NMR as a Tool to Investigate the Processes of Mitochondrial and Cytosolic Iron-Sulfur Cluster Biosynthesis
Abstract
1. Introduction
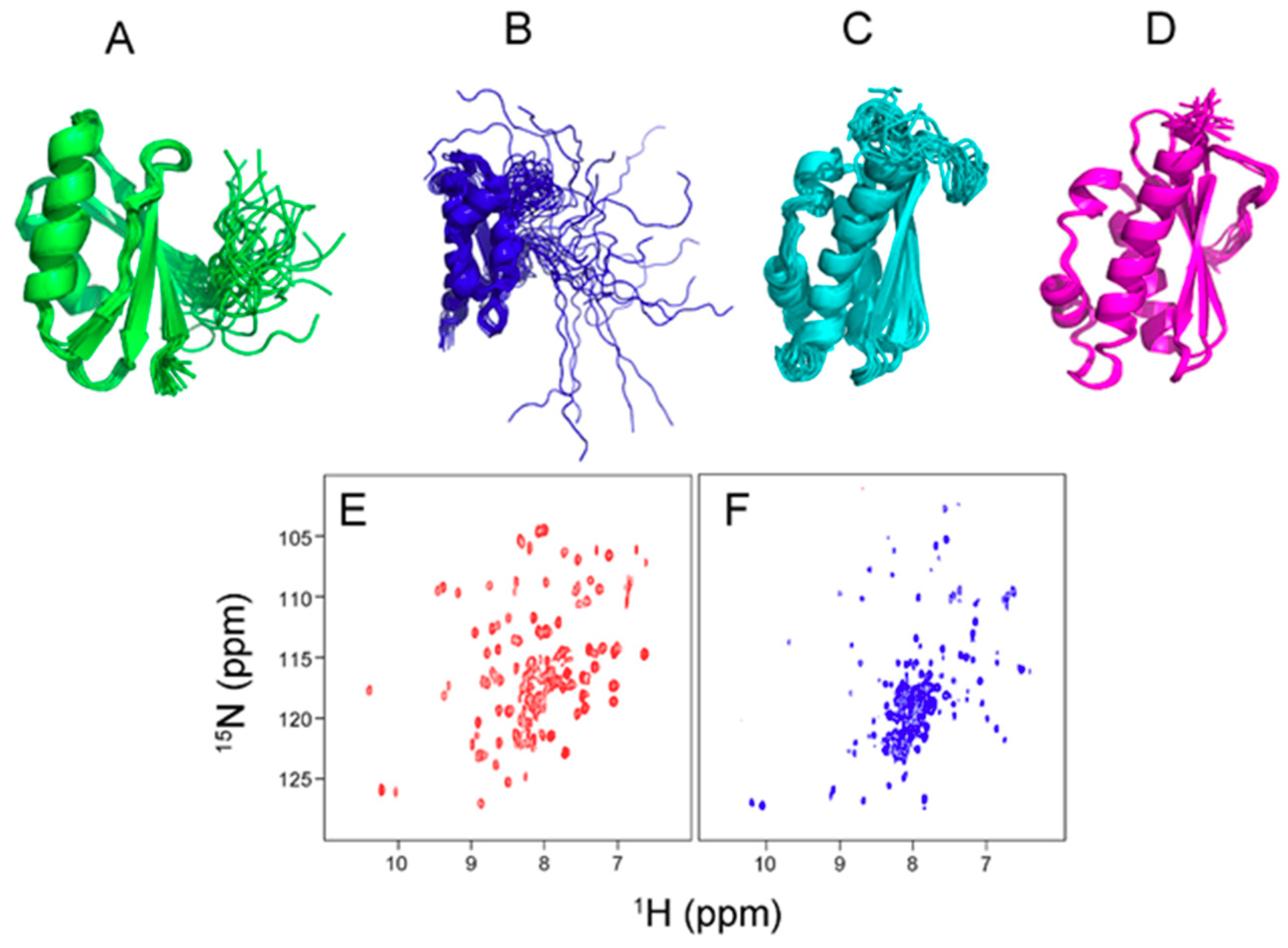
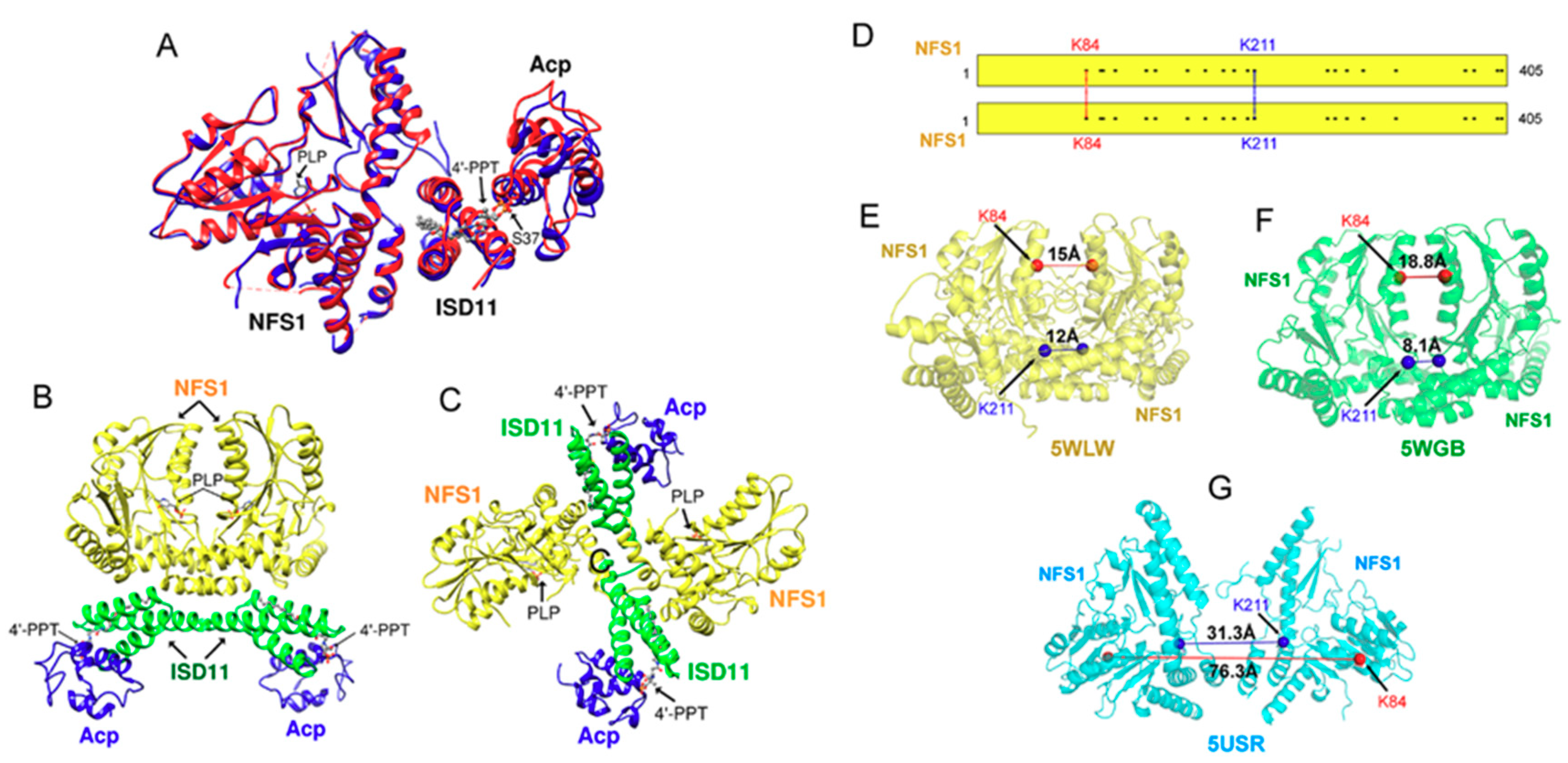
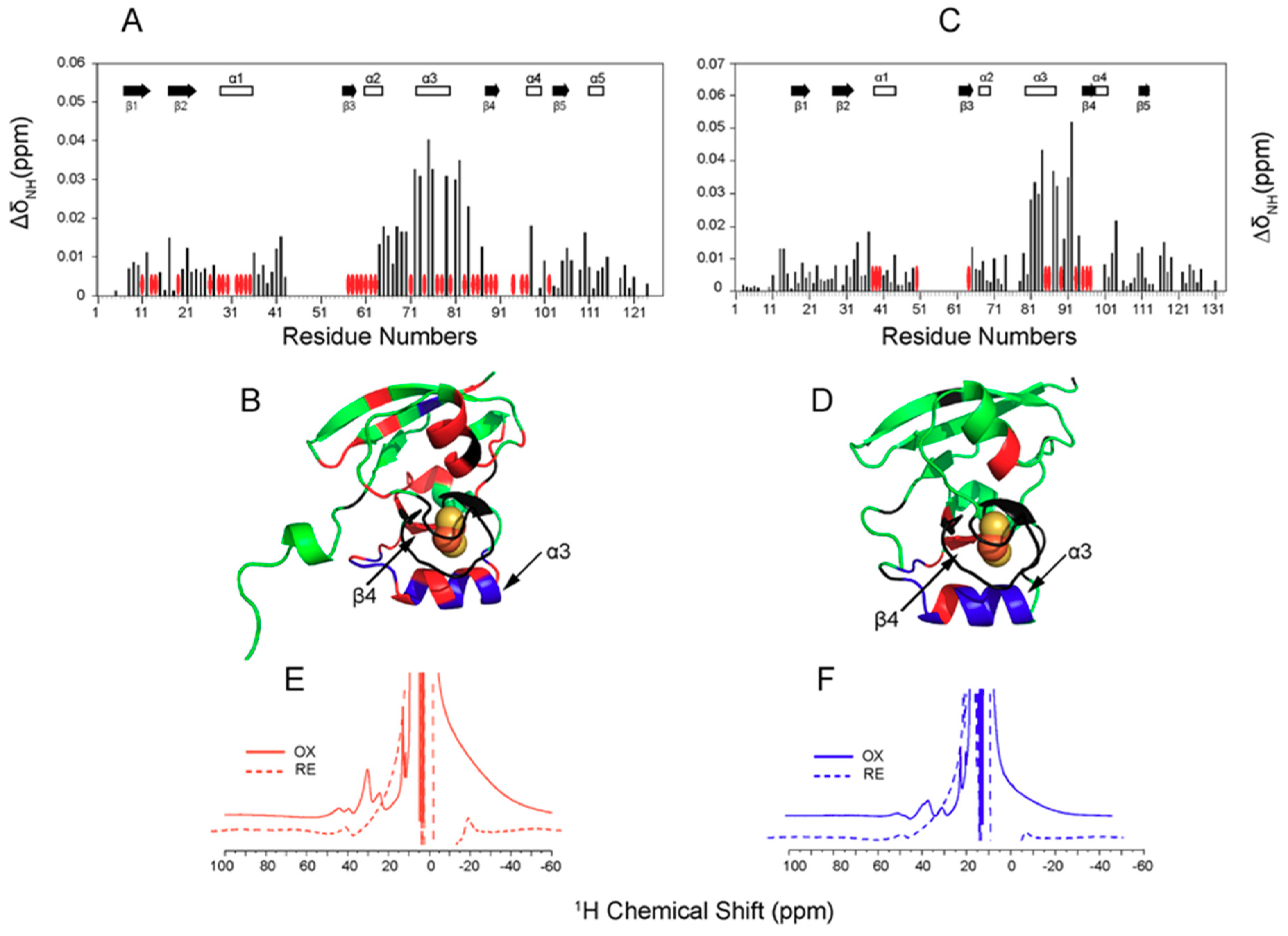
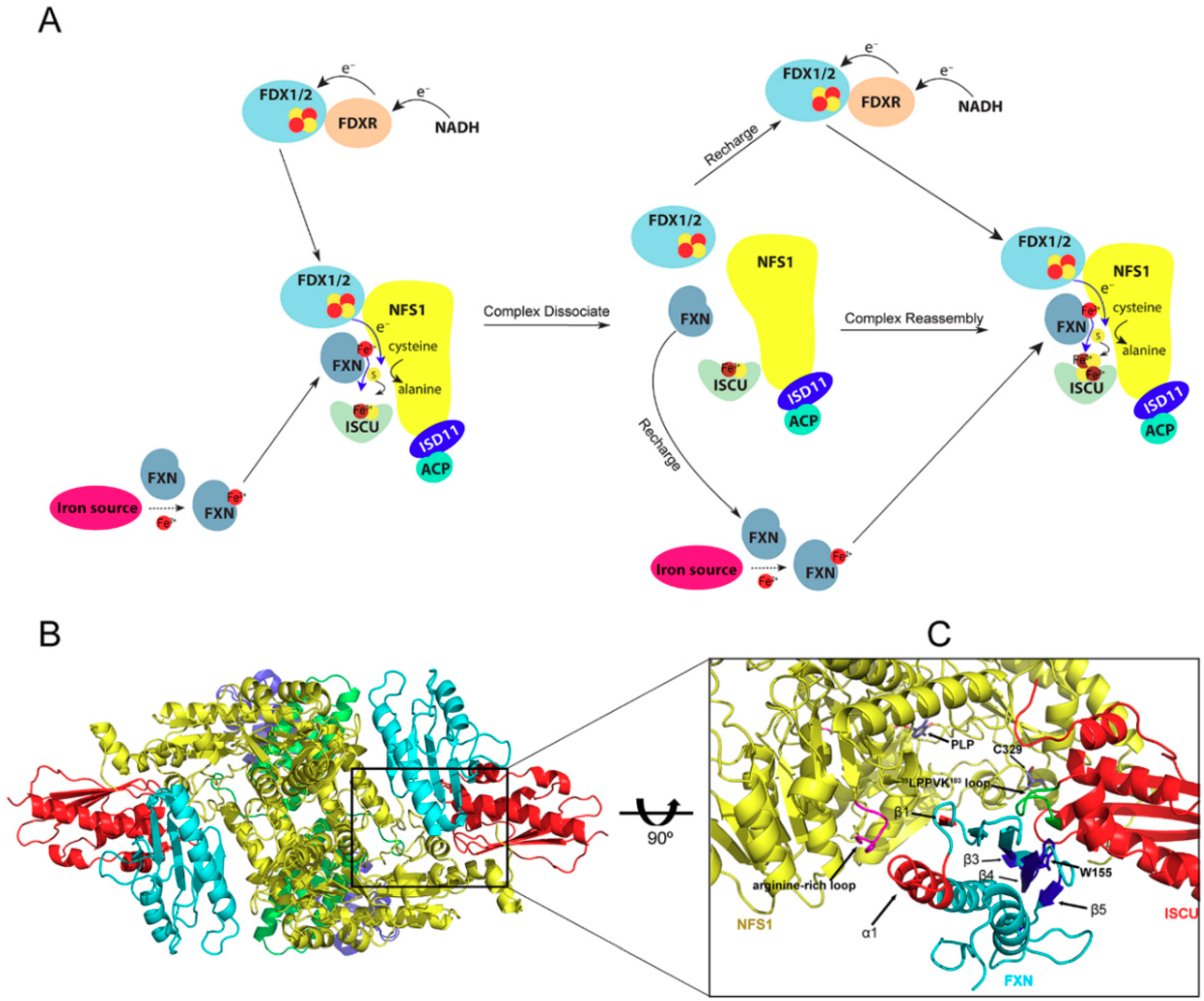
2. Fe-S Cluster Assembly on A Scaffold Protein as Studied by NMR Spectroscopy
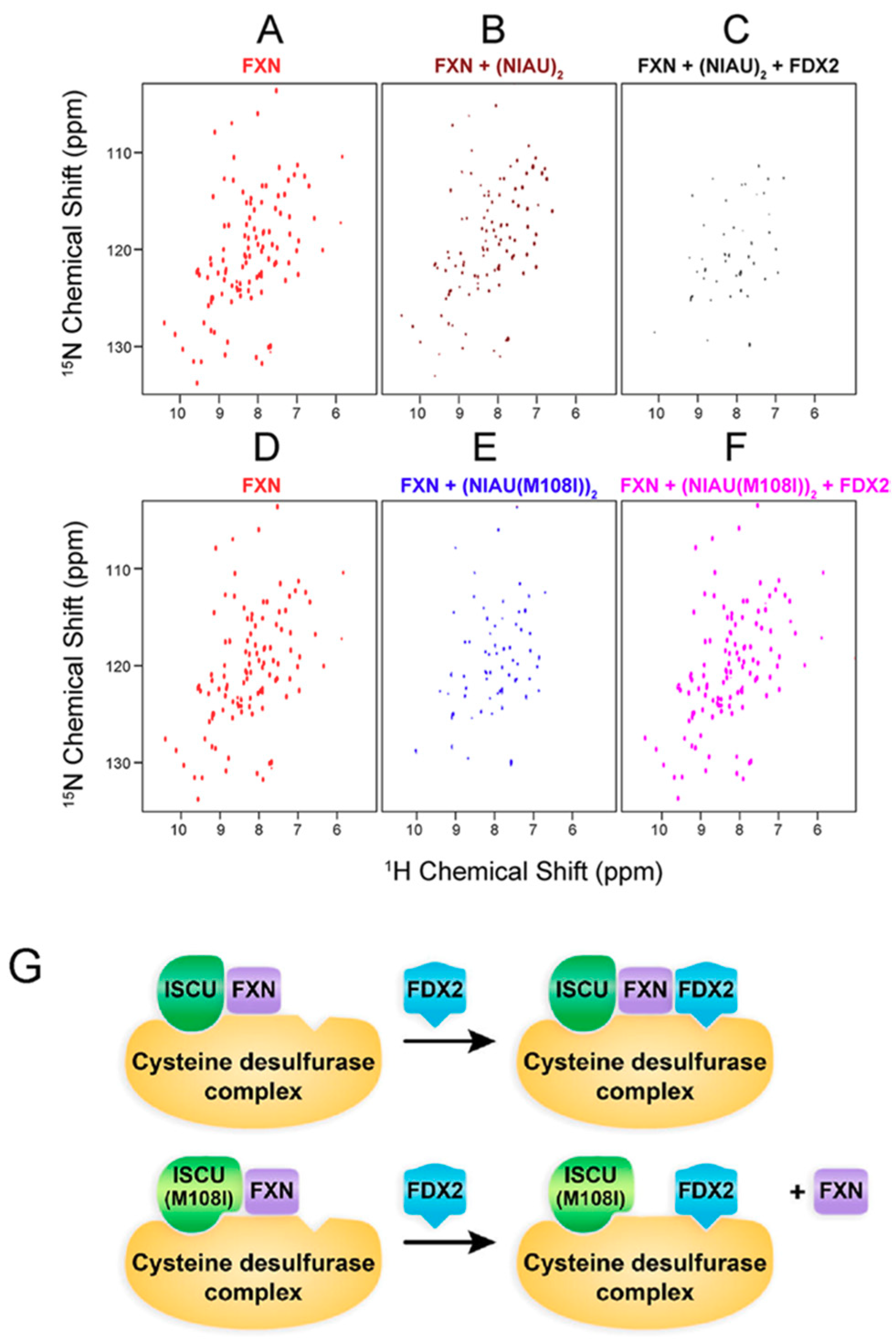
3. [2Fe-2S] Cluster Transfer as Studied by NMR Spectroscopy
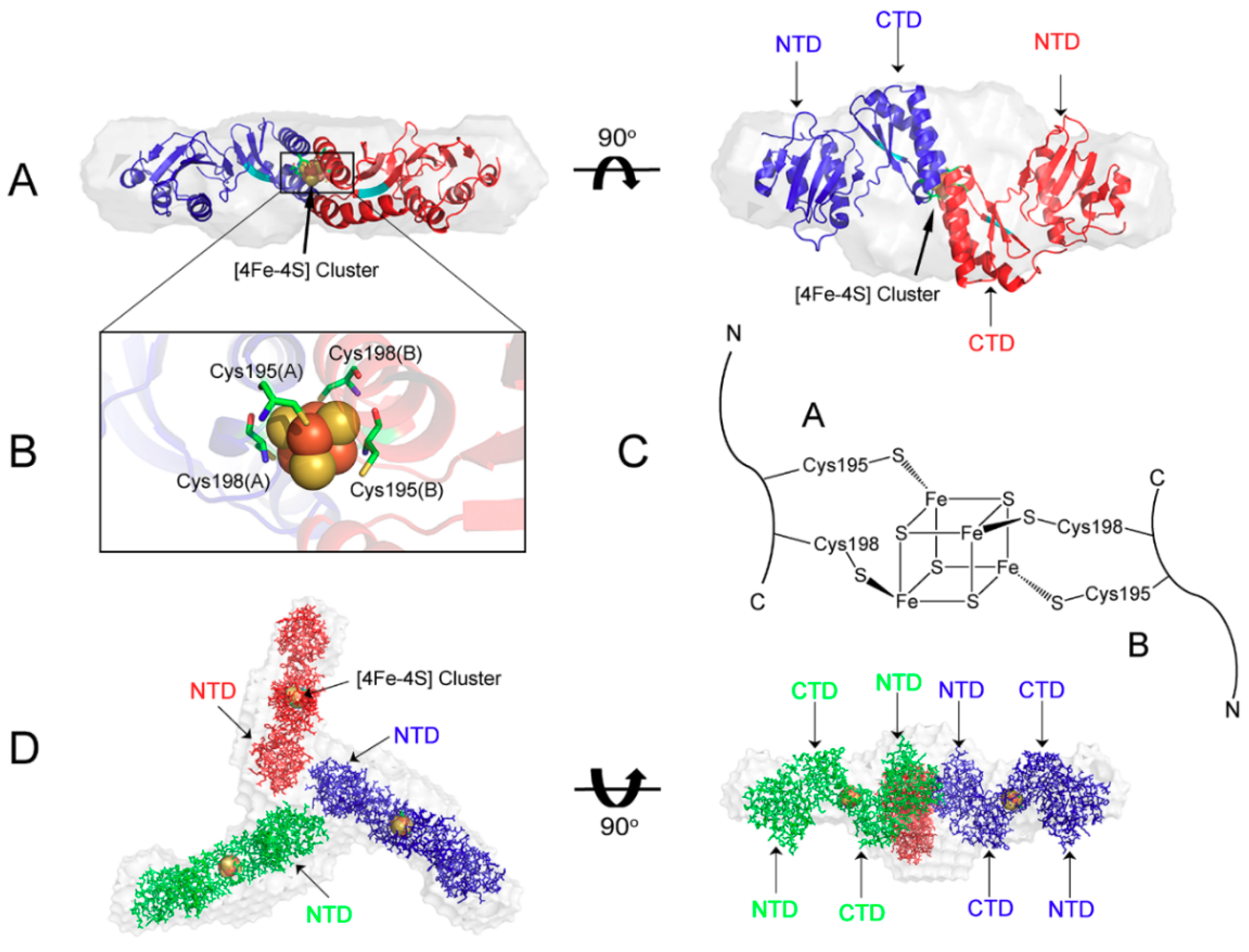
4. Maturation of [4Fe-4S] Cluster by NMR
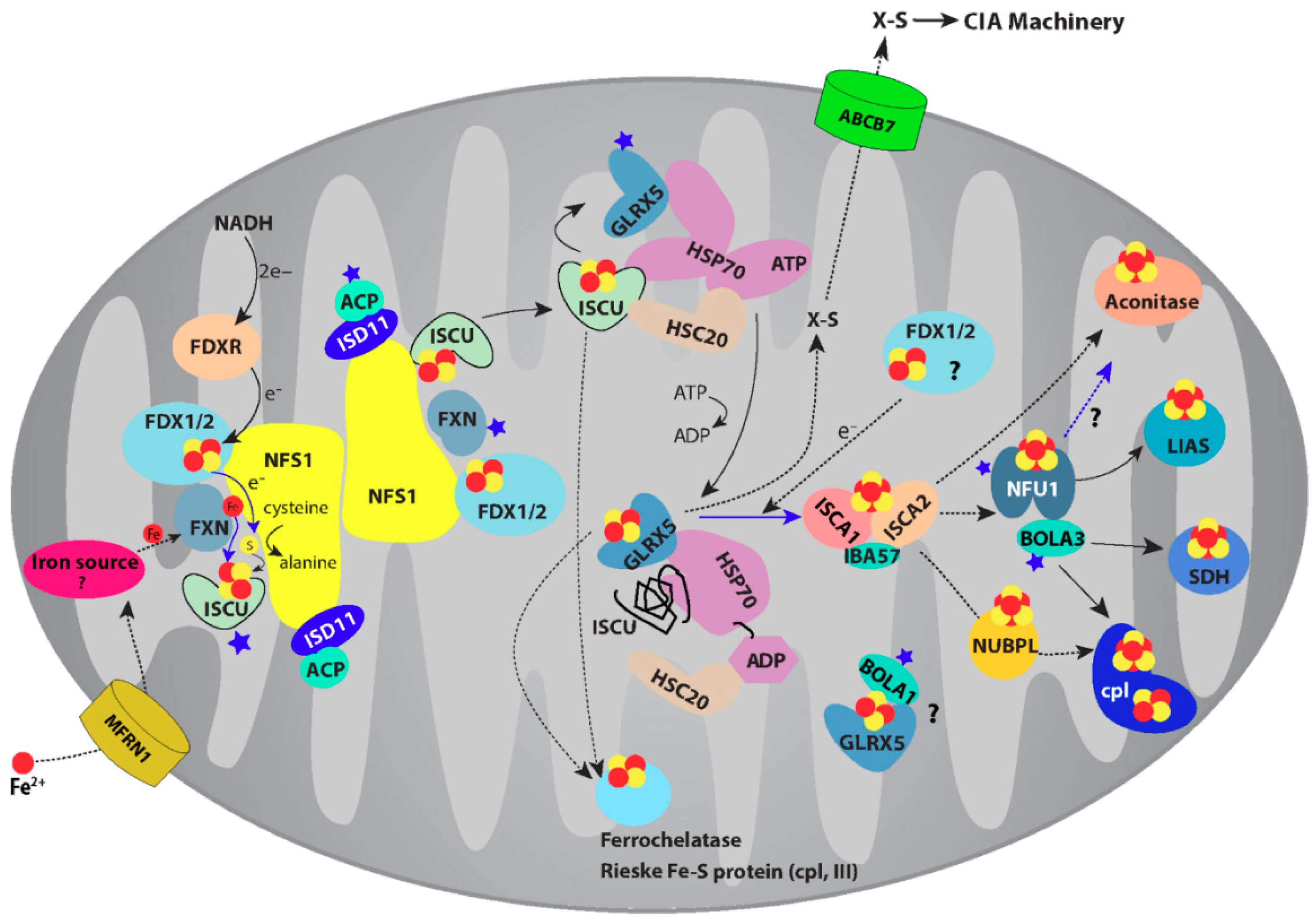
5. Trafficking of [4Fe-4S] Cluster Studied by NMR
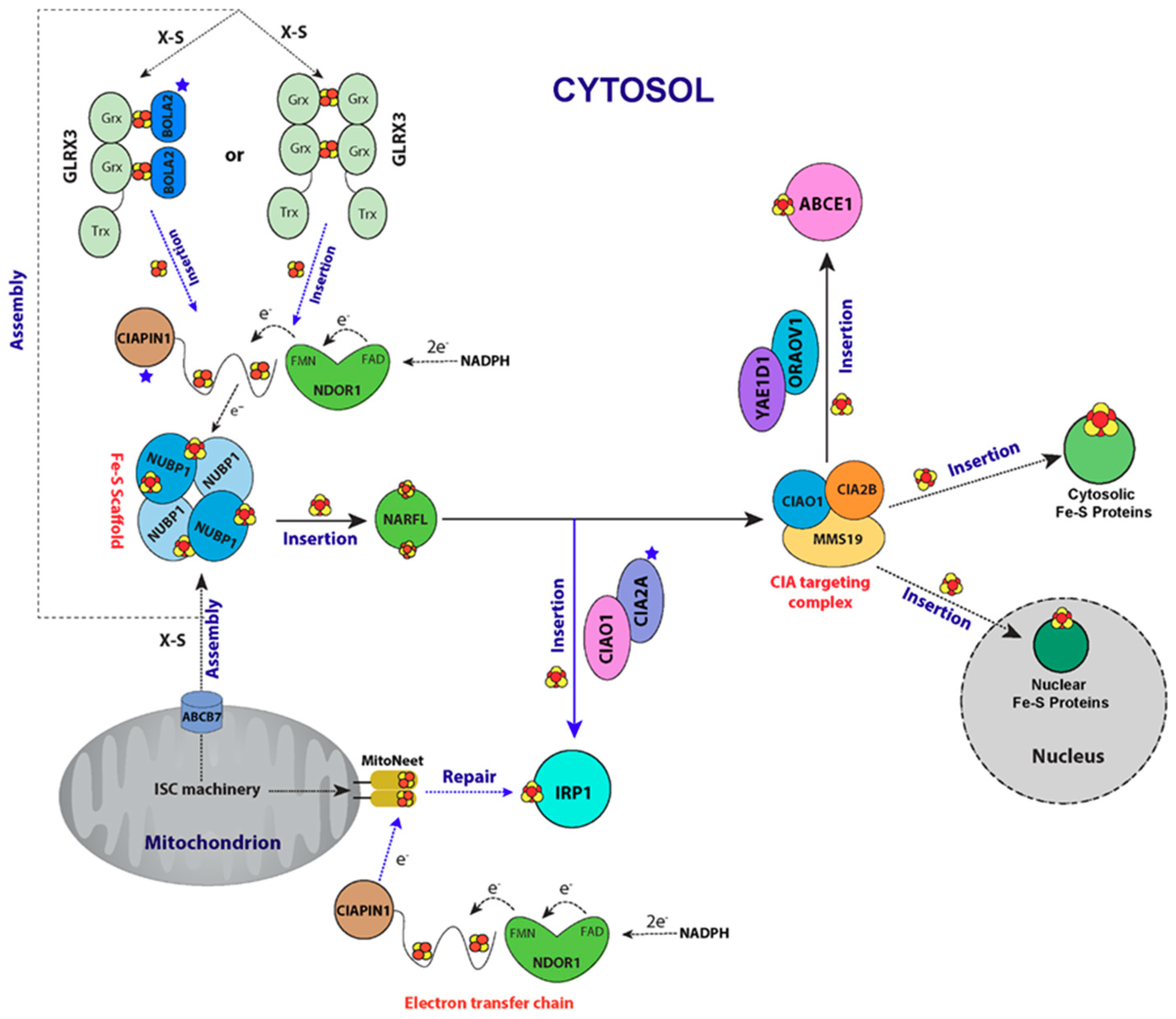
6. CIA Machinery as Studied by NMR Spectroscopy
7. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Waldron, K.J.; Rutherford, J.C.; Ford, D.; Robinson, N.J. Metalloproteins and metal sensing. Nature 2009, 460, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Finney, L.A.; O′Halloran, T.V. Transition metal speciation in the cell: Insights from the chemistry of metal ion receptors. Science 2003, 300, 931–936. [Google Scholar] [CrossRef] [PubMed]
- Beinert, H.; Holm, R.H.; Munck, E. Iron-sulfur clusters: Nature’s modular, multipurpose structures. Science 1997, 277, 653–659. [Google Scholar] [CrossRef] [PubMed]
- Frazzon, J.; Dean, D.R. Formation of iron-sulfur clusters in bacteria: An emerging field in bioinorganic chemistry. Curr. Opin. Chem. Biol. 2003, 7, 166–173. [Google Scholar] [CrossRef]
- Johnson, D.C.; Dean, D.R.; Smith, A.D.; Johnson, M.K. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 2005, 74, 247–281. [Google Scholar] [CrossRef] [PubMed]
- Lill, R. Function and biogenesis of iron-sulphur proteins. Nature 2009, 460, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Py, B.; Barras, F. Building Fe-S proteins: Bacterial strategies. Nat. Rev. Microbiol. 2010, 8, 436–446. [Google Scholar] [CrossRef] [PubMed]
- Beinert, H. Iron-sulfur proteins: Ancient structures, still full of surprises. J. Biol. Inorg. Chem. 2000, 5, 2–15. [Google Scholar] [CrossRef] [PubMed]
- Lill, R.; Dutkiewicz, R.; Elsasser, H.P.; Hausmann, A.; Netz, D.J.A.; Pierik, A.J.; Stehling, O.; Urzica, E.; Muhlenhoff, U. Mechanisms of iron-sulfur protein maturation in mitochondria, cytosol and nucleus of eukaryotes. Biochim. Biophys. Acta 2006, 1763, 652–667. [Google Scholar] [CrossRef] [PubMed]
- Ayala-Castro, C.; Saini, A.; Outten, F.W. Fe-S cluster assembly pathways in bacteria. Microbiol. Mol. Biol. Rev. 2008, 72, 110–125. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chakraborty, S.; Hosseinzadeh, P.; Yu, Y.; Tian, S.; Petrik, I.; Bhagi, A.; Lu, Y. Metalloproteins containing cytochrome, iron-sulfur, or copper redox centers. Chem. Rev. 2014, 114, 4366–4469. [Google Scholar] [CrossRef] [PubMed]
- Flint, D.H.; Allen, R.M. Iron-sulfur proteins with nonredox functions. Chem. Rev. 1996, 96, 2315–2334. [Google Scholar] [CrossRef] [PubMed]
- Rouault, T.A. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat. Chem. Biol. 2006, 2, 406–414. [Google Scholar] [CrossRef] [PubMed]
- Volz, K. The functional duality of iron regulatory protein 1. Curr. Opin. Struct. Biol. 2008, 18, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Fleischhacker, A.S.; Kiley, P.J. Iron-containing transcription factors and their roles as sensors. Curr. Opin. Chem. Biol. 2011, 15, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Tong, W.H.; Rouault, T.A. Metabolic regulation of citrate and iron by aconitases: Role of iron-sulfur cluster biogenesis. Biometals 2007, 20, 549–564. [Google Scholar] [CrossRef] [PubMed]
- White, M.F.; Dillingham, M.S. Iron-sulphur clusters in nucleic acid processing enzymes. Curr. Opin. Struct. Biol. 2012, 22, 94–100. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, E.; Holt, M.E.; Thompson, M.K.; Salay, L.E.; Ehlinger, A.C.; Chazin, W.J.; Barton, J.K. The [4Fe4S] cluster of human DNA primase functions as a redox switch using DNA charge transport. Science 2017, 355, eaag1789. [Google Scholar] [CrossRef] [PubMed]
- Veatch, J.R.; McMurray, M.A.; Nelson, Z.W.; Gottschling, D.E. Mitochondrial dysfunction leads to nuclear genome instability via an iron-sulfur cluster defect. Cell 2009, 137, 1247–1258. [Google Scholar] [CrossRef] [PubMed]
- Netz, D.J.A.; Stith, C.M.; Stumpfig, M.; Kopf, G.; Vogel, D.; Genau, H.M.; Stodola, J.L.; Lill, R.; Burgers, P.M.J.; Pierik, A.J. Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat. Chem. Biol. 2012, 8, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Rudolf, J.; Makrantoni, V.; Ingledew, W.J.; Stark, M.J.R.; White, M.F. The DNA repair helicases XPD and FancJ have essential iron-sulfur domains. Mol. Cell 2006, 23, 801–808. [Google Scholar] [CrossRef] [PubMed]
- Schnackerz, K.D.; Dobritzsch, D.; Lindqvist, Y.; Cook, P.F. Dihydropyrimidine dehydrogenase: A flavoprotein with four iron-sulfur clusters. Biochim. Biophys. Acta 2004, 1701, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Kispal, G.; Sipos, K.; Lange, H.; Fekete, Z.; Bedekovics, T.; Janaky, T.; Bassler, J.; Aguilar Netz, D.J.; Balk, J.; Rotte, C.; et al. Biogenesis of cytosolic ribosomes requires the essential iron-sulphur protein Rli1p and mitochondria. EMBO J. 2005, 24, 589–598. [Google Scholar] [CrossRef] [PubMed]
- Maio, N.; Rouault, T.A. Iron-sulfur cluster biogenesis in mammalian cells: New insights into the molecular mechanisms of cluster delivery. Biochim. Biophys. Acta 2015, 1853, 1493–1512. [Google Scholar] [CrossRef] [PubMed]
- Ollagnier-De Choudens, S.; Sanakis, Y.; Hewitson, K.S.; Roach, P.; Baldwin, J.E.; Munck, E.; Fontecave, M. Iron-sulfur center of biotin synthase and lipoate synthase. Biochemistry 2000, 39, 4165–4173. [Google Scholar] [CrossRef] [PubMed]
- Lanz, N.D.; Booker, S.J. Auxiliary iron-sulfur cofactors in radical SAM enzymes. Biochim. Biophys. Acta 2015, 1853, 1316–1334. [Google Scholar] [CrossRef] [PubMed]
- Landgraf, B.J.; McCarthy, E.L.; Booker, S.J. Radical S-Adenosylmethionine Enzymes in Human Health and Disease. Annu. Rev. Biochem. 2016, 85, 485–514. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Yin, H.; Yang, C.; Dou, Y.; Liu, Z.; Zhang, P.; Yu, H.; Huang, Y.; Feng, J.; Hao, J.; et al. A magnetic protein biocompass. Nat. Mater. 2015, 15, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Roche, B.; Aussel, L.; Ezraty, B.; Mandin, P.; Py, B.; Barras, F. Iron/sulfur proteins biogenesis in prokaryotes: Formation, regulation and diversity. Biochim. Biophys. Acta 2013, 1827, 455–469. [Google Scholar] [CrossRef] [PubMed]
- Blanc, B.; Gerez, C.; Ollagnier de Choudens, S. Assembly of Fe/S proteins in bacterial systems: Biochemistry of the bacterial ISC system. Biochim. Biophys. Acta 2015, 1853, 1436–1447. [Google Scholar] [CrossRef] [PubMed]
- Prischi, F.; Konarev, P.V.; Iannuzzi, C.; Pastore, C.; Adinolfi, S.; Martin, S.R.; Svergun, D.I.; Pastore, A. Structural bases for the interaction of frataxin with the central components of iron-sulphur cluster assembly. Nat. Commun. 2010, 1, 95. [Google Scholar] [CrossRef] [PubMed]
- Adinolfi, S.; Iannuzzi, C.; Prischi, F.; Pastore, C.; Iametti, S.; Martin, S.R.; Bonomi, F.; Pastore, A. Bacterial frataxin CyaY is the gatekeeper of iron-sulfur cluster formation catalyzed by IscS. Nat. Struct. Mol. Biol. 2009, 16, 390–396. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Bothe, J.R.; Alderson, T.R.; Markley, J.L. Tangled web of interactions among proteins involved in iron-sulfur cluster assembly as unraveled by NMR, SAXS, chemical crosslinking, and functional studies. Biochim. Biophys. Acta 2015, 1853, 1416–1428. [Google Scholar] [CrossRef] [PubMed]
- Gray, M.W. Mitochondrial evolution. Cold Spring Harb. Perspect. Biol. 2012, 4, a011403. [Google Scholar] [CrossRef] [PubMed]
- Lill, R.; Muhlenhoff, U. Iron-sulfur-protein biogenesis in eukaryotes. Trends Biochem. Sci. 2005, 30, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Stehling, O.; Lill, R. The role of mitochondria in cellular iron-sulfur protein biogenesis: Mechanisms, connected processes, and diseases. Cold Spring Harb. Perspect. Biol. 2013, 5, a011312. [Google Scholar] [CrossRef] [PubMed]
- Braymer, J.J.; Lill, R. Iron-sulfur cluster biogenesis and trafficking in mitochondria. J. Biol. Chem. 2017, 292, 12754–12763. [Google Scholar] [CrossRef] [PubMed]
