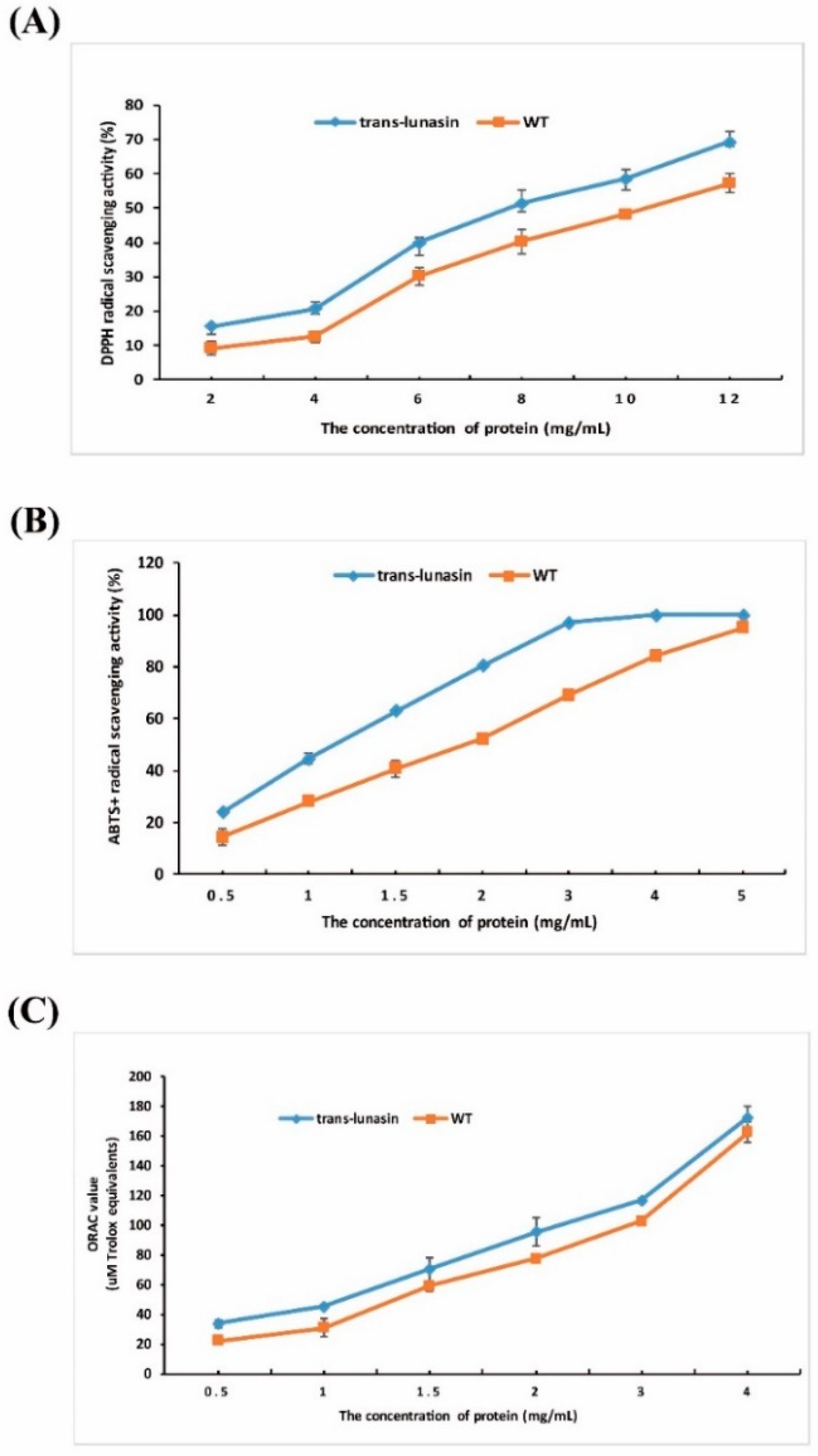
3.9. Antioxidant Activity Assay
3.10. Anti-Inflammatory Activities Assay
3.11. Statistical Analysis
4. Results and Discussion
4.1. Presence of Lunasin in Transgenic Rice
4.2. Lunasin Content Detection in Transgenic Rice Extract
4.3. Flavone, Total Polyphenol, and Phenolic Acid Analysis
4.4. Antioxidant Activity Assay
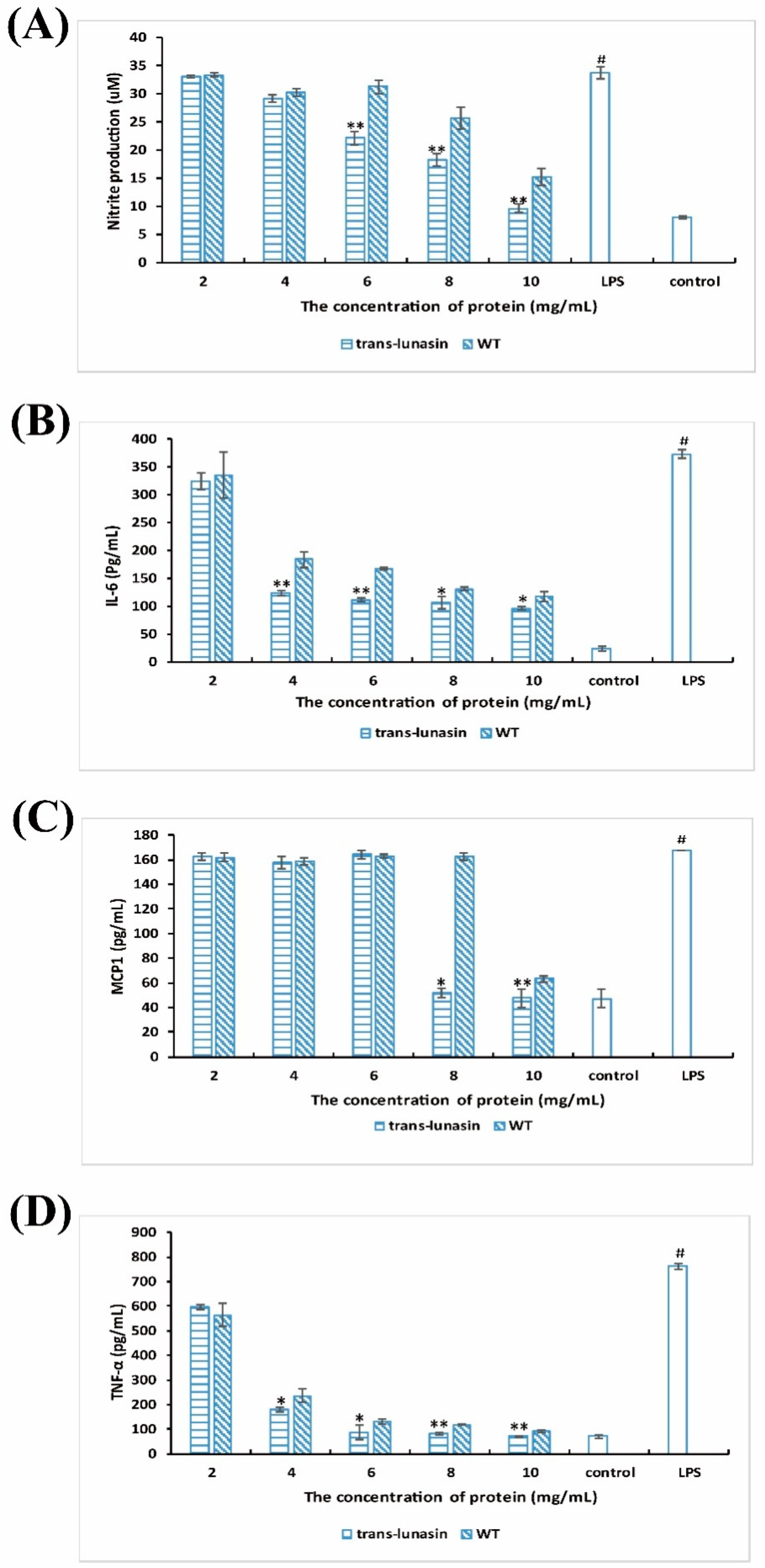
4.5. Anti-Inflammatory Activity Assay
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| LET | Lunasin-enriched fraction from trans-lunasin rice |
| PEW | Peptide-enriched fraction from wild type rice |
| PCR | Polymerase chain reaction |
| MCP1/CCL2 | Monocyte chemoattractant protein chemokine (C-C motif) ligand 2 |
| TNF-α | Tumor cell necrosis factor-α |
| IL-6 | Interleukin-6 |
| UPLC-MS/MS | Ultra Performance Liquid Chromatography with Tandem Mass Spectrometry |
| XEVO TQ-S | Triple Quadrupole |
| AAPH | 2,2′-Azobis(2-methylpropionamidine)dihydrochloride |
| RPMI-1640 | Roswell Park Memorial Institute 1640 |
| FBS | Fetal Bovine Serum |
| UPLC | Ultra Performance Liquid Chromatography |
References
- Lule, V.K.; Garg, S.; Pophaly, S.D.; Hitesh; Tomar, S.K. Potential health benefits of lunasin: A multifaceted soy-derived bioactive peptide. J. Food Sci. 2015, 80, R485–R494. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Bisetty, K. A molecular dynamics study of lunasin. S. Afr. J. Chem. 2012, 65, 115–124. [Google Scholar]
- Galvez, A.F.; Chen, N.; Macasieb, J.; de Lumen, B. Chemopreventive property of a soybean peptide (lunasin) that binds to deacetylated histones and inhibits acetylation. Cancer Res. 2001, 61, 7473–7478. [Google Scholar] [PubMed]
- Hernández-Ledesma, B.; Hsieh, C.C.; De Lumen, B. Relationship between lunasin’s sequence and its inhibitory activity of histones H3 and H4 acetylation. Mol. Nutr. Food Res. 2011, 55, 989–998. [Google Scholar] [CrossRef] [PubMed]
- De Lumen, B. Lunasin: A cancer-preventive soy peptide. Nutr. Rev. 2005, 63, 16–21. [Google Scholar] [CrossRef] [PubMed]
- De Mejia, E.G.; Dia, V.P. Lunasin and lunasin-like peptides inhibit inflammation through suppression of NF-κB pathway in the macrophage. Peptides 2009, 30, 2388–2398. [Google Scholar] [CrossRef] [PubMed]
