Genome-Wide Identification and Characterization of Aquaporins and Their Role in the Flower Opening Processes in Carnation (Dianthus caryophyllus)
Abstract
1. Introduction
2. Results
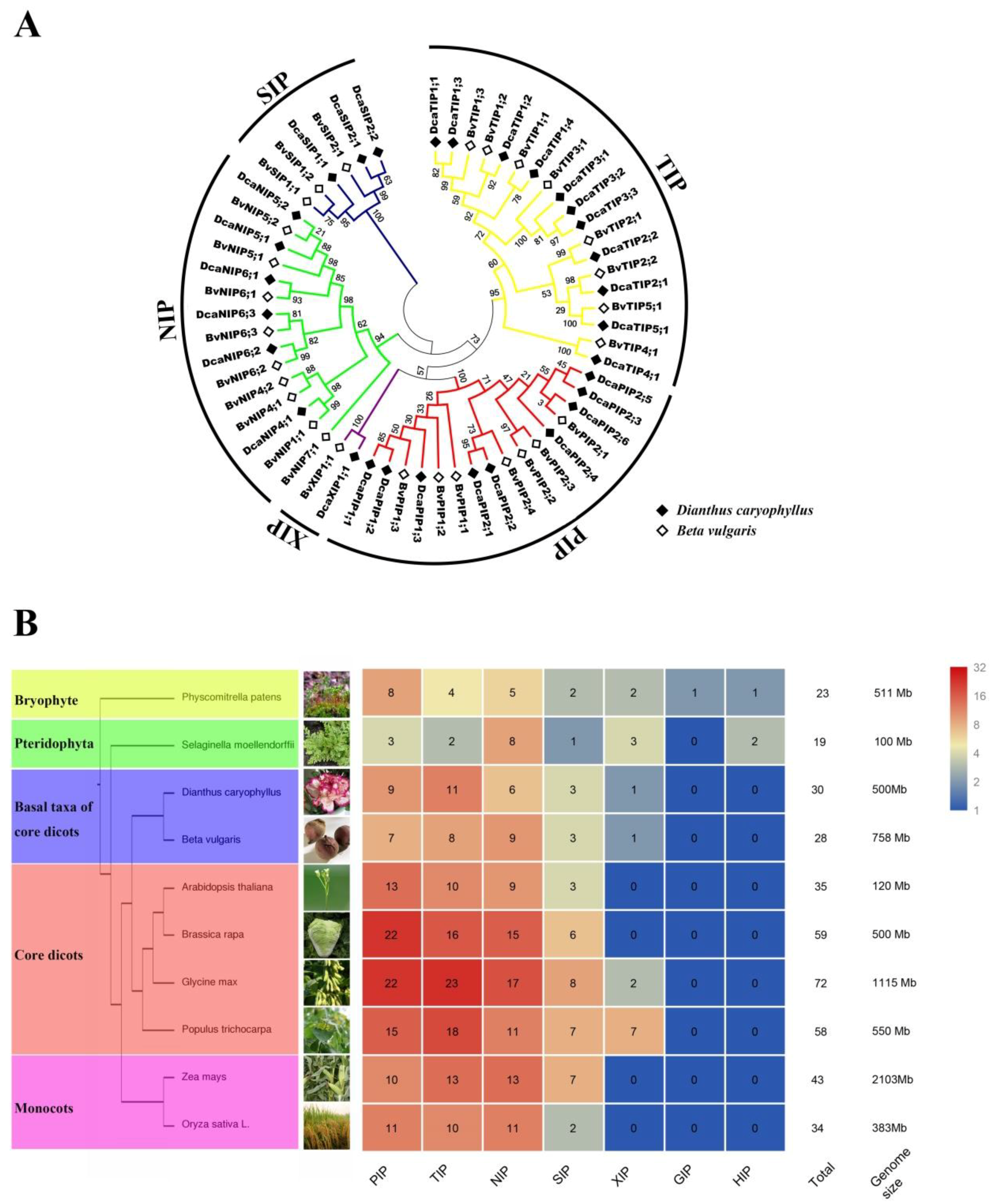
2.1. Identification, Classification, Nomenclature of DcaAQPs, and Properties of DcaAQPs
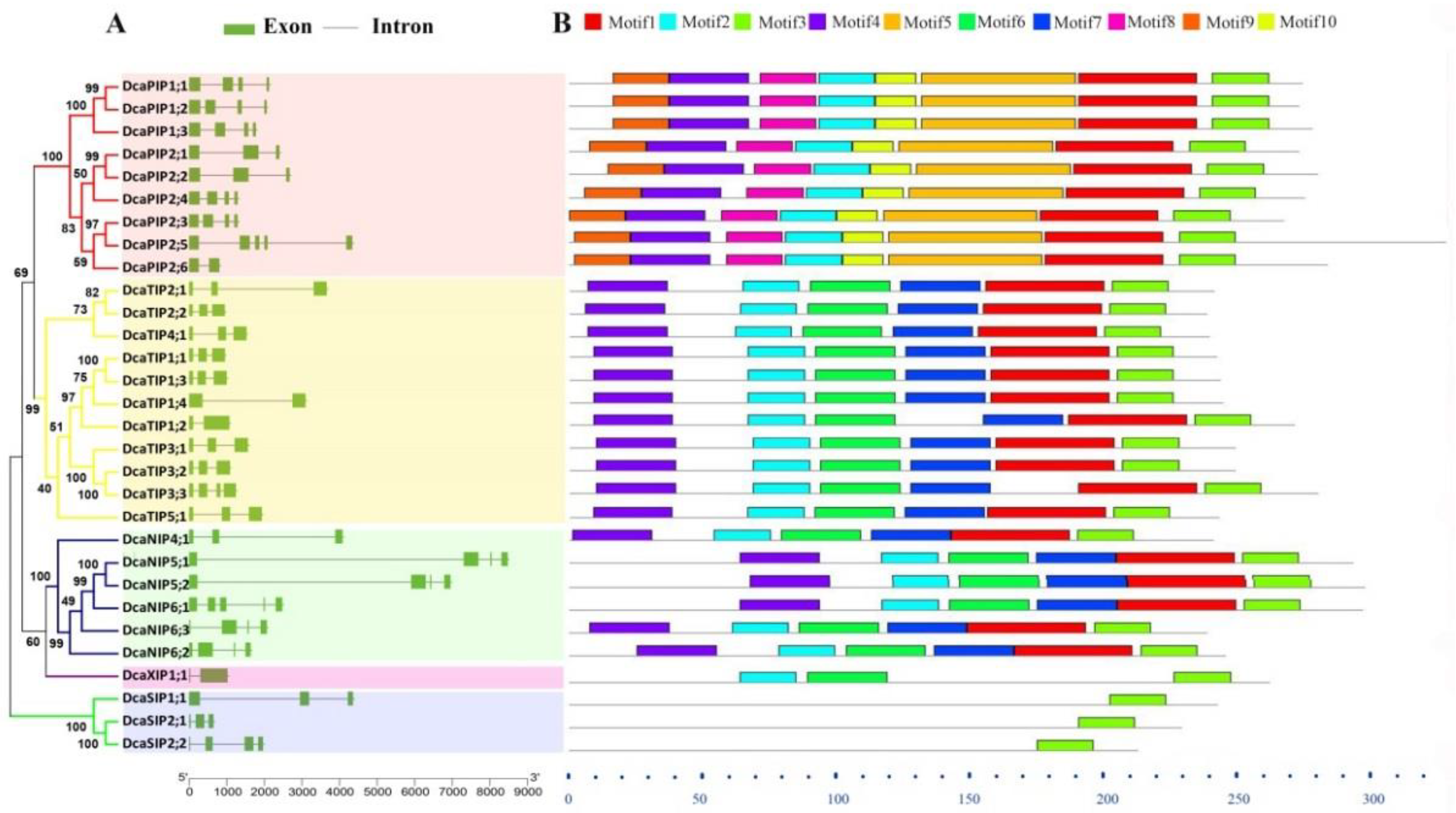
2.2. Gene Structure and Conserved Motif Analysis of DcaAQPs
2.3. Conserved Domain Analysis and Functional Prediction of DcaAQPs
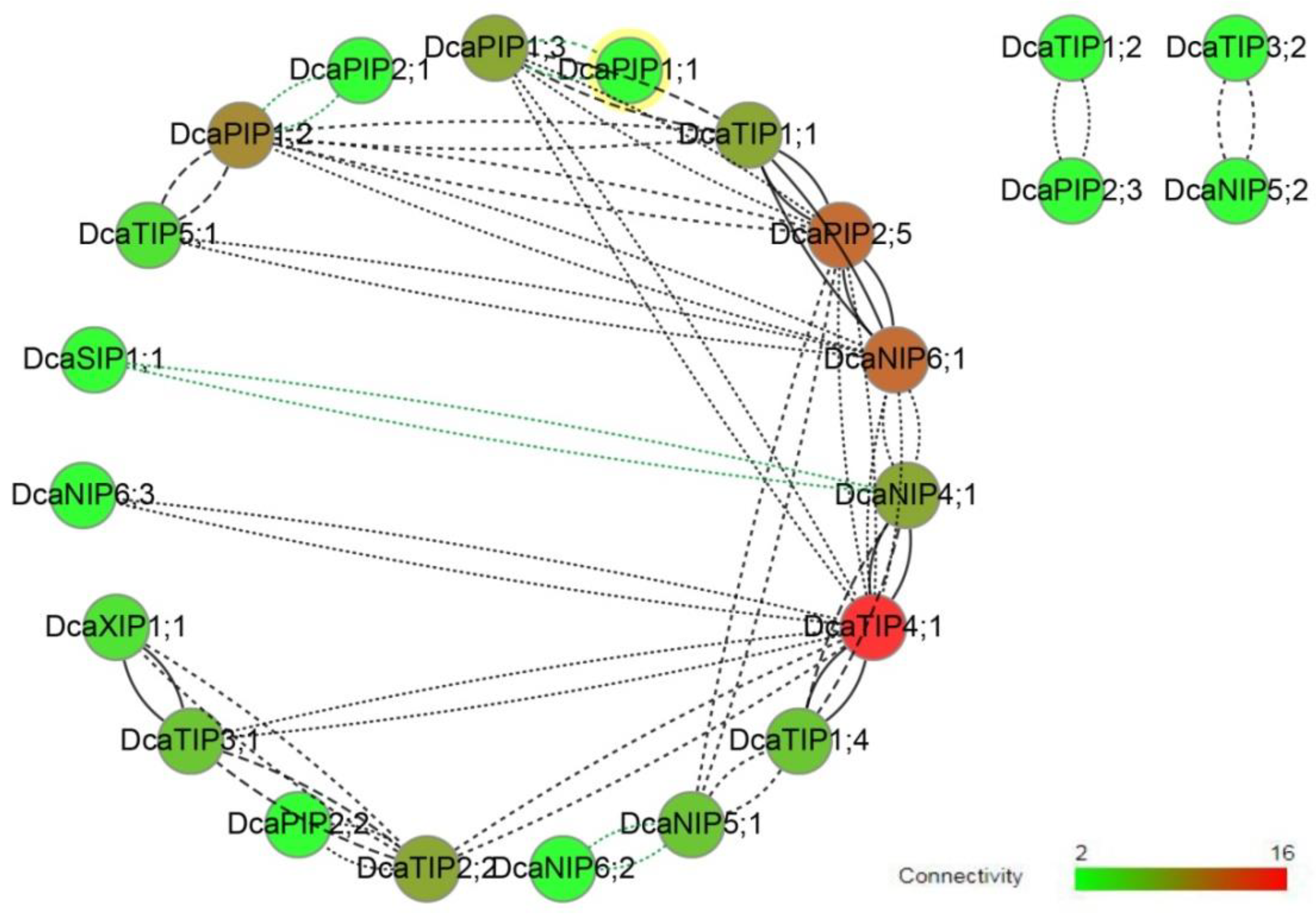
2.4. Co-Expression Network
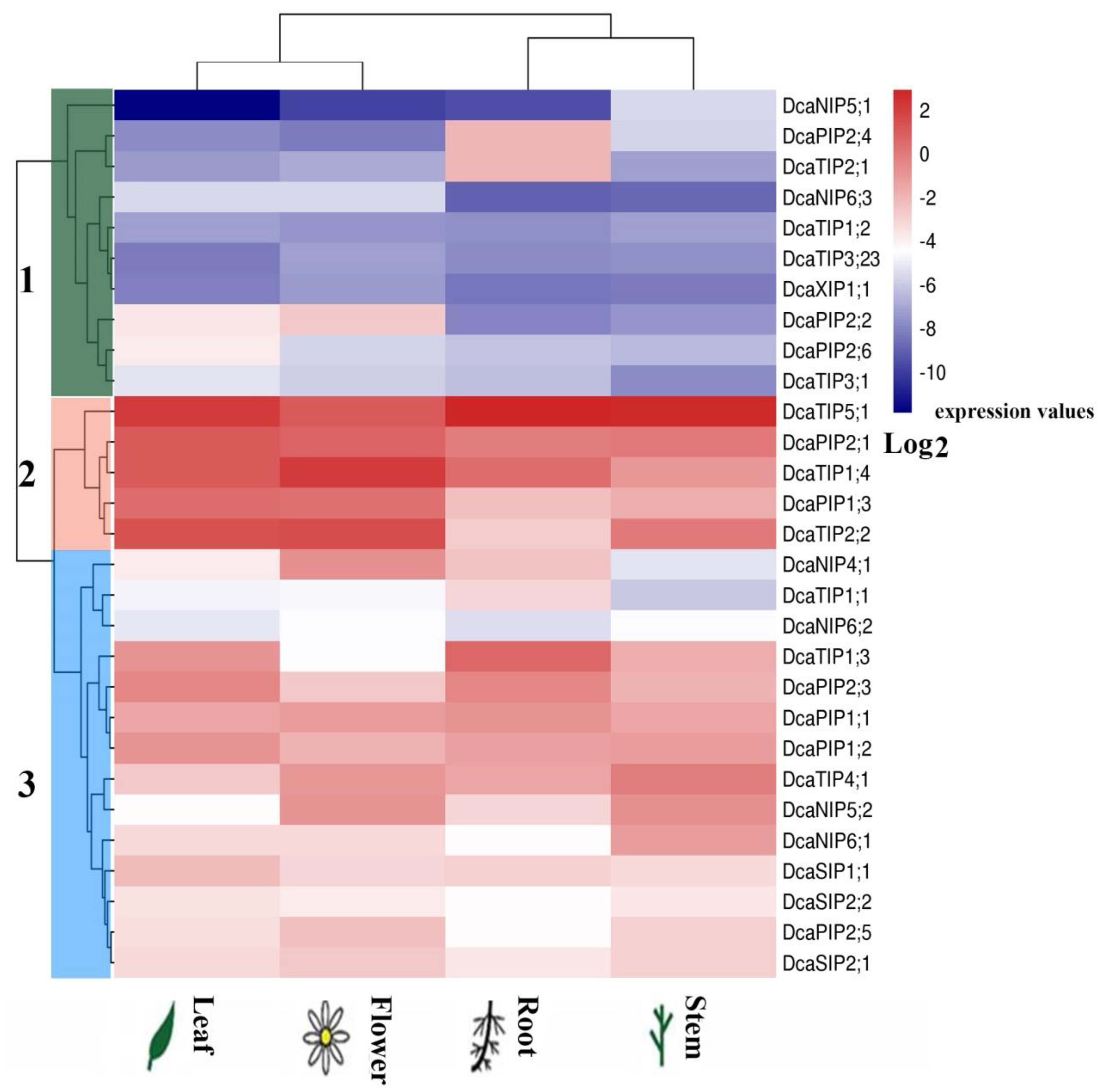
2.5. Expression of DcaAQP Genes in Different Tissues
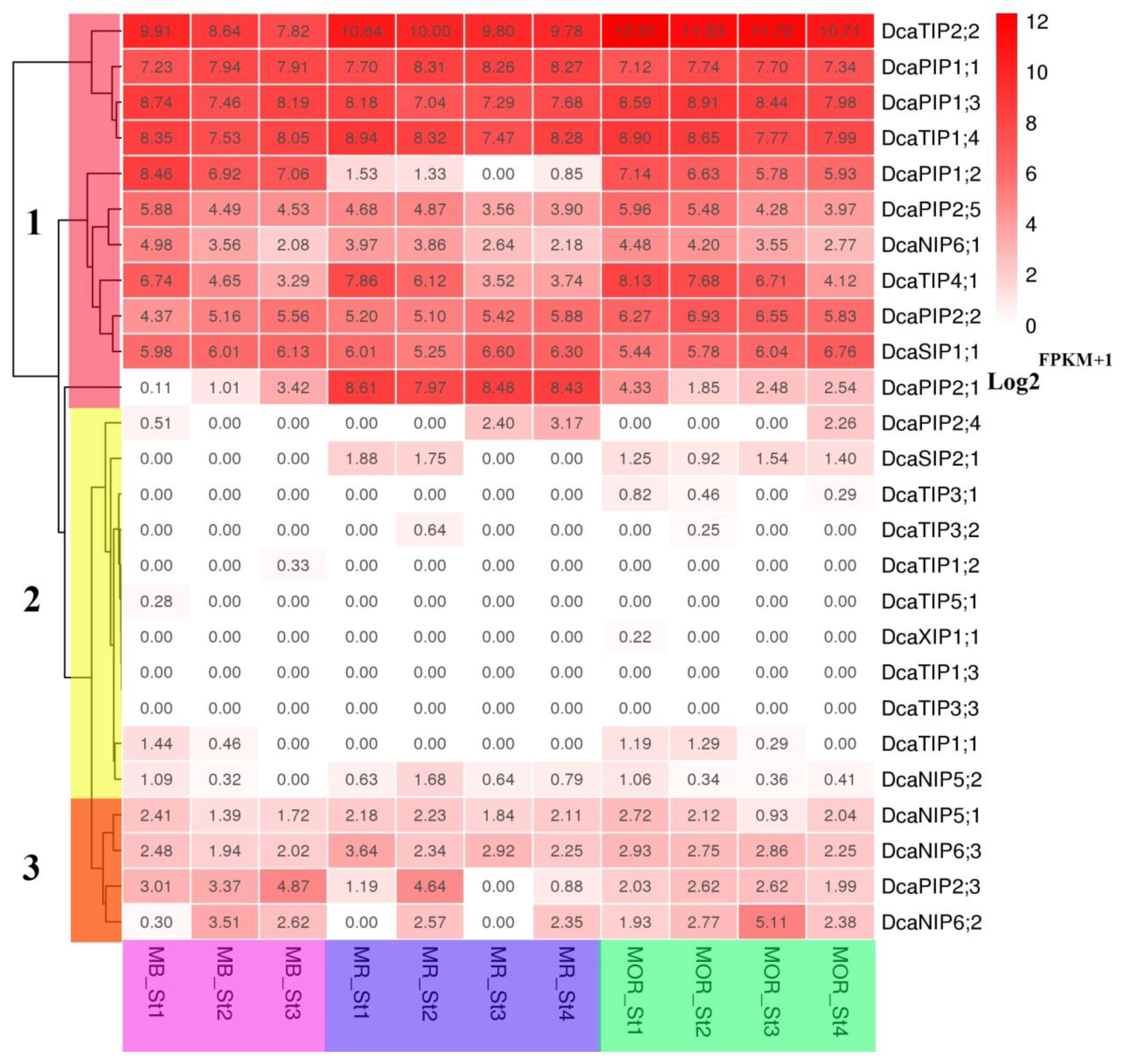
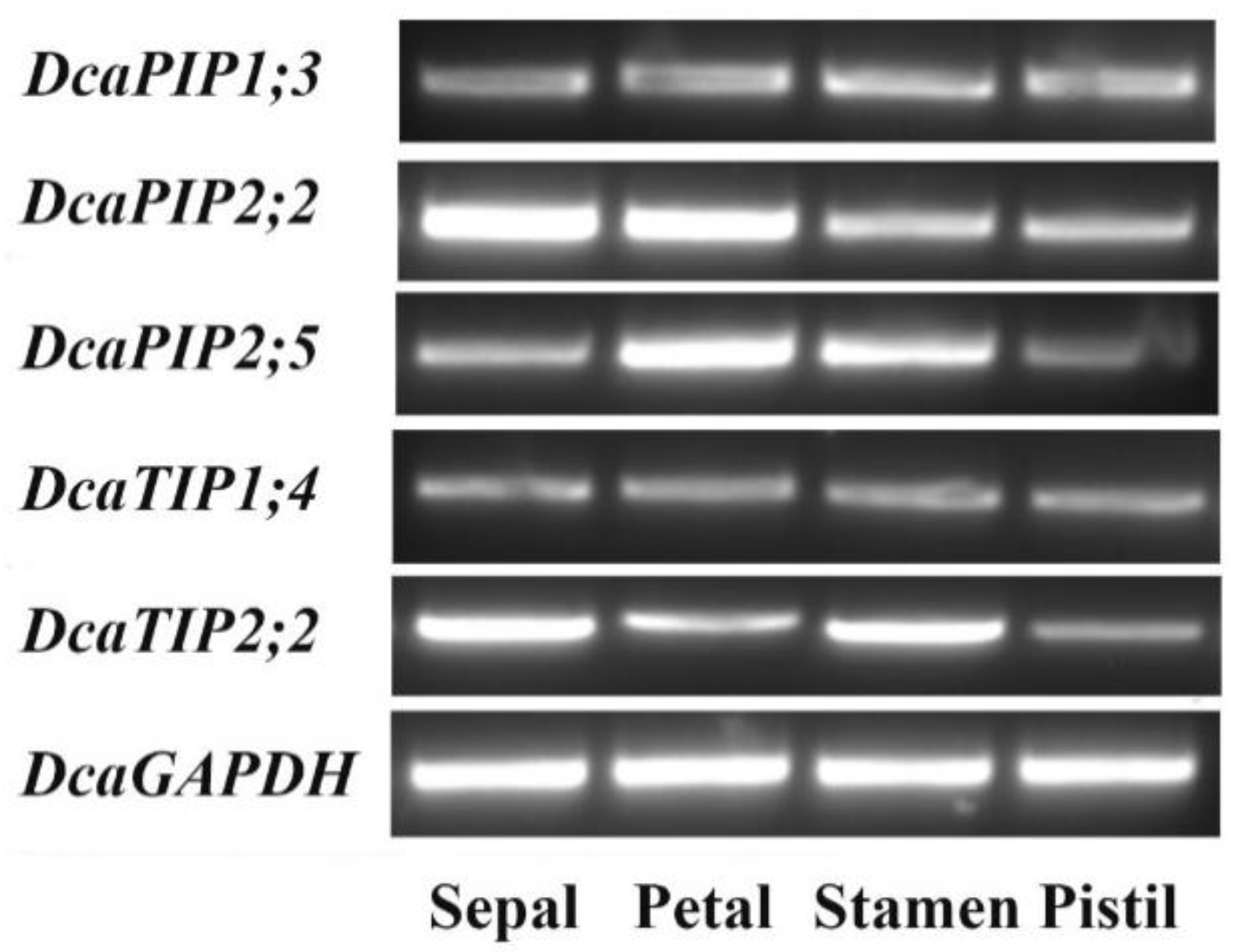
2.6. Expression Patterns of DcaAQP Genes with RNA-Seq and Semi-qRTPCR
3. Discussion