- Uzarska, M.A.; Dutkiewicz, R.; Freibert, S.A.; Lill, R.; Muehlenhoff, U. The mitochondrial Hsp70 chaperone Ssq1 facilitates Fe/S cluster transfer from Isu1 to Grx5 by complex formation. Mol. Biol. Cell 2013, 24, 1830–1841. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Manzaneque, M.T.; Tamarit, J.; Belli, G.; Ros, J.; Herrero, E. Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol. Biol. Cell 2002, 13, 1109–1121. [Google Scholar] [CrossRef] [PubMed]
- Muhlenhoff, U.; Hoffmann, B.; Richter, N.; Rietzschel, N.; Spantgar, F.; Stehling, O.; Uzarska, M.A.; Lill, R. Compartmentalization of iron between mitochondria and the cytosol and its regulation. Eur. J. Cell Biol. 2015, 94, 292–308. [Google Scholar] [CrossRef] [PubMed]
- Lill, R.; Dutkiewicz, R.; Freibert, S.A.; Heidenreich, T.; Mascarenhas, J.; Netz, D.J.; Paul, V.D.; Pierik, A.J.; Richter, N.; Stumpfig, M.; et al. The role of mitochondria and the CIA machinery in the maturation of cytosolic and nuclear iron-sulfur proteins. Eur. J. Cell Biol. 2015, 94, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Netz, D.J.; Mascarenhas, J.; Stehling, O.; Pierik, A.J.; Lill, R. Maturation of cytosolic and nuclear iron-sulfur proteins. Trends Cell Biol. 2014, 24, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Beilschmidt, L.K.; Ollagnier de Choudens, S.; Fournier, M.; Sanakis, I.; Hograindleur, M.A.; Clemancey, M.; Blondin, G.; Schmucker, S.; Eisenmann, A.; Weiss, A.; et al. ISCA1 is essential for mitochondrial Fe4S4 biogenesis in vivo. Nat. Commun. 2017, 8, 15124. [Google Scholar] [CrossRef] [PubMed]
- Sheftel, A.D.; Wilbrecht, C.; Stehling, O.; Niggemeyer, B.; Elsasser, H.P.; Muhlenhoff, U.; Lill, R. The human mitochondrial ISCA1, ISCA2, and IBA57 proteins are required for [4Fe-4S] protein maturation. Mol. Biol. Cell 2012, 23, 1157–1166. [Google Scholar] [CrossRef] [PubMed]
- Muhlenhoff, U.; Richter, N.; Pines, O.; Pierik, A.J.; Lill, R. Specialized function of yeast Isa1 and Isa2 proteins in the maturation of mitochondrial [4Fe-4S] proteins. J. Biol. Chem. 2011, 286, 41205–41216. [Google Scholar] [CrossRef] [PubMed]
- Melber, A.; Na, U.; Vashisht, A.; Weiler, B.D.; Lill, R.; Wohlschlegel, J.A.; Winge, D.R. Role of Nfu1 and Bol3 in iron-sulfur cluster transfer to mitochondrial clients. Elife 2016, 5, e15991. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Sastre, A.; Tort, F.; Stehling, O.; Uzarska, M.A.; Arranz, J.A.; Del Toro, M.; Labayru, M.T.; Landa, J.; Font, A.; Garcia-Villoria, J.; et al. A fatal mitochondrial disease is associated with defective NFU1 function in the maturation of a subset of mitochondrial Fe-S proteins. Am. J. Hum. Genet. 2011, 89, 656–667. [Google Scholar] [CrossRef] [PubMed]
- Cameron, J.M.; Janer, A.; Levandovskiy, V.; Mackay, N.; Rouault, T.A.; Tong, W.H.; Ogilvie, I.; Shoubridge, E.A.; Robinson, B.H. Mutations in iron-sulfur cluster scaffold genes NFU1 and BOLA3 cause a fatal deficiency of multiple respiratory chain and 2-oxoacid dehydrogenase enzymes. Am. J. Hum. Genet. 2011, 89, 486–495. [Google Scholar] [CrossRef] [PubMed]
- Rouault, T.A. Biogenesis of iron-sulfur clusters in mammalian cells: New insights and relevance to human disease. Dis. Model. Mech. 2012, 5, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Stehling, O.; Wilbrecht, C.; Lill, R. Mitochondrial iron-sulfur protein biogenesis and human disease. Biochimie 2014, 100, 61–77. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Rouault, T.A. Human iron-sulfur cluster assembly, cellular iron homeostasis, and disease. Biochemistry 2010, 49, 4945–4956. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Markley, J.L. NMR Spectroscopic Studies of Paramagnetic Proteins—Iron-Sulfur Proteins. Annu. Rev. Biophys. Biomol. Struct. 1995, 24, 209–237. [Google Scholar] [CrossRef] [PubMed]
- Bertini, I.; Couture, M.M.J.; Donaire, A.; Eltis, L.D.; Felli, I.C.; Luchinat, C.; Piccioli, M.; Rosato, A. The solution structure refinement of the paramagnetic reduced high-potential iron-sulfur protein I from Ectothiorhodospira halophila by using stable isotope labeling and nuclear relaxation. Eur. J. Biochem. 1996, 241, 440–452. [Google Scholar] [CrossRef] [PubMed]
- Xia, B.; Jenk, D.; LeMaster, D.M.; Westler, W.M.; Markley, J.L. Electron-nuclear interactions in two prototypical [2Fe-2S] proteins: Selective (chiral) deuteration and analysis of H-1 and H-2 NMR signals from the α and β hydrogens of cysteinyl residues that ligate the iron in the active sites of human ferredoxin and Anabaena 7120 vegetative ferredoxin. Arch. Biochem. Biophys. 2000, 373, 328–334. [Google Scholar] [PubMed]
- Cheng, H.; Westler, W.M.; Xia, B.; Oh, B.H.; Markley, J.L. Protein expression, selective isotopic labeling, and analysis of hyperfine-shifted NMR signals of Anabaena 7120 vegetative [2Fe-2S] ferredoxin. Arch. Biochem. Biophys. 1995, 316, 619–634. [Google Scholar] [CrossRef] [PubMed]
- Bertini, I.; Donaire, A.; Luchinat, C.; Rosato, A. Paramagnetic relaxation as a tool for solution structure determination: Clostridium pasteurianum ferredoxin as an example. Proteins-Struct. Funct. Genet. 1997, 29, 348–358. [Google Scholar] [CrossRef]
- Machonkin, T.E.; Westler, W.M.; Markley, J.L. 13C{13C} 2D NMR: A novel strategy for the study of paramagnetic proteins with slow electronic relaxation rates. J. Am. Chem. Soc. 2002, 124, 3204–3205. [Google Scholar] [CrossRef] [PubMed]
- Bermel, W.; Bertini, I.; Felli, I.C.; Kummerle, R.; Pierattelli, R. Novel 13C direct detection experiments, including extension to the third dimension, to perform the complete assignment of proteins. J. Magn. Reson. 2006, 178, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Banci, L.; Camponeschi, F.; Ciofi-Baffoni, S.; Piccioli, M. The NMR contribution to protein-protein networking in Fe-S protein maturation. J. Biol. Inorg. Chem. 2018, 23, 665–685. [Google Scholar] [CrossRef] [PubMed]
- Ciofi-Baffoni, S.; Gallo, A.; Muzzioli, R.; Piccioli, M. The IR-15N-HSQC-AP experiment: A new tool for NMR spectroscopy of paramagnetic molecules. J. Biomol. NMR 2014, 58, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Lian, L.Y. NMR studies of weak protein-protein interactions. Prog. Nucl. Magn. Reson. Spectrosc. 2013, 71, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Piccioli, M.; Turano, P. Transient iron coordination sites in proteins: Exploiting the dual nature of paramagnetic NMR. Coord. Chem. Rev. 2015, 284, 313–328. [Google Scholar] [CrossRef]
- Ciofi-Baffoni, S.; Nasta, V.; Banci, L. Protein networks in the maturation of human iron-sulfur proteins. Metallomics 2017, 10, 49–72. [Google Scholar] [CrossRef] [PubMed]
- Campuzano, V.; Montermini, L.; Molto, M.D.; Pianese, L.; Cossee, M.; Cavalcanti, F.; Monros, E.; Rodius, F.; Duclos, F.; Monticelli, A.; et al. Friedreich’s ataxia: Autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 1996, 271, 1423–1427. [Google Scholar] [CrossRef] [PubMed]
- Santos, R.; Lefevre, S.; Sliwa, D.; Seguin, A.; Camadro, J.M.; Lesuisse, E. Friedreich ataxia: Molecular mechanisms, redox considerations, and therapeutic opportunities. Antioxid. Redox Signal. 2010, 13, 651–690. [Google Scholar] [CrossRef] [PubMed]
- Crooks, D.R.; Jeong, S.Y.; Tong, W.H.; Ghosh, M.C.; Olivierre, H.; Haller, R.G.; Rouault, T.A. Tissue specificity of a human mitochondrial disease: Differentiation-enhanced mis-splicing of the Fe-S scaffold gene ISCU renders patient cells more sensitive to oxidative stress in ISCU myopathy. J. Biol. Chem. 2012, 287, 40119–40130. [Google Scholar] [CrossRef] [PubMed]
- Olsson, A.; Lind, L.; Thornell, L.E.; Holmberg, M. Myopathy with lactic acidosis is linked to chromosome 12q23.3-24.11 and caused by an intron mutation in the ISCU gene resulting in a splicing defect. Hum. Mol. Genet. 2008, 17, 1666–1672. [Google Scholar] [CrossRef] [PubMed]
- Mochel, F.; Knight, M.A.; Tong, W.H.; Hernandez, D.; Ayyad, K.; Taivassalo, T.; Andersen, P.M.; Singleton, A.; Rouault, T.A.; Fischbeck, K.H.; et al. Splice mutation in the iron-sulfur cluster scaffold protein ISCU causes myopathy with exercise intolerance. Am. J. Hum. Genet. 2008, 82, 652–660. [Google Scholar] [CrossRef] [PubMed]
- Spiegel, R.; Saada, A.; Halvardson, J.; Soiferman, D.; Shaag, A.; Edvardson, S.; Horovitz, Y.; Khayat, M.; Shalev, S.A.; Feuk, L.; et al. Deleterious mutation in FDX1L gene is associated with a novel mitochondrial muscle myopathy. Eur. J. Hum. Genet. 2014, 22, 902–906. [Google Scholar] [CrossRef] [PubMed]