- Hernándezledesma, B.; Hsieh, C.C.; De Lumen, B. Antioxidant and anti-inflammatory properties of cancer preventive peptide lunasin in RAW 264.7 macrophages. Biochem. Biophys. Res. Commun. 2009, 390, 803–808. [Google Scholar] [CrossRef] [PubMed]
- Galvez, A.F. Methods for Using Soy Peptides to Inhibit H3 Acetylation, Reduce Expression of HMG CoA Reductase, and Increase LDL Receptor and Sp1 Expression in a Mammal. U.S. Patent 7731995, 8 June 2010. [Google Scholar]
- Galvez, A.F. Identification of lunasin as the active component in soy protein responsible for reducing LDL cholesterol and risk of cardiovascular disease. Circulation 2012, 126, A10693. [Google Scholar]
- Ren, G.X.; Zhu, Y.Y.; Shi, Z.X. Detection of lunasin in quinoa (Chenopodium quinoa, Willd) and the in vitro evaluation of its antioxidant and anti-inflammatory activities. J. Sci. Food Agric. 2017, 97, 4110–4116. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.J.; Jeong, J.B.; Kim, D.S.; Park, J.H.; Lee, J.B.; Kweon, D.H.; Chung, G.; Seo, E.W.; De Lumen, B. The cancer preventive peptide lunasin from wheat inhibits core histone acetylation. Cancer Lett. 2007, 255, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.J.; Lam, Y.; de Lumen, B. Barley lunasin suppresses ras-induced colony formation and inhibits core histone acetylation in mammalian cells. J. Agric. Food Chem. 2002, 50, 5903–5908. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.J.; Lee, J.R.; Jeong, J.B.; Park, J.H.; Cheong, Y.K.; de Lumen, B. The cancer preventive seed peptide lunasin from rye is bioavailable and bioactive. Nutr. Cancer 2009, 61, 680–686. [Google Scholar] [CrossRef] [PubMed]
- Nakurte, I.; Kirhnere, I.; Namniece, J.; Saleniece, K.; Krigere, L.; Mekss, P.; Vicupee, Z.; Bleideree, M.; Legzdinad, L.; Muceniecec, R. Detection of the lunasin peptide in oats (Avena sativa L). J. Cereal Sci. 2013, 57, 319–324. [Google Scholar] [CrossRef]
- De la Rosa, A.P.B.; Fomsgaard, I.S.; Laursen, B.; Mortensen, A.G.; Olvera-Martínez, L.; Silva-Sánchez, C.; Mendoza-Herrera, A.; González-Castañeda, J.; De León-Rodríguez, A. Amaranth (Amaranthus hypochondriacus) as an alternative crop for sustainable food production: Phenolic acids and flavonoids with potential impact on its nutraceutical quality. J. Cereal Sci. 2009, 49, 117–121. [Google Scholar] [CrossRef]