4. Materials and Methods
4.1. Plant Materials, RNA and DNA Extraction
4.2. Identification of DcaAQPs
4.3. Phylogenetic Analysis and Gene Duplication of DcaAQPs
4.4. Bioinformatics Analysis of DcaAQPs
4.5. Ka/Ks Analysis and Co-Expression Network
4.6. qRT-PCR Analysis in Different Tissues
4.7. RNA-Seq Analysis during Flower Opening Stages and Semi-qRT-PCR in Individual Flower Organs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AQPs | Aquaporins |
| DcaAQPs | AQPs in D. caryophyllus |
| PIPs | Plasma membrane intrinsic proteins |
| TIPs | Tonoplast intrinsic proteins |
| NIPs | NOD26-like intrinsic proteins |
| SIPs | Small basic intrinsic proteins |
| XIPs | X-intrinsic proteins |
| GIPs | GlpF-like intrinsic proteins |
| HIPs | Intrinsic hybrid proteins |
| TM | Trans-membranes |
| ar/R | selectivity filter Aromatic/arginine selectivity filter |
| HMM | The Hidden Markov model |
| qRT-PCR | Quantitative real-time RT-PCR |
| RNA-seq | RNA Sequencing |
| Semi-qRTRCR | Semi-quantitative RT-PCR |
| MW | Molecular weight |
| pI | Isoelectric point |
| Plas | Plasma membrane |
| Vacu | Vacuolar membrane |
| AA | Amino acid |
| TMD | Transmembrane domain |
| ORF | Opening reading frame |
| DcaGADPH | Glyceraldehyde-3-phosphate dehydrogenase |
| FPKM | The fragment per kilobase of exon per million fragments mapped |
References
- Deshmukh, R.K.; Sonah, H.; Bélanger, R.R. Plant aquaporins: Genome-wide identification, transcriptomics, proteomics, and advanced analytical tools. Front. Plant Sci. 2016, 7, 1896. [Google Scholar] [CrossRef] [PubMed]