- Farhan, S.M.; Wang, J.; Robinson, J.F.; Lahiry, P.; Siu, V.M.; Prasad, C.; Kronick, J.B.; Ramsay, D.A.; Rupar, C.A.; Hegele, R.A. Exome sequencing identifies NFS1 deficiency in a novel Fe-S cluster disease, infantile mitochondrial complex II/III deficiency. Mol. Genet. Genom. Med. 2014, 2, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Invernizzi, F.; Tigano, M.; Dallabona, C.; Donnini, C.; Ferrero, I.; Cremonte, M.; Ghezzi, D.; Lamperti, C.; Zeviani, M. A homozygous mutation in LYRM7/MZM1L associated with early onset encephalopathy, lactic acidosis, and severe reduction of mitochondrial complex III activity. Hum. Mutat. 2013, 34, 1619–1622. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Jeong, S.Y.; Ghosh, M.C.; Kovtunovych, G.; Silvestri, L.; Ortillo, D.; Uchida, N.; Tisdale, J.; Camaschella, C.; Rouault, T.A. Glutaredoxin 5 deficiency causes sideroblastic anemia by specifically impairing heme biosynthesis and depleting cytosolic iron in human erythroblasts. J. Clin. Investig. 2010, 120, 1749–1761. [Google Scholar] [CrossRef] [PubMed]
- Wingert, R.A.; Galloway, J.L.; Barut, B.; Foott, H.; Fraenkel, P.; Axe, J.L.; Weber, G.J.; Dooley, K.; Davidson, A.J.; Schmid, B.; et al. Deficiency of glutaredoxin 5 reveals Fe-S clusters are required for vertebrate haem synthesis. Nature 2005, 436, 1035–1039. [Google Scholar] [CrossRef] [PubMed]
- Allikmets, R.; Raskind, W.H.; Hutchinson, A.; Schueck, N.D.; Dean, M.; Koeller, D.M. Mutation of a putative mitochondrial iron transporter gene (ABC7) in X-linked sideroblastic anemia and ataxia (XLSA/A). Hum. Mol. Genet. 1999, 8, 743–749. [Google Scholar] [CrossRef] [PubMed]
- Shaw, G.C.; Cope, J.J.; Li, L.; Corson, K.; Hersey, C.; Ackermann, G.E.; Gwynn, B.; Lambert, A.J.; Wingert, R.A.; Traver, D.; et al. Mitoferrin is essential for erythroid iron assimilation. Nature 2006, 440, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Calvo, S.E.; Tucker, E.J.; Compton, A.G.; Kirby, D.M.; Crawford, G.; Burtt, N.P.; Rivas, M.; Guiducci, C.; Bruno, D.L.; Goldberger, O.A.; et al. High-throughput, pooled sequencing identifies mutations in NUBPL and FOXRED1 in human complex I deficiency. Nat. Genet. 2010, 42, 851–858. [Google Scholar] [CrossRef] [PubMed]
- Torraco, A.; Stehling, O.; Stumpfig, C.; Rosser, R.; De Rasmo, D.; Fiermonte, G.; Verrigni, D.; Rizza, T.; Vozza, A.; Di Nottia, M.; et al. ISCA1 Mutation in a Patient with Infantile-Onset Leukodystrophy Causes Defects in Mitochondrial [4Fe-4S] Proteins. Hum. Mol. Genet. 2018, 27, 2739–2754. [Google Scholar] [CrossRef] [PubMed]
- Shukla, A.; Hebbar, M.; Srivastava, A.; Kadavigere, R.; Upadhyai, P.; Kanthi, A.; Brandau, O.; Bielas, S.; Girisha, K.M. Homozygous p.(Glu87Lys) variant in ISCA1 is associated with a multiple mitochondrial dysfunctions syndrome. J. Hum. Genet. 2017, 62, 723–727. [Google Scholar] [CrossRef] [PubMed]
- Al-Hassnan, Z.N.; Al-Dosary, M.; Alfadhel, M.; Faqeih, E.A.; Alsagob, M.; Kenana, R.; Almass, R.; Al-Harazi, O.S.; Al-Hindi, H.; Malibari, O.I.; et al. ISCA2 mutation causes infantile neurodegenerative mitochondrial disorder. J. Med. Genet. 2015, 52, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Torraco, A.; Ardissone, A.; Invernizzi, F.; Rizza, T.; Fiermonte, G.; Niceta, M.; Zanetti, N.; Martinelli, D.; Vozza, A.; Verrigni, D.; et al. Novel mutations in IBA57 are associated with leukodystrophy and variable clinical phenotypes. J. Neurol. 2017, 264, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Lossos, A.; Stumpfig, C.; Stevanin, G.; Gaussen, M.; Zimmerman, B.; Mundwiller, E.; Asulin, M.; Chamma, L.; Sheffer, R.; Misk, A.; et al. Fe/S protein assembly gene IBA57 mutation causes hereditary spastic paraplegia. Neurology 2015, 84, 659–667. [Google Scholar] [CrossRef] [PubMed]
- De Mena, L.; Coto, E.; Sanchez-Ferrero, E.; Ribacoba, R.; Guisasola, L.M.; Salvador, C.; Blazquez, M.; Alvarez, V. Mutational screening of the mortalin gene (HSPA9) in Parkinson’s disease. J. Neural. Transm. (Vienna) 2009, 116, 1289–1293. [Google Scholar] [CrossRef] [PubMed]
- Markley, J.L.; Kim, J.H.; Dai, Z.; Bothe, J.R.; Cai, K.; Frederick, R.O.; Tonelli, M. Metamorphic protein IscU alternates conformations in the course of its role as the scaffold protein for iron-sulfur cluster biosynthesis and delivery. FEBS Lett. 2013, 587, 1172–1179. [Google Scholar] [CrossRef] [PubMed]
- Cai, K.; Frederick, R.O.; Kim, J.H.; Reinen, N.M.; Tonelli, M.; Markley, J.L. Human mitochondrial chaperone (mtHSP70) and cysteine desulfurase (NFS1) bind preferentially to the disordered conformation, whereas co-chaperone (HSC20) binds to the structured conformation of the iron-sulfur cluster scaffold protein (ISCU). J. Biol. Chem. 2013, 288, 28755–28770. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Tonelli, M.; Markley, J.L. Disordered form of the scaffold protein IscU is the substrate for iron-sulfur cluster assembly on cysteine desulfurase. Proc. Nat. Acad. Sci. USA 2012, 109, 454–459. [Google Scholar] [CrossRef] [PubMed]
- Bothe, J.R.; Tonelli, M.; Ali, I.K.; Dai, Z.; Frederick, R.O.; Westler, W.M.; Markley, J.L. The Complex Energy Landscape of the Protein IscU. Biophys. J. 2015, 109, 1019–1025. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Tonelli, M.; Kim, T.; Markley, J.L. Three-Dimensional Structure and Determinants of Stability of the Iron-Sulfur Cluster Scaffold Protein IscU from Escherichia coli. Biochemistry 2012, 51, 5557–5563. [Google Scholar] [CrossRef] [PubMed]
- Ramelot, T.A.; Cort, J.R.; Goldsmith-Fischman, S.; Kornhaber, G.J.; Xiao, R.; Shastry, R.; Acton, T.B.; Honig, B.; Montelione, G.T.; Kennedy, M.A. Solution NMR structure of the iron-sulfur cluster assembly protein U (IscU) with zinc bound at the active site. J. Mol. Biol. 2004, 344, 567–583. [Google Scholar] [CrossRef] [PubMed]
- Iannuzzi, C.; Adrover, M.; Puglisi, R.; Yan, R.; Temussi, P.A.; Pastore, A. The role of zinc in the stability of the marginally stable IscU scaffold protein. Protein Sci. 2014, 23, 1208–1219. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.L.; Barondeau, D.P. Human Frataxin Is an Allosteric Switch That Activates the Fe-S Cluster Biosynthetic Complex. Biochemistry 2010, 49, 9132–9139. [Google Scholar] [CrossRef] [PubMed]
- Fox, N.G.; Martelli, A.; Nabhan, J.F.; Janz, J.; Borkowska, O.; Bulawa, C.; Yue, W.W. Zinc(II) binding on human wild-type ISCU and Met140 variants modulates NFS1 desulfurase activity. Biochimie 2018, 152, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Boniecki, M.T.; Freibert, S.A.; Muhlenhoff, U.; Lill, R.; Cygler, M. Structure and functional dynamics of the mitochondrial Fe/S cluster synthesis complex. Nat. Commun. 2017, 8, 1287. [Google Scholar] [CrossRef] [PubMed]
- Sensi, S.L.; Ton-That, D.; Sullivan, P.G.; Jonas, E.A.; Gee, K.R.; Kaczmarek, L.K.; Weiss, J.H. Modulation of mitochondrial function by endogenous Zn2+ pools. Proc. Nat. Acad. Sci. USA 2003, 100, 6157–6162. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.M.; Kristal, B.S.; Effron, M.S.; Shestopalov, A.I.; Ullucci, P.A.; Sheu, K.F.; Blass, J.P.; Cooper, A.J. Zn2+ inhibits α-ketoglutarate-stimulated mitochondrial respiration and the isolated α-ketoglutarate dehydrogenase complex. J. Biol. Chem. 2000, 275, 13441–13447. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Tonelli, M.; Markley, J.L. Metamorphic protein IscU changes conformation by cis-trans isomerizations of two peptidyl-prolyl peptide bonds. Biochemistry 2012, 51, 9595–9602. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Tonelli, M.; Frederick, R.O.; Chow, D.C.; Markley, J.L. Specialized Hsp70 chaperone (HscA) binds preferentially to the disordered form, whereas J-protein (HscB) binds preferentially to the structured form of the iron-sulfur cluster scaffold protein (IscU). J. Biol. Chem. 2012, 287, 31406–31413. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Kelly, G.; Pastore, A. The scaffold protein IscU retains a structured conformation in the Fe-S cluster assembly complex. ChemBioChem 2014, 15, 1682–1686. [Google Scholar] [CrossRef] [PubMed]
- Muhlenhoff, U.; Balk, J.; Richhardt, N.; Kaiser, J.T.; Sipos, K.; Kispal, G.; Lill, R. Functional characterization of the eukaryotic cysteine desulfurase Nfs1p from Saccharomyces cerevisiae. J. Biol. Chem. 2004, 279, 36906–36915. [Google Scholar] [CrossRef] [PubMed]