- Panda, P.K.; Mukhopadhyay, S.; Behera, B.; Bhol, C.S.; Dey, S.; Das, D.N.; Sinha, N.; Bissoyi, A.; Pramanik, K.; Maiti, T.K.; et al. Antitumor effect of soybean lectin mediated through reactive oxygen species-dependent pathway. Life Sci. 2014, 111, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Zaizen, Y.; Higuchi, Y.; Matsuo, N.; Shirabe, K.; Tokuda, H.; Takeshita, M. Antitumor effects of soybean hypocotyls and soybeans on the mammary tumor induction by N-methyl-N-nitrosourea in F344 rats. Anticancer Res. 2000, 20, 1439–1444. [Google Scholar] [PubMed]
- Dia, V.P.; Wang, W.; Oh, V.L.; Lumen, B.; Mejia, E. Isolation, purification and characterisation of lunasin from defatted soybean flour and in vitro evaluation of its anti-inflammatory activity. Food Chem. 2009, 114, 108–115. [Google Scholar] [CrossRef]
- Schmidt, F.R. Recombinant expression systems in the pharmaceutical industry. Appl. Microbiol. Biotechnol. 2004, 65, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.F.; Pan, T.M. Recombinant expression of bioactive peptide lunasin in Escherichia coli. Appl. Microbiol. Biotechnol. 2010, 88, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Davis, K.R.; Seber, L.E.; Mcconnell, E.J.; Boles, K. Production of recombinant lunasin peptides with enhanced anticancer activity using transient expression in tobacco. Cancer Res. 2012, 72, 3850. [Google Scholar] [CrossRef]
- Ren, G.X.; Zhu, Y.Y.; Dun, B.Q. The Expression, Purification, and Activity Identification of Recombinant Lunasin in Pichia. CN Patent 105647960A, 8 June 2016. [Google Scholar]
- Huang, Y.Y.; Rong, G.Z.; Han, N.; Sun, J.Y.; Weng, X.Y. Expression of exogenous xylanase gene (ATX) in transgenic rice. Chin. J. Rice Sci. 2009, 23, 604–610. [Google Scholar]
- Shu, Q.Y.; Ye, G.Y.; Cui, H.; Cheng, X.Y.; Xiang, Y.B.; Wu, D.X. Transgenic rice plants with a synthetic cry1Ab gene from Bacillus thuringiensis were highly resistant to eight lepidopteran rice pest species. Mol. Breed. 2000, 6, 433–439. [Google Scholar] [CrossRef]