- Maurel, C.; Boursiac, Y.; Luu, D.T.; Santoni, V.; Shahzad, Z.; Verdoucq, L. Aquaporins in plants. Physiol. Rev. 2015, 95, 1321–1358. [Google Scholar] [CrossRef] [PubMed]
- Preston, G.M.; Agre, P. Isolation of the cDNA for erythrocyte integral membrane protein of 28 kilodaltons: Member of an ancient channel family. Proc. Natl. Acad. Sci. USA 1991, 88, 11110–11114. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, R.K.; Vivancos, J.; Ramakrishnan, G.; Guérin, V.; Carpentier, G.; Sonah, H.; Labbé, C.; Isenring, P.; Belzile, F.; Bélanger, R.R. A precise spacing between NPA domains of aquaporins is essential for silicon permeability in plants. Plant J. 2015, 83, 489–500. [Google Scholar] [CrossRef] [PubMed]
- Sonah, H.; Deshmukh, R.K.; Labbé, C.; Bélanger, R.R. Analysis of aquaporins in Brassicaceae species reveals high-level of conservation and dynamic role against biotic and abiotic stress in canola. Sci. Rep. 2017, 7, 2771. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; Yang, S.; Wang, Y.; Bendahmane, M.; Fu, X. Genome-wide identification and characterization of aquaporin gene family in Beta vulgaris. PeerJ 2017, 5, e3747. [Google Scholar] [CrossRef] [PubMed]
- Danielson, J.Å.; Johanson, U. Unexpected complexity of the aquaporin gene family in the moss Physcomitrella patens. BMC Plant Biol. 2008, 8, 45. [Google Scholar] [CrossRef] [PubMed]
- Tornroth-Horsefield, S.; Wang, Y.; Hedfalk, K.; Johanson, U.; Karlsson, M.; Tajkhorshid, E.; Neutze, R.; Kjellbom, P. Structural mechanism of plant aquaporin gating. Nature 2006, 439, 688–694. [Google Scholar] [CrossRef] [PubMed]