- Rocha, A.G.; Knight, S.A.B.; Pandey, A.; Yoon, H.; Pain, J.; Pain, D.; Dancis, A. Cysteine desulfurase is regulated by phosphorylation of Nfs1 in yeast mitochondria. Mitochondrion 2018, 40, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Adam, A.C.; BornhÖvd, C.; Prokisch, H.; Neupert, W.; Hell, K. The Nfs1 interacting protein Isd11 has an essential role in Fe/S cluster biogenesis in mitochondria. EMBO J. 2006, 25, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Ghosh, M.C.; Tong, W.H.; Rouault, T.A. Human ISD11 is essential for both iron-sulfur cluster assembly and maintenance of normal cellular iron homeostasis. Hum. Mol. Genet. 2009, 18, 3014–3025. [Google Scholar] [CrossRef] [PubMed]
- Wiedemann, N.; Urzica, E.; Guiard, B.; Muller, H.; Lohaus, C.; Meyer, H.E.; Ryan, M.T.; Meisinger, C.; Muhlenhoff, U.; Lill, R.; et al. Essential role of Isd11 in mitochondrial iron-sulfur cluster synthesis on Isu scaffold proteins. EMBO J. 2006, 25, 184–195. [Google Scholar] [CrossRef] [PubMed]
- Angerer, H. The superfamily of mitochondrial Complex1_LYR motif-containing (LYRM) proteins. Biochem. Soc. Trans. 2013, 41, 1335–1341. [Google Scholar] [CrossRef] [PubMed]
- Angerer, H. Eukaryotic LYR Proteins Interact with Mitochondrial Protein Complexes. Biology 2015, 4, 133–150. [Google Scholar] [CrossRef] [PubMed]
- Kastaniotis, A.J.; Autio, K.J.; Keratar, J.M.; Monteuuis, G.; Makela, A.M.; Nair, R.R.; Pietikainen, L.P.; Shvetsova, A.; Chen, Z.; Hiltunen, J.K. Mitochondrial fatty acid synthesis, fatty acids and mitochondrial physiology. Biochim. Biophys. Acta 2017, 1862, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Hiltunen, J.K.; Autio, K.J.; Schonauer, M.S.; Kursu, V.A.; Dieckmann, C.L.; Kastaniotis, A.J. Mitochondrial fatty acid synthesis and respiration. Biochim. Biophys. Acta 2010, 1797, 1195–1202. [Google Scholar] [CrossRef] [PubMed]
- Byers, D.M.; Gong, H. Acyl carrier protein: Structure-function relationships in a conserved multifunctional protein family. Biochem. Cell Biol. 2007, 85, 649–662. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.N.; Zhang, Y.M.; Rock, C.O.; Zheng, J.J. Structural modification of acyl carrier protein by butyryl group. Protein Sci. 2009, 18, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Zornetzer, G.A.; White, R.D.; Markley, J.L.; Fox, B.G. Preparation of isotopically labeled spinach acyl-acyl carrier protein for NMR structural studies. Protein Expr. Purif. 2006, 46, 446–455. [Google Scholar] [CrossRef] [PubMed]
- Findlow, S.C.; Winsor, C.; Simpson, T.J.; Crosby, J.; Crump, M.P. Solution structure and dynamics of oxytetracycline polyketide synthase acyl carrier protein from Streptomyces rimosus. Biochemistry 2003, 42, 8423–8433. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.Y.; Tam, A.; Lin, L.; Hixon, J.; Fritz, C.C.; Powers, R. Solution structure of B. subtilis acyl carrier protein. Structure 2001, 9, 277–287. [Google Scholar] [CrossRef]
- Mayo, K.H.; Prestegard, J.H. Acyl carrier protein from Escherichia coli Structural characterization of short-chain acylated acyl carrier proteins by NMR. Biochemistry 1985, 24, 7834–7838. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, C.; Haushalter, R.W.; Lee, D.J.; Markwick, P.R.; Bruegger, J.; Caldara-Festin, G.; Finzel, K.; Jackson, D.R.; Ishikawa, F.; O’Dowd, B.; et al. Trapping the dynamic acyl carrier protein in fatty acid biosynthesis. Nature 2014, 505, 427–431. [Google Scholar] [CrossRef] [PubMed]
- Cronan, J.E. The chain-flipping mechanism of ACP (acyl carrier protein)-dependent enzymes appears universal. Biochem. J. 2014, 460, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Beld, J.; Cang, H.; Burkart, M.D. Visualizing the chain-flipping mechanism in fatty-acid biosynthesis. Angew. Chem. 2014, 53, 14456–14461. [Google Scholar] [CrossRef] [PubMed]
- Herrera, M.G.; Pignataro, M.F.; Noguera, M.E.; Cruz, K.M.; Santos, J. Rescuing the Rescuer: On the Protein Complex between the Human Mitochondrial Acyl Carrier Protein and ISD11. ACS Chem. Biol. 2018, 13, 1455–1462. [Google Scholar] [CrossRef] [PubMed]
- Cai, K.; Frederick, R.O.; Tonelli, M.; Markley, J.L. Mitochondrial cysteine desulfurase and ISD11 coexpressed in Escherichia coli yield complex containing acyl carrier protein. ACS Chem. Biol. 2017, 12, 918–921. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Friemel, M.; Aloisi, C.; Huynen, M.; Taylor, I.A.; Leimkuhler, S.; Pastore, A. The Eukaryotic-Specific ISD11 Is a Complex-Orphan Protein with Ability to Bind the Prokaryotic IscS. PLoS ONE 2016, 11, e0157895. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Kovrigin, E.L.; Eletr, Z. NMR studies of Escherichia coli acyl carrier protein: Dynamic and structural differences of the apo- and holo-forms. Biochem. Biophys. Res. Commun. 2006, 341, 776–783. [Google Scholar] [CrossRef] [PubMed]
- Gully, D.; Moinier, D.; Loiseau, L.; Bouveret, E. New partners of acyl carrier protein detected in Escherichia coli by tandem affinity purification. FEBS Lett. 2003, 548, 90–96. [Google Scholar] [CrossRef]
- Marinoni, E.N.; de Oliveira, J.S.; Nicolet, Y.; Raulfs, E.C.; Amara, P.; Dean, D.R.; Fontecilla-Camps, J.C. (IscS-IscU)2 complex structures provide insights into Fe2S2 biogenesis and transfer. Angew. Chem. Int. Ed. Engl. 2012, 51, 5439–5442. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Proteau, A.; Villarroya, M.; Moukadiri, I.; Zhang, L.; Trempe, J.F.; Matte, A.; Armengod, M.E.; Cygler, M. Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions. PLoS Biol. 2010, 8, e1000354. [Google Scholar] [CrossRef] [PubMed]
- Cupp-Vickery, J.R.; Urbina, H.; Vickery, L.E. Crystal structure of IscS, a cysteine desulfurase from Escherichia coli. J. Mol. Biol. 2003, 330, 1049–1059. [Google Scholar] [CrossRef]
- Van Vranken, J.G.; Jeong, M.Y.; Wei, P.; Chen, Y.C.; Gygi, S.P.; Winge, D.R.; Rutter, J. The mitochondrial acyl carrier protein (ACP) coordinates mitochondrial fatty acid synthesis with iron sulfur cluster biogenesis. Elife 2016, 5, e17828. [Google Scholar] [CrossRef] [PubMed]
- Cory, S.A.; Van Vranken, J.G.; Brignole, E.J.; Patra, S.; Winge, D.R.; Drennan, C.L.; Rutter, J.; Barondeau, D.P. Structure of human Fe-S assembly subcomplex reveals unexpected cysteine desulfurase architecture and acyl-ACP-ISD11 interactions. Proc. Nat. Acad. Sci. USA 2017, 114, E5325–E5334. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Vinothkumar, K.R.; Hirst, J. Structure of mammalian respiratory complex I. Nature 2016, 536, 354–358. [Google Scholar] [CrossRef] [PubMed]
- Fiedorczuk, K.; Letts, J.A.; Degliesposti, G.; Kaszuba, K.; Skehel, M.; Sazanov, L.A. Atomic structure of the entire mammalian mitochondrial complex I. Nature 2016, 538, 406–410. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.; Rathore, S.; Kimanius, D.; Aibara, S.; Bai, X.C.; Rorbach, J.; Amunts, A.; Ramakrishnan, V. Structures of the human mitochondrial ribosome in native states of assembly. Nat. Struct. Mol. Biol. 2017, 24, 866–869. [Google Scholar] [CrossRef] [PubMed]
- Cai, K.; Frederick, R.O.; Dashti, H.; Markley, J.L. Architectural Features of Human Mitochondrial Cysteine Desulfurase Complexes from Crosslinking Mass Spectrometry and Small Angle X-ray Scattering. Structure 2018, 26, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Muhlenhoff, U.; Richhardt, N.; Gerber, J.; Lill, R. Characterization of iron-sulfur protein assembly in isolated mitochondria: A requirement for ATP, NADH, and reduced iron. J. Biol. Chem. 2002, 277, 29810–29816. [Google Scholar] [CrossRef] [PubMed]
- Muhlenhoff, U.; Gerber, J.; Richhardt, N.; Lill, R. Components involved in assembly and dislocation of iron-sulfur clusters on the scaffold protein Isu1p. EMBO J. 2003, 22, 4815–4825. [Google Scholar] [CrossRef] [PubMed]
- Webert, H.; Freibert, S.A.; Gallo, A.; Heidenreich, T.; Linne, U.; Amlacher, S.; Hurt, E.; Muhlenhoff, U.; Banci, L.; Lill, R. Functional reconstitution of mitochondrial Fe/S cluster synthesis on Isu1 reveals the involvement of ferredoxin. Nat. Commun. 2014, 5, 5013. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Adinolfi, S.; Pastore, A. Ferredoxin, in conjunction with NADPH and ferredoxin-NADP reductase, transfers electrons to the IscS/IscU complex to promote iron-sulfur cluster assembly. Biochim. Biophys. Acta 2015, 1854, 1113–1117. [Google Scholar] [CrossRef] [PubMed]
- Ewen, K.M.; Ringle, M.; Bernhardt, R. Adrenodoxin—A versatile ferredoxin. IUBMB Life 2012, 64, 506–512. [Google Scholar] [CrossRef] [PubMed]