- Weng, X.Y.; Huang, Y.Y.; Hou, C.X.; Jiang, D.A. Effects of an exogenous xylanase gene expression on the growth of transgenic rice and the expression level of endogenous xylanase inhibitor gene RIXI. J. Sci. Food Agric. 2012, 93, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Nandia, S.; Suzukib, Y.A.; Huanga, J.; Yaldaa, D.; Phama, P.; Wua, L.; Bartleya, G.; Huanga, N.; Lönnerdalb, B. Expression of human lactoferrin in transgenic rice grains for the application in infant formula. Plant Sci. 2002, 163, 713–722. [Google Scholar] [CrossRef]
- Supartana, P.; Shimizu, T.; Shioiri, H.; Nogawa, M.; Nozue, M.; Kojima, M. Development of simple and efficient in planta transformation method for rice (Oryza sativa L.) using Agrobacterium tumefaciens. J. Biosci. Bioeng. 2005, 100, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J.D. Cetyltrimethyl ammonium bromide (CTAB) DNA miniprep for plant DNA isolation. Cold Spring Harb. Protoc. 2009. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.Q.; Mahmood, T.; Yang, P.C. Western Blot: Technique, Theory and Trouble Shooting. N. Am. J. Med. Sci. 2012, 4, 429–434. [Google Scholar] [CrossRef]
- Wu, J.-L.; Ren, B.-R.; Li, W.-L.; Zhang, H.-Q. Comparison analysis on content of total flavonoids from above-ground part of Stenoloma chusanum. J. Plant Resour. Environ. 2007, 16, 77–78. [Google Scholar]
- Cai, W.G.; Wu, W.; Shao, J.F.; Chen, Q.; Wang, Y.B.; Liu, Z.Q. Determination of polyphenol content in houttuynia cordata thunb by folin-ciocalteu colorimetric method. Food Sci. 2010, 31, 201–204. [Google Scholar]
- Kumar, G.S.; Nayaka, H.; Dharmesh, S.M.; Salimatha, P.V. Free and bound phenolic antioxidants in amla (Emblica officinalis) and turmeric (Curcuma longa). J. Food Compost. Anal. 2006, 19, 446–452. [Google Scholar] [CrossRef]
- Menghini, L.; Leporini, L.; Vecchiotti, G.; Locatelli, M.; Carradori, S.; Ferrante, C.; Zengin, G.; Recinella, L.; Chiavarolia, A.; Leone, S.; et al. Crocus sativus, L. stigmas and byproducts: Qualitative fingerprint, antioxidant potentials and enzyme inhibitory activities. Food Res. Int. 2018, 109, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Sersen, F.; Gregan, F.; Kotora, P.; Kmetova, J.; Filo, J.; Loos, D.; Gregan, J. Synthesis and Free Radical Scavenging Activity of New Hydroxybenzylidene Hydrazines. Molecules 2017, 22, 894. [Google Scholar] [CrossRef] [PubMed]