- Savage, D.F.; O’Connell, J.D., 3rd; Miercke, L.J.; Finer-Moore, J.; Stroud, R.M. Structural context shapes the aquaporin selectivity filter. Proc. Natl. Acad. Sci. USA 2010, 107, 17164–17169. [Google Scholar] [CrossRef] [PubMed]
- Hove, R.M.; Bhave, M. Plant aquaporins with non-aqua functions: Deciphering the signature sequences. Plant Mol. Biol. 2011, 75, 413–430. [Google Scholar] [CrossRef] [PubMed]
- Azad, A.K.; Ahmed, J.; Alum, M.A.; Hasan, M.M.; Ishikawa, T.; Sawa, Y.; Katsuhara, M. Genome-wide characterization of major intrinsic proteins in four grass plants and their non-aqua transport selectivity profiles with comparative perspective. PLoS ONE 2016, 11, e0157735. [Google Scholar] [CrossRef] [PubMed]
- Siefritz, F.; Otto, B.; Bienert, G.P.; van der Krol, A.; Kaldenhoff, R. The plasma membrane aquaporin NtAQP1 is a key component of the leaf unfolding mechanism in tobacco. Plant J. 2004, 37, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Vera-Estrella, R.; Barkla, B.J.; Amezcua-Romero, J.C.; Pantoja, O. Day/night regulation of aquaporins during the CAM cycle in Mesembryanthemum crystallinum. Plant Cell Environ. 2012, 35, 485–501. [Google Scholar] [CrossRef] [PubMed]
- Uehlein, N.; Kaldenhoff, R. Aquaporins and plant leaf movements. Ann. Bot. 2007, 101, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Secchi, F.; Zwieniecki, M.A. Down-regulation of plasma intrinsic protein1 aquaporin in Poplar trees is detrimental to recovery from embolism. Plant Physiol. 2014, 164, 1789–1799. [Google Scholar] [CrossRef] [PubMed]
- Martre, P.; Morillon, R.; Barrieu, F.; North, G.B.; Nobel, P.S.; Chrispeels, M.J. Plasma membrane aquaporins play a significant role during recovery from water deficit. Plant Physiol. 2002, 130, 2101–2110. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, T.; Suga, S.; Tsuchiya, T.; Hisada, H.; Morishima, S.; Okada, Y.; Maeshima, M. Molecular cloning, water channel activity and tissue specific expression of two isoforms of radish vacuolar aquaporin. Plant Cell Physiol. 1998, 39, 905–913. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, K.; Kondo, S.; Hara, S.; Morishita, Y. The evolutionary aspects of aquaporin family. Am. J. Physiol.-Reg. I 2011, 300, R566–R576. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Santoni, V.; Maurel, C. Plant aquaporins: Roles in plant physiology. Biochim. Biophys. Acta (BBA) Gen. Subj. 2014, 1840, 1574–1582. [Google Scholar] [CrossRef] [PubMed]
- Birlanga, V.; Villanova, J.; Cano, A.; Cano, E.A.; Acosta