- Sheftel, A.D.; Stehling, O.; Pierik, A.J.; Elsasser, H.P.; Muhlenhoff, U.; Webert, H.; Hobler, A.; Hannemann, F.; Bernhardt, R.; Lill, R. Humans possess two mitochondrial ferredoxins, Fdx1 and Fdx2, with distinct roles in steroidogenesis, heme, and Fe/S cluster biosynthesis. Proc. Natl. Acad. Sci. USA 2010, 107, 11775–11780. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Ghosh, M.; Kovtunovych, G.; Crooks, D.R.; Rouault, T.A. Both human ferredoxins 1 and 2 and ferredoxin reductase are important for iron-sulfur cluster biogenesis. Biochim. Biophys. Acta 2012, 1823, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Cai, K.; Tonelli, M.; Frederick, R.O.; Markley, J.L. Human mitochondrial ferredoxin 1 (FDX1) and ferredoxin 2 (FDX2) both bind cysteine desulfurase and donate electrons for iron-sulfur cluster biosynthesis. Biochemistry 2017, 56, 487–499. [Google Scholar] [CrossRef] [PubMed]
- Kurisu, G.; Kusunoki, M.; Katoh, E.; Yamazaki, T.; Teshima, K.; Onda, Y.; Kimata-Ariga, Y.; Hase, T. Structure of the electron transfer complex between ferredoxin and ferredoxin-NADP+ reductase. Nat. Struct. Biol. 2001, 8, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Yoon, T.; Cowan, J.A. Iron-sulfur cluster biosynthesis. Characterization of frataxin as an iron donor for assembly of [2Fe-2S] clusters in ISU-type proteins. J. Am. Chem. Soc. 2003, 125, 6078–6084. [Google Scholar] [CrossRef] [PubMed]
- Kondapalli, K.C.; Kok, N.M.; Dancis, A.; Stemmler, T.L. Drosophila frataxin: An iron chaperone during cellular Fe-S cluster bioassembly. Biochemistry 2008, 47, 6917–6927. [Google Scholar] [CrossRef] [PubMed]
- Melber, A.; Winge, D.R. Steps Toward Understanding Mitochondrial Fe/S Cluster Biogenesis. Methods Enzymol. 2018, 599, 265–292. [Google Scholar] [PubMed]
- Lindahl, P.A.; Moore, M.J. Labile Low-Molecular-Mass Metal Complexes in Mitochondria: Trials and Tribulations of a Burgeoning Field. Biochemistry 2016, 55, 4140–4153. [Google Scholar] [CrossRef] [PubMed]
- Anzovino, A.; Lane, D.J.; Huang, M.L.; Richardson, D.R. Fixing frataxin: ‘Ironing out’ the metabolic defect in Friedreich’s ataxia. Br. J. Pharmacol. 2014, 171, 2174–2190. [Google Scholar] [CrossRef] [PubMed]
- Vaubel, R.A.; Isaya, G. Iron-sulfur cluster synthesis, iron homeostasis and oxidative stress in Friedreich ataxia. Mol. Cell. Neurosci. 2013, 55, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Nair, M.; Adinolfi, S.; Pastore, C.; Kelly, G.; Temussi, P.; Pastore, A. Solution structure of the bacterial frataxin ortholog, CyaY: Mapping the iron binding sites. Structure 2004, 12, 2037–2048. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Alam, S.L.; Proteasa, S.V.; Zhang, Y.; Lesuisse, E.; Dancis, A.; Stemmler, T.L. Yeast frataxin solution structure, iron binding, and ferrochelatase interaction. Biochemistry 2004, 43, 16254–16262. [Google Scholar] [CrossRef] [PubMed]
- Musco, G.; Stier, G.; Kolmerer, B.; Adinolfi, S.; Martin, S.; Frenkiel, T.; Gibson, T.; Pastore, A. Towards a structural understanding of Friedreich’s ataxia: The solution structure of frataxin. Structure 2000, 8, 695–707. [Google Scholar] [CrossRef]
- Parent, A.; Elduque, X.; Cornu, D.; Belot, L.; Le Caer, J.P.; Grandas, A.; Toledano, M.B.; D’Autreaux, B. Mammalian frataxin directly enhances sulfur transfer of NFS1 persulfide to both ISCU and free thiols. Nat. Commun. 2015, 6, 5686. [Google Scholar] [CrossRef] [PubMed]
- Bridwell-Rabb, J.; Fox, N.G.; Tsai, C.L.; Winn, A.M.; Barondeau, D.P. Human frataxin activates Fe-S cluster biosynthesis by facilitating sulfur transfer chemistry. Biochemistry 2014, 53, 4904–4913. [Google Scholar] [CrossRef] [PubMed]
- Bridwell-Rabb, J.; Iannuzzi, C.; Pastore, A.; Barondeau, D.P. Effector role reversal during evolution: The case of frataxin in Fe-S cluster biosynthesis. Biochemistry 2012, 51, 2506–2514. [Google Scholar] [CrossRef] [PubMed]
- Cook, J.D.; Bencze, K.Z.; Jankovic, A.D.; Crater, A.K.; Busch, C.N.; Bradley, P.B.; Stemmler, A.J.; Spaller, M.R.; Stemmler, T.L. Monomeric yeast frataxin is an iron-binding protein. Biochemistry 2006, 45, 7767–7777. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.; Knight, S.A.B.; Pandey, A.; Pain, J.; Turkarslan, S.; Pain, D.; Dancis, A. Turning Saccharomyces cerevisiae into a Frataxin-Independent Organism. PLoS Genet. 2015, 11, e1005135. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.; Knight, S.A.; Pandey, A.; Pain, J.; Zhang, Y.; Pain, D.; Dancis, A. Frataxin-bypassing Isu1: Characterization of the bypass activity in cells and mitochondria. Biochem. J. 2014, 459, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.; Golla, R.; Lesuisse, E.; Pain, J.; Donald, J.E.; Lyver, E.R.; Pain, D.; Dancis, A. Mutation in the Fe-S scaffold protein Isu bypasses frataxin deletion. Biochem. J. 2012, 441, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Colin, F.; Martelli, A.; Clemancey, M.; Latour, J.M.; Gambarelli, S.; Zeppieri, L.; Birck, C.; Page, A.; Puccio, H.; Ollagnier de Choudens, S. Mammalian frataxin controls sulfur production and iron entry during de novo Fe4S4 cluster assembly. J. Am. Chem. Soc. 2013, 135, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Cai, K.; Frederick, R.O.; Tonelli, M.; Markley, J.L. Interactions of iron-bound frataxin with ISCU and ferredoxin on the cysteine desulfurase complex leading to Fe-S cluster assembly. J. Inorg. Biochem. 2018, 183, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Cai, K.; Frederick, R.O.; Tonelli, M.; Markley, J.L. ISCU(M108I) and ISCU(D39V) Differ from Wild-Type ISCU in Their Failure to Form Cysteine Desulfurase Complexes Containing Both Frataxin and Ferredoxin. Biochemistry 2018, 57, 1491–1500. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Konarev, P.V.; Iannuzzi, C.; Adinolfi, S.; Roche, B.; Kelly, G.; Simon, L.; Martin, S.R.; Py, B.; Barras, F.; et al. Ferredoxin competes with bacterial frataxin in binding to the desulfurase IscS. J. Biol. Chem. 2013, 288, 24777–24787. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Frederick, R.O.; Reinen, N.M.; Troupis, A.T.; Markley, J.L. [2Fe-2S]-Ferredoxin binds directly to cysteine desulfurase and supplies an electron for iron-sulfur cluster assembly but is displaced by the scaffold protein or bacterial frataxin. J. Am. Chem. Soc. 2013, 135, 8117–8120. [Google Scholar] [CrossRef] [PubMed]
- Pastore, C.; Adinolfi, S.; Huynen, M.A.; Rybin, V.; Martin, S.; Mayer, M.; Bukau, B.; Pastore, A. YfhJ, a molecular adaptor in iron-sulfur cluster formation or a frataxin-like protein? Structure 2006, 14, 857–867. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Bothe, J.R.; Frederick, R.O.; Holder, J.C.; Markley, J.L. Role of IscX in iron-sulfur cluster biogenesis in Escherichia coli. J. Am. Chem. Soc. 2014, 136, 7933–7942. [Google Scholar] [CrossRef] [PubMed]
- Adinolfi, S.; Puglisi, R.; Crack, J.C.; Iannuzzi, C.; Dal Piaz, F.; Konarev, P.V.; Svergun, D.I.; Martin, S.; Le Brun, N.E.; Pastore, A. The Molecular Bases of the Dual Regulation of Bacterial Iron Sulfur Cluster Biogenesis by CyaY and IscX. Front. Mol. Biosci. 2018, 4, 97. [Google Scholar] [CrossRef] [PubMed]
- De Vries, S.J.; van Dijk, M.; Bonvin, A.M. The HADDOCK web server for data-driven biomolecular docking. Nat. Protoc. 2010, 5, 883–897. [Google Scholar] [CrossRef] [PubMed]
- Manicki, M.; Majewska, J.; Ciesielski, S.; Schilke, B.; Blenska, A.; Kominek, J.; Marszalek, J.; Craig, E.A.; Dutkiewicz, R. Overlapping binding sites of the frataxin homologue assembly factor and the heat shock protein 70 transfer factor on the Isu iron-sulfur cluster scaffold protein. J. Biol. Chem. 2014, 289, 30268–30278. [Google Scholar] [CrossRef] [PubMed]
- Kato, S.; Mihara, H.; Kurihara, T.; Takahashi, Y.; Tokumoto, U.; Yoshimura, T.; Esaki, N. Cys-328 of IscS and Cys-63 of IscU are the sites of disulfide bridge formation in a covalently bound IscS/IscU complex: Implications for the mechanism of iron-sulfur cluster assembly. Proc. Natl. Acad. Sci. USA 2002, 99, 5948–5952. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.D.; Frazzon, J.; Dean, D.R.; Johnson, M.K. Role of conserved cysteines in mediating sulfur transfer from IscS to IscU. FEBS Lett. 2005, 579, 5236–5240. [Google Scholar] [CrossRef] [PubMed]
- Vickery, L.E.; Cupp-Vickery, J.R. Molecular chaperones HscA/Ssq1 and HscB/Jac1 and their roles in iron-sulfur protein maturation. Crit. Rev. Biochem. Mol. Biol. 2007, 42, 95–111. [Google Scholar] [CrossRef] [PubMed]