- Kotora, P.; Šeršeň, F.; Filo, J.; Loos, D.; Gregáň, J.; Gregáň, F. The Scavenging of DPPH, Galvinoxyl and ABTS Radicals by Imine Analogs of Resveratrol. Molecules 2016, 21, 127. [Google Scholar] [CrossRef] [PubMed]
- Khajehei, F.; Merkt, N.; Claupein, W.; Graeff-Hoenninger, S. Yacon (Smallanthus sonchifolius Poepp. & Endl.) as a Novel Source of Health Promoting Compounds: Antioxidant Activity, Phytochemicals and Sugar Content in Flesh, Peel, and Whole Tubers of Seven Cultivars. Molecules 2018, 23, 278. [Google Scholar] [CrossRef]
- Yang, B.; Wang, J.S.; Zhao, M.M.; Liu, Y.; Wang, W.; Jiang, Y.M. Identification of polysaccharides from pericarp tissues of litchi (Litchi chinensis Sonn.) fruit in relation to their antioxidant activities. Carbohydr. Res. 2006, 341, 634–638. [Google Scholar] [CrossRef] [PubMed]
- Thaipong, K.; Boonprakob, U.; Crosby, K.; Cisneros-zevallos, L.; Byrne, D.H. Comparison of ABTS, DPPH, FRAP, and ORAC assays for estimating antioxidant activity from guava fruit extracts. J. Food Compost. Anal. 2006, 19, 669–675. [Google Scholar] [CrossRef]
- Yao, Y.; Yang, X.S.; Tian, J.; Liu, C.Y.; Cheng, X.Z.; Ren, G.X. Antioxidant and Antidiabetic Activities of Black Mung Bean (Vigna radiata L.). J. Agric. Food Chem. 2013, 61, 8104–8109. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Yang, X.S.; Shi, Z.X.; Ren, G.X. Anti-inflammatory activity of saponins from quinoa (Chenopodium quinoa Willd.) seeds in lipopolysaccharide-stimulated RAW 264.7 macrophages cells. J. Food Sci. 2014, 79, H1018–H1023. [Google Scholar] [CrossRef] [PubMed]
- Dinelli, G.; Bregola, V.; Bosi, S.; Fiori, J.; Gotti, R.; Simonetti, E.; Trozzi, C.; Leoncini, E.; Prata, C.; Massaccesi, L.; et al. Lunasin in wheat: A chemical and molecular study on its presence or absence. Food Chem. 2014, 151, 520–525. [Google Scholar] [CrossRef] [PubMed]
- Gungor, S.S.U.; Ozay, S.G.; Ilcim, A.; Kökdil, G. Composition and content of total phenolics and flavonoids, and antioxidant activity of Trigonella isthmocarpa. Eur. J. Chem. 2013, 4, 7–9. [Google Scholar] [CrossRef]