- Rouault, T.A. Mammalian iron-sulphur proteins: Novel insights into biogenesis and function. Nat. Rev. Mol. Cell Biol. 2015, 16, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Londono, C.; Osorio, C.; Gama, V.; Alzate, O. Mortalin, apoptosis, and neurodegeneration. Biomolecules 2012, 2, 143–164. [Google Scholar] [CrossRef] [PubMed]
- Kaul, S.C.; Deocaris, C.C.; Wadhwa, R. Three faces of mortalin: A housekeeper, guardian and killer. Exp. Gerontol. 2007, 42, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Craig, E.A.; Marszalek, J. How Do J-Proteins Get Hsp70 to Do So Many Different Things? Trends Biochem. Sci. 2017, 42, 355–368. [Google Scholar] [CrossRef] [PubMed]
- Kampinga, H.H.; Craig, E.A. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 2010, 11, 579–592. [Google Scholar] [CrossRef] [PubMed]
- Majewska, J.; Ciesielski, S.J.; Schilke, B.; Kominek, J.; Blenska, A.; Delewski, W.; Song, J.Y.; Marszalek, J.; Craig, E.A.; Dutkiewicz, R. Binding of the chaperone Jac1 protein and cysteine desulfurase Nfs1 to the iron-sulfur cluster scaffold Isu protein is mutually exclusive. J. Biol. Chem. 2013, 288, 29134–29142. [Google Scholar] [CrossRef] [PubMed]
- Maio, N.; Kim, K.S.; Singh, A.; Rouault, T.A. A Single Adaptable Cochaperone-Scaffold Complex Delivers Nascent Iron-Sulfur Clusters to Mammalian Respiratory Chain Complexes I-III. Cell Metab. 2017, 25, 945–953. [Google Scholar] [CrossRef] [PubMed]
- Maio, N.; Singh, A.; Uhrigshardt, H.; Saxena, N.; Tong, W.H.; Rouault, T.A. Cochaperone binding to LYR motifs confers specificity of iron sulfur cluster delivery. Cell Metab. 2014, 19, 445–457. [Google Scholar] [CrossRef] [PubMed]
- Hoff, K.G.; Cupp-Vickery, J.R.; Vickery, L.E. Contributions of the LPPVK motif of the iron-sulfur template protein IscU to interactions with the Hsc66-Hsc20 chaperone system. J. Biol. Chem. 2003, 278, 37582–37589. [Google Scholar] [CrossRef] [PubMed]
- Dutkiewicz, R.; Nowak, M.; Craig, E.A.; Marszalek, J. Fe-S Cluster Hsp70 Chaperones: The ATPase Cycle and Protein Interactions. Methods Enzymol. 2017, 595, 161–184. [Google Scholar] [PubMed]
- Miao, B.J.; Davis, J.E.; Craig, E.A. Mge1 functions as a nucleotide release factor for Ssc1, a mitochondrial Hsp70 of Saccharomyces cerevisiae. J. Mol. Biol. 1997, 265, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Shakamuri, P.; Zhang, B.; Johnson, M.K. Monothiol glutaredoxins function in storing and transporting [Fe2S2] clusters assembled on IscU scaffold proteins. J. Am. Chem. Soc. 2012, 134, 15213–15216. [Google Scholar] [CrossRef] [PubMed]
- Johansson, C.; Roos, A.K.; Montano, S.J.; Sengupta, R.; Filippakopoulos, P.; Guo, K.; von Delft, F.; Holmgren, A.; Oppermann, U.; Kavanagh, K.L. The crystal structure of human GLRX5: Iron-sulfur cluster co-ordination, tetrameric assembly and monomer activity. Biochem. J. 2011, 433, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Banci, L.; Brancaccio, D.; Ciofi-Baffoni, S.; Del Conte, R.; Gadepalli, R.; Mikolajczyk, M.; Neri, S.; Piccioli, M.; Winkelmann, J. [2Fe-2S] cluster transfer in iron-sulfur protein biogenesis. Proc. Natl. Acad. Sci. USA 2014, 111, 6203–6208. [Google Scholar] [CrossRef] [PubMed]
- Mapolelo, D.T.; Zhang, B.; Randeniya, S.; Albetel, A.N.; Li, H.; Couturier, J.; Outten, C.E.; Rouhier, N.; Johnson, M.K. Monothiol glutaredoxins and A-type proteins: Partners in Fe-S cluster trafficking. Dalton Trans. 2013, 42, 3107–3115. [Google Scholar] [CrossRef] [PubMed]
- Maio, N.; Rouault, T.A. Mammalian Fe-S proteins: Definition of a consensus motif recognized by the co-chaperone HSC20. Metallomics 2016, 8, 1032–1046. [Google Scholar] [CrossRef] [PubMed]
- Agar, J.N.; Krebs, C.; Frazzon, J.; Huynh, B.H.; Dean, D.R.; Johnson, M.K. IscU as a scaffold for iron-sulfur cluster biosynthesis: Sequential assembly of [2Fe-2S] and [4Fe-4S] clusters in IscU. Biochemistry 2000, 39, 7856–7862. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.D.; Jameson, G.N.; Dos Santos, P.C.; Agar, J.N.; Naik, S.; Krebs, C.; Frazzon, J.; Dean, D.R.; Huynh, B.H.; Johnson, M.K. NifS-mediated assembly of [4Fe-4S] clusters in the N- and C-terminal domains of the NifU scaffold protein. Biochemistry 2005, 44, 12955–12969. [Google Scholar] [CrossRef] [PubMed]
- Unciuleac, M.C.; Chandramouli, K.; Naik, S.; Mayer, S.; Huynh, B.H.; Johnson, M.K.; Dean, D.R. In vitro activation of apo-aconitase using a [4Fe-4S] cluster-loaded form of the IscU [Fe-S] cluster scaffolding protein. Biochemistry 2007, 46, 6812–6821. [Google Scholar] [CrossRef] [PubMed]
- Ajit Bolar, N.; Vanlander, A.V.; Wilbrecht, C.; Van der Aa, N.; Smet, J.; De Paepe, B.; Vandeweyer, G.; Kooy, F.; Eyskens, F.; De Latter, E.; et al. Mutation of the iron-sulfur cluster assembly gene IBA57 causes severe myopathy and encephalopathy. Hum. Mol. Genet. 2013, 22, 2590–2602. [Google Scholar] [CrossRef] [PubMed]
- Brancaccio, D.; Gallo, A.; Mikolajczyk, M.; Zovo, K.; Palumaa, P.; Novellino, E.; Piccioli, M.; Ciofi-Baffoni, S.; Banci, L. Formation of [4Fe-4S] clusters in the mitochondrial iron-sulfur cluster assembly machinery. J. Am. Chem. Soc. 2014, 136, 16240–16250. [Google Scholar] [CrossRef] [PubMed]
- Brancaccio, D.; Gallo, A.; Piccioli, M.; Novellino, E.; Ciofi-Baffoni, S.; Banci, L. [4Fe-4S] Cluster Assembly in Mitochondria and Its Impairment by Copper. J. Am. Chem. Soc. 2017, 139, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Sheftel, A.D.; Stehling, O.; Pierik, A.J.; Netz, D.J.A.; Kerscher, S.; Elsasser, H.P.; Wittig, I.; Balk, J.; Brandt, U.; Lill, R. Human Ind1, an Iron-Sulfur Cluster Assembly Factor for Respiratory Complex I. Mol. Cell. Biol. 2009, 29, 6059–6073. [Google Scholar] [CrossRef] [PubMed]
- Bych, K.; Kerscher, S.; Netz, D.J.A.; Pierik, A.J.; Zwicker, K.; Huynen, M.A.; Lill, R.; Brandt, U.; Balk, J. The iron-sulphur protein Ind1 is required for effective complex I assembly. EMBO J. 2008, 27, 1736–1746. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, S.; Naik, S.G.; O′Carroll, I.P.; Huynh, B.H.; Dean, D.R.; Johnson, M.K.; Dos Santos, P.C. A proposed role for the Azotobacter vinelandii NfuA protein as an intermediate iron-sulfur cluster carrier. J. Biol. Chem. 2008, 283, 14092–14099. [Google Scholar] [CrossRef] [PubMed]
- Angelini, S.; Gerez, C.; Ollagnier-de Choudens, S.; Sanakis, Y.; Fontecave, M.; Barras, F.; Py, B. NfuA, a new factor required for maturing Fe/S proteins in Escherichia coli under oxidative stress and iron starvation conditions. J. Biol. Chem. 2008, 283, 14084–14091. [Google Scholar] [CrossRef] [PubMed]
- Tong, W.H.; Jameson, G.N.; Huynh, B.H.; Rouault, T.A. Subcellular compartmentalization of human Nfu, an iron-sulfur cluster scaffold protein, and its ability to assemble a [4Fe-4S] cluster. Proc. Nat. Acad. Sci. USA 2003, 100, 9762–9767. [Google Scholar] [CrossRef] [PubMed]
- Ahting, U.; Mayr, J.A.; Vanlander, A.V.; Hardy, S.A.; Santra, S.; Makowski, C.; Alston, C.L.; Zimmermann, F.A.; Abela, L.; Plecko, B.; et al. Clinical, biochemical, and genetic spectrum of seven patients with NFU1 deficiency. Front. Genet. 2015, 6, 123. [Google Scholar] [CrossRef] [PubMed]
- Cai, K.; Liu, G.; Frederick, R.O.; Xiao, R.; Montelione, G.T.; Markley, J.L. Structural/Functional Properties of Human NFU1, an Intermediate [4Fe-4S] Carrier in Human Mitochondrial Iron-Sulfur Cluster Biogenesis. Structure 2016, 24, 2080–2091. [Google Scholar] [CrossRef] [PubMed]
- Yabe, T.; Yamashita, E.; Kikuchi, A.; Morimoto, K.; Nakagawa, A.; Tsukihara, T.; Nakai, M. Structural analysis of Arabidopsis CnfU protein: An iron-sulfur cluster biosynthetic scaffold in chloroplasts. J. Mol. Biol. 2008, 381, 160–173. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Subramanian, S.; Couturier, J.; Naik, S.G.; Kim, S.K.; Leustek, T.; Knaff, D.B.; Wu, H.C.; Vignols, F.; Huynh, B.H.; et al. Arabidopsis thaliana Nfu2 accommodates [2Fe-2S] or [4Fe-4S] clusters and is competent for in vitro maturation of chloroplast [2Fe-2S] and [4Fe-4S] cluster-containing proteins. Biochemistry 2013, 52, 6633–6645. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, E.L.; Booker, S.J. Destruction and reformation of an iron-sulfur cluster during catalysis by lipoyl synthase. Science 2017, 358, 373–377. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Outten, C.E. Monothiol CGFS glutaredoxins and BolA-like proteins: [2Fe-2S] Binding partners in iron homeostasis. Biochemistry 2012, 51, 4377–4389. [Google Scholar] [CrossRef] [PubMed]
- Willems, P.; Wanschers, B.F.J.; Esseling, J.; Szklarczyk, R.; Kudla, U.; Duarte, I.; Forkink, M.; Nooteboom, M.; Swarts, H.; Gloerich, J.; et al. BOLA1 Is an Aerobic Protein That Prevents Mitochondrial Morphology Changes Induced by Glutathione Depletion. Antioxid. Redox Signal. 2013, 18, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Uzarska, M.A.; Nasta, V.; Weiler, B.D.; Spantgar, F.; Ciofi-Baffoni, S.; Saviello, M.R.; Gonnelli, L.; Muhlenhoff, U.; Banci, L.; Lill, R. Mitochondrial Bol1 and Bol3 function as assembly factors for specific iron-sulfur proteins. Elife 2016, 5, e16673. [Google Scholar] [CrossRef] [PubMed]
- Roret, T.; Tsan, P.; Couturier, J.; Zhang, B.; Johnson, M.K.; Rouhier, N.; Didierjean, C. Structural and spectroscopic insights into BolA-glutaredoxin complexes. J. Biol. Chem. 2014, 289, 24588–24598. [Google Scholar] [CrossRef] [PubMed]
- Kasai, T.; Inoue, M.; Koshiba, S.; Yabuki, T.; Aoki, M.; Nunokawa, E.; Seki, E.; Matsuda, T.; Matsuda, N.; Tomo, Y.; et al. Solution structure of a BolA-like protein from Mus musculus. Protein Sci. 2004, 13, 545–548. [Google Scholar] [CrossRef] [PubMed]
- Nasta, V.; Giachetti, A.; Ciofi-Baffoni, S.; Banci, L. Structural insights into the molecular function of human [2Fe-2S] BOLA1-GRX5 and [2Fe-2S] BOLA3-GRX5 complexes. Biochim. Biophys. Acta 2017, 1861, 2119–2131. [Google Scholar] [CrossRef] [PubMed]
- Muhlenhoff, U.; Molik, S.; Godoy, J.R.; Uzarska, M.A.; Richter, N.; Seubert, A.; Zhang, Y.; Stubbe, J.; Pierrel, F.; Herrero, E.; et al. Cytosolic monothiol glutaredoxins function in intracellular iron sensing and trafficking via their bound iron-sulfur cluster. Cell Metab. 2010, 12, 373–385. [Google Scholar] [CrossRef] [PubMed]
- Paul, V.D.; Lill, R. Biogenesis of cytosolic and nuclear iron-sulfur proteins and their role in genome stability. Biochim. Biophys. Acta 2015, 1853, 1528–1539. [Google Scholar] [CrossRef] [PubMed]
- Lill, R.; Srinivasan, V.; Muhlenhoff, U. The role of mitochondria in cytosolic-nuclear iron-sulfur protein biogenesis and in cellular iron regulation. Curr. Opin. Microbiol. 2014, 22, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.K.; Pallesen, L.J.; Spang, R.J.; Walden, W.E. Cytosolic iron-sulfur cluster assembly (CIA) system: Factors, mechanism, and relevance to cellular iron regulation. J. Biol. Chem. 2010, 285, 26745–26751. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.; Pain, J.; Dziuba, N.; Pandey, A.K.; Dancis, A.; Lindahl, P.A.; Pain, D. Mitochondria Export Sulfur Species Required for Cytosolic tRNA Thiolation. Cell Chem. Biol. 2018, 25, 738–748. [Google Scholar] [CrossRef] [PubMed]
- Stehling, O.; Netz, D.J.; Niggemeyer, B.; Rosser, R.; Eisenstein, R.S.; Puccio, H.; Pierik, A.J.; Lill, R. Human Nbp35 is essential for both cytosolic iron-sulfur protein assembly and iron homeostasis. Mol. Cell. Biol. 2008, 28, 5517–5528. [Google Scholar] [CrossRef] [PubMed]
- Netz, D.J.; Pierik, A.J.; Stumpfig, M.; Muhlenhoff, U.; Lill, R. The Cfd1-Nbp35 complex acts as a scaffold for iron-sulfur protein assembly in the yeast cytosol. Nat. Chem. Biol. 2007, 3, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Lee, F.S. A role for IOP1 in mammalian cytosolic iron-sulfur protein biogenesis. J. Biol. Chem. 2008, 283, 9231–9238. [Google Scholar] [CrossRef] [PubMed]
- Balk, J.; Netz, D.J.A.; Tepper, K.; Pierik, A.J.; Lill, R. The essential WD40 protein Cia1 is involved in a late step of cytosolic and nuclear iron-sulfur protein assembly. Mol. Cell. Biol. 2005, 25, 10833–10841. [Google Scholar] [CrossRef] [PubMed]
- Stehling, O.; Mascarenhas, J.; Vashisht, A.A.; Sheftel, A.D.; Niggemeyer, B.; Rosser, R.; Pierik, A.J.; Wohlschlegel, J.A.; Lill, R. Human CIA2A-FAM96A and CIA2B-FAM96B integrate iron homeostasis and maturation of different subsets of cytosolic-nuclear iron-sulfur proteins. Cell Metab. 2013, 18, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Seki, M.; Takeda, Y.; Iwai, K.; Tanaka, K. IOP1 Protein Is an External Component of the Human Cytosolic Iron-Sulfur Cluster Assembly (CIA) Machinery and Functions in the MMS19 Protein-dependent CIA Pathway. J. Biol. Chem. 2013, 288, 16680–16689. [Google Scholar] [CrossRef] [PubMed]
- Stehling, O.; Vashisht, A.A.; Mascarenhas, J.; Jonsson, Z.O.; Sharma, T.; Netz, D.J.; Pierik, A.J.; Wohlschlegel, J.A.; Lill, R. MMS19 assembles iron-sulfur proteins required for DNA metabolism and genomic integrity. Science 2012, 337, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, B.J.; Wang, L.; Wan, S.; Luo, Y.; Wang, L.; Lin, J.; Xia, B. Solution structure of monomeric human FAM96A. J. Biomol. NMR 2013, 56, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.E.; Richards, A.A.; Ariffin, J.K.; Ross, I.L.; Sweet, M.J.; Kellie, S.; Kobe, B.; Martin, J.L. The mammalian DUF59 protein Fam96a forms two distinct types of domain-swapped dimer. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Maione, V.; Cantini, F.; Severi, M.; Banci, L. Investigating the role of the human CIA2A-CIAO1 complex in the maturation of aconitase. Biochim. Biophys. Acta 2018, 1862, 1980–1987. [Google Scholar] [CrossRef] [PubMed]
- Paul, V.D.; Muhlenhoff, U.; Stumpfig, M.; Seebacher, J.; Kugler, K.G.; Renicke, C.; Taxis, C.; Gavin, A.C.; Pierik, A.J.; Lill, R. The deca-GX3 proteins Yae1-Lto1 function as adaptors recruiting the ABC protein Rli1 for iron-sulfur cluster insertion. Elife 2015, 4, e08231. [Google Scholar] [CrossRef] [PubMed]
- Gari, K.; Leon Ortiz, A.M.; Borel, V.; Flynn, H.; Skehel, J.M.; Boulton, S.J. MMS19 links cytoplasmic iron-sulfur cluster assembly to DNA metabolism. Science 2012, 337, 243–245. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.S.; Maio, N.; Singh, A.; Rouault, T.A. Cytosolic HSC20 integrates de novo iron-sulfur cluster biogenesis with the CIAO1-mediated transfer to recipients. Hum. Mol. Genet. 2018, 27, 837–852. [Google Scholar] [CrossRef] [PubMed]
- Banci, L.; Bertini, I.; Calderone, V.; Ciofi-Baffoni, S.; Giachetti, A.; Jaiswal, D.; Mikolajczyk, M.; Piccioli, M.; Winkelmann, J. Molecular view of an electron transfer process essential for iron-sulfur protein biogenesis. Proc. Natl. Acad. Sci. USA 2013, 110, 7136–7141. [Google Scholar] [CrossRef] [PubMed]
- Netz, D.J.; Stumpfig, M.; Dore, C.; Muhlenhoff, U.; Pierik, A.J.; Lill, R. Tah18 transfers electrons to Dre2 in cytosolic iron-sulfur protein biogenesis. Nat. Chem. Biol. 2010, 6, 758–765. [Google Scholar] [CrossRef] [PubMed]
- Frey, A.G.; Palenchar, D.J.; Wildemann, J.D.; Philpott, C.C. A Glutaredoxin center dot BolA Complex Serves as an Iron-Sulfur Cluster Chaperone for the Cytosolic Cluster Assembly Machinery. J. Biol. Chem. 2016, 291, 22344–22356. [Google Scholar] [CrossRef] [PubMed]
- Ojeda, L.; Keller, G.; Muhlenhoff, U.; Rutherford, J.C.; Lill, R.; Winge, D.R. Role of glutaredoxin-3 and glutaredoxin-4 in the iron regulation of the Aft1 transcriptional activator in Saccharomyces cerevisiae. J. Biol. Chem. 2006, 281, 17661–17669. [Google Scholar] [CrossRef] [PubMed]
- Banci, L.; Camponeschi, F.; Ciofi-Baffoni, S.; Muzzioli, R. Elucidating the Molecular Function of Human BOLA2 in GRX3-Dependent Anamorsin Maturation Pathway. J. Am. Chem. Soc. 2015, 137, 16133–16143. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Lee, F.S. Mouse knock-out of IOP1 protein reveals its essential role in mammalian cytosolic iron-sulfur protein biogenesis. J. Biol. Chem. 2011, 286, 15797–15805. [Google Scholar] [CrossRef] [PubMed]
- Banci, L.; Bertini, I.; Ciofi-Baffoni, S.; Boscaro, F.; Chatzi, A.; Mikolajczyk, M.; Tokatlidis, K.; Winkelmann, J. Anamorsin Is a [2Fe-2S] Cluster-Containing Substrate of the Mia40-Dependent Mitochondrial Protein Trapping Machinery. Chem. Biol. 2011, 18, 794–804. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, C.; Dancis, A.; Nakamaru-Ogiso, E. EPR studies of wild type and mutant Dre2 identify essential [2Fe-2S] and [4Fe-4S] clusters and their cysteine ligands. J. Biochem. 2017, 161, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Banci, L.; Ciofi-Baffoni, S.; Mikolajczyk, M.; Winkelmann, J.; Bill, E.; Pandelia, M.E. Human anamorsin binds [2Fe-2S] clusters with unique electronic properties. J. Biol. Inorg. Chem. 2013, 18, 883–893. [Google Scholar] [CrossRef] [PubMed]