- Fang, S.C.; Hsu, C.L.; Yen, G.C. Anti-inflammatory effects of phenolic compounds isolated from the fruits of Artocarpus heterophyllus. J. Agric. Food Chem. 2008, 56, 4463–4468. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.B.; De Lumen, B.; Jeong, H.J. Lunasin peptide purified from Solanum nigrum L. protects DNA from oxidative damage by suppressing the generation of hydroxyl radical via blocking fenton reaction. Cancer Lett. 2010, 293, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Seber, L.E.; Barnett, B.W.; Mcconnell, E.J.; Hume, S.D.; Cai, J.; Boles, K.; Davis, K.R. Scalable purification and characterization of the anticancer lunasin peptide from soybean. PLoS ONE 2012, 7, e35409. [Google Scholar] [CrossRef] [PubMed]
- Medvedev, A.E.; Kopydlowski, K.M.; Vogel, S.N. Inhibition of lipopolysaccharide-induced signal transduction in endotoxin-tolerized mouse macrophages: Dysregulation of cytokine, chemokine, and toll-like receptor 2 and 4 gene expression. J. Immunol. 2000, 164, 5564–5574. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Verma, I.M. NF-κB regulation in the immune system. Nat. Rev. Immunol. 2002, 2, 725–734. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, G.; Hao, Y.; Zhu, Y.; Shi, Z.; Zhao, G. Expression of Bioactive Lunasin Peptide in Transgenic Rice Grains for the Application in Functional Food. Molecules 2018, 23, 2373. https://doi.org/10.3390/molecules23092373
Ren G, Hao Y, Zhu Y, Shi Z, Zhao G. Expression of Bioactive Lunasin Peptide in Transgenic Rice Grains for the Application in Functional Food. Molecules. 2018; 23(9):2373. https://doi.org/10.3390/molecules23092373
Chicago/Turabian StyleRen, Guixing, Yuqiong Hao, Yingying Zhu, Zhenxing Shi, and Gang Zhao. 2018. "Expression of Bioactive Lunasin Peptide in Transgenic Rice Grains for the Application in Functional Food" Molecules 23, no. 9: 2373. https://doi.org/10.3390/molecules23092373
APA StyleRen, G., Hao, Y., Zhu, Y., Shi, Z., & Zhao, G. (2018). Expression of Bioactive Lunasin Peptide in Transgenic Rice Grains for the Application in Functional Food. Molecules, 23(9), 2373. https://doi.org/10.3390/molecules23092373