- Outten, C.E.; Albetel, A.N. Iron sensing and regulation in Saccharomyces cerevisiae: Ironing out the mechanistic details. Curr. Opin. Microbiol. 2013, 16, 662–668. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Mapolelo, D.T.; Randeniya, S.; Johnson, M.K.; Outten, C.E. Human glutaredoxin 3 forms [2Fe-2S] -bridged complexes with human BolA2. Biochemistry 2012, 51, 1687–1696. [Google Scholar] [CrossRef] [PubMed]
- Banci, L.; Ciofi-Baffoni, S.; Gajda, K.; Muzzioli, R.; Peruzzini, R.; Winkelmann, J. N-terminal domains mediate [2Fe-2S] cluster transfer from glutaredoxin-3 to anamorsin. Nat. Chem. Biol. 2015, 11, 772–778. [Google Scholar] [CrossRef] [PubMed]
- Tamir, S.; Paddock, M.L.; Darash-Yahana-Baram, M.; Holt, S.H.; Sohn, Y.S.; Agranat, L.; Michaeli, D.; Stofleth, J.T.; Lipper, C.H.; Morcos, F.; et al. Structure-function analysis of NEET proteins uncovers their role as key regulators of iron and ROS homeostasis in health and disease. Biochim. Biophys. 2015, 1853, 1294–1315. [Google Scholar] [CrossRef] [PubMed]
- Geldenhuys, W.J.; Leeper, T.C.; Carroll, R.T. MitoNEET as a novel drug target for mitochondrial dysfunction. Drug Discov. Today 2014, 19, 1601–1606. [Google Scholar] [CrossRef] [PubMed]
- Bai, F.; Morcos, F.; Sohn, Y.S.; Darash-Yahana, M.; Rezende, C.O.; Lipper, C.H.; Paddock, M.L.; Song, L.H.; Luo, Y.T.; Holt, S.H.; et al. The Fe-S cluster-containing NEET proteins mitoNEET and NAF-1 as chemotherapeutic targets in breast cancer. Proc. Nat. Acad. Sci. USA 2015, 112, 3698–3703. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Zhou, T.; Ye, K.; Wang, J. Crystal structure of human mitoNEET reveals distinct groups of iron sulfur proteins. Proc. Nat. Acad. Sci. USA 2007, 104, 14640–14645. [Google Scholar] [CrossRef] [PubMed]
- Ferecatu, I.; Goncalves, S.; Golinelli-Cohen, M.P.; Clemancey, M.; Martelli, A.; Riquier, S.; Guittet, E.; Latour, J.M.; Puccio, H.; Drapier, J.C.; et al. The diabetes drug target MitoNEET governs a novel trafficking pathway to rebuild an Fe-S cluster into cytosolic aconitase/iron regulatory protein 1. J. Biol. Chem. 2014, 289, 28070–28086. [Google Scholar] [CrossRef] [PubMed]
- Golinelli-Cohen, M.P.; Lescop, E.; Mons, C.; Goncalves, S.; Clemancey, M.; Santolini, J.; Guittet, E.; Blondin, G.; Latour, J.M.; Bouton, C. Redox Control of the Human Iron-Sulfur Repair Protein MitoNEET Activity via Its Iron-Sulfur Cluster. J. Biol. Chem. 2016, 291, 7583–7593. [Google Scholar] [CrossRef] [PubMed]
- Andreini, C.; Banci, L.; Rosato, A. Exploiting Bacterial Operons to Illuminate Human Iron-Sulfur Proteins. J. Proteome Res. 2016, 15, 1308–1322. [Google Scholar] [CrossRef] [PubMed]
- Camponeschi, F.; Ciofi-Baffoni, S.; Banci, L. Anamorsin/Ndor1 Complex Reduces [2Fe-2S]-MitoNEET via a Transient Protein-Protein Interaction. J. Am. Chem. Soc. 2017, 139, 9479–9482. [Google Scholar] [CrossRef] [PubMed]
- Luchinat, E.; Banci, L. In-Cell NMR in Human Cells: Direct Protein Expression Allows Structural Studies of Protein Folding and Maturation. Acc. Chem. Res. 2018, 51, 1550–1557. [Google Scholar] [CrossRef] [PubMed]
- Luchinat, E.; Banci, L. A Unique Tool for Cellular Structural Biology: In-cell NMR. J. Biol. Chem. 2016, 291, 3776–3784. [Google Scholar] [CrossRef] [PubMed]
- Freedberg, D.I.; Selenko, P. Live cell NMR. Annu. Rev. Biophys. 2014, 43, 171–192. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, A.Y.; Burz, D.S.; Shekhtman, A. In-cell NMR spectroscopy. Prog. Nucl. Magn. Reson. Spectrosc. 2011, 59, 197–212. [Google Scholar] [CrossRef] [PubMed]
- Sakakibara, D.; Sasaki, A.; Ikeya, T.; Hamatsu, J.; Hanashima, T.; Mishima, M.; Yoshimasu, M.; Hayashi, N.; Mikawa, T.; Walchli, M.; et al. Protein structure determination in living cells by in-cell NMR spectroscopy. Nature 2009, 458, 102–105. [Google Scholar] [CrossRef] [PubMed]
- Inomata, K.; Ohno, A.; Tochio, H.; Isogai, S.; Tenno, T.; Nakase, I.; Takeuchi, T.; Futaki, S.; Ito, Y.; Hiroaki, H.; et al. High-resolution multi-dimensional NMR spectroscopy of proteins in human cells. Nature 2009, 458, 106–109. [Google Scholar] [CrossRef] [PubMed]

| Human Protein | UNIPROT ID | Yeast Ortholog | Diseases | Cofactors | Putative Functions |
|---|---|---|---|---|---|
| FXN | Q16595 | Yfh1 | Friedreich’s ataxia (FRDA) [64,65] | Fe2+ | Proximal iron donor, controls iron entry and sulfur transfer to ISCU |
| ISCU | Q9H1K1 | Isu1 and Isu2 | ISCU myopathy [66,67,68] | [2Fe-2S] | Scaffold for Fe-S cluster assembly |
| FDX1 | P10109 | [2Fe-2S] | Electron donor in cluster assembly | ||
| FDX2 (FDX1L) | Q6P4F2 | Yah1 | Myopathy [69] | [2Fe-2S] | Electron donor in cluster assembly |
| NFS1 | Q8WV90 | Nfs1 | Infantile mitochondrial complex II/III deficiency [70] | PLP | Cysteine desulfurase |
| ISD11 (LYRM4) | Q9HD34 | Isd11 | Respiratory chain deficiency [71] | Stabilization of NFS1 in cluster assembly | |
| ACP (NDUFAB1) | O14561 | Acp1 | 4′-phospho-pantetheine | Stabilization of NFS1-ISD11 complex in cluster assembly | |
| GLRX5 | Q86SX6 | Grx5 | Microcytic anemia and sideroblastic anemia [72,73] | [2Fe-2S] | Fe-S cluster carrier protein |
| ABCB7 | O75027 | Atm1 | Sideroblastic anemia and ataxia [74] | Mitochondrial export | |
| Mitoferrin1 (MFRN1) | Q9NYZ2 | Mrs3 and Mrs4 | Variant erythropoietic protoporphyria [75] | Mitochondrial iron importer | |
| NUBPL | Q8TB37 | Ind1 | Mitochondrial encephalomyopathy [76] | [4Fe-4S] | Involved in Fe-S cluster transfer to Complex I |
| NFU1 | Q9UMS0 | Nfu1 | Multiple mitochondrial dysfunction syndrome 1 (MMDS1) [47,48] | [4Fe-4S] | Fe-S delivery to specific recipients |
| ISCA1 | Q9BUE6 | Isa1 | Multiple mitochondrial dysfunction syndrome 5 (MMDS5) [77,78] | [2Fe-2S], [4Fe-4S] | [4Fe-4S] cluster assembly |
| ISCA2 | Q86U28 | Isa2 | Multiple mitochondrial dysfunction syndrome 4 (MMDS4) [79] | [2Fe-2S], [4Fe-4S] | [4Fe-4S] cluster assembly |
| BOLA1 | Q9Y3E2 | Bol1 | Iron sensing/[2Fe-2S] delivery | ||
| BOLA3 | Q53S33 | Aim1 | Multiple mitochondrial dysfunction syndrome 2 (MMDS2) [48] | Fe-S delivery to specific recipients | |
| IBA57 | Q5T440 | Iba57 | Multiple mitochondrial dysfunction syndrome 3 (MMDS3) [80,81] | [4Fe-4S] cluster assembly | |
| HSC20 | Q8IWL3 | Jac1 | J-type co-chaperone | ||
| HSC70 | P38646 | Ssq1 | Parkinson’s disease [82] | ATP, ADP | Hsp70-type chaperone involved in cluster delivery |
| Human Protein | UNIPROT ID | Yeast Ortholog | Cofactors | Putative Functions |
|---|---|---|---|---|
| NUBP1 | P53384 | Nbp35 | [4Fe-4S] | Scaffold protein for formation of a [4Fe-4S] cluster [211,212] |
| NUBP2 | Q9Y5Y2 | Cfd1 | [4Fe-4S] | Scaffold protein for formation of a [4Fe-4S] cluster [211,212] |
| NDOR1 | Q9UHB4 | Tah18 | FAD, FMN, NADPH | Electron transfer [224,225] |
| Anamorsin (CIAPIN1) | Q6FI81 | Dre2 | [2Fe-2S], [4Fe-4S] | Electron transfer [224,225] |
| GLRX3 | O76003 | Grx3 and Grx4 | [2Fe-2S], GSH | Fe-S cluster transfer, iron trafficking [206,226,227] |
| BOLA2 | Q9H3K6 | Isd11 | [4Fe-4S] | Fe-S cluster transfer [226,228] |
| NARFL (IOP1) | Q9H6Q4 | Nar1 | Adaptor protein to connect early and late-acting CIA components [216,229] | |
| CIAO1 | O76071 | Cia1 | CIA targeting complex [214] | |
| MMS19 | Q96T76 | Met18 | CIA targeting complex [217,222] | |
| CIA2A (FAM96A) | Q9H5X1 | Specific maturation factor of IRP1 [215] | ||
| CIA2B (FAM96B) | Q9Y3D0 | Cia2 | CIA targeting complex [215] | |
| YAE1D1 | Q9NRH1 | Yae1 | Specific maturation factor of the cytosolic ABCE1 [221] | |
| ORAOV1 | Q8WV07 | YNL260C | Specific maturation factor of the cytosolic ABCE1 [221] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, K.; Markley, J.L. NMR as a Tool to Investigate the Processes of Mitochondrial and Cytosolic Iron-Sulfur Cluster Biosynthesis. Molecules 2018, 23, 2213. https://doi.org/10.3390/molecules23092213
Cai K, Markley JL. NMR as a Tool to Investigate the Processes of Mitochondrial and Cytosolic Iron-Sulfur Cluster Biosynthesis. Molecules. 2018; 23(9):2213. https://doi.org/10.3390/molecules23092213
Chicago/Turabian StyleCai, Kai, and John L. Markley. 2018. "NMR as a Tool to Investigate the Processes of Mitochondrial and Cytosolic Iron-Sulfur Cluster Biosynthesis" Molecules 23, no. 9: 2213. https://doi.org/10.3390/molecules23092213
APA StyleCai, K., & Markley, J. L. (2018). NMR as a Tool to Investigate the Processes of Mitochondrial and Cytosolic Iron-Sulfur Cluster Biosynthesis. Molecules, 23(9), 2213. https://doi.org/10.3390/molecules23092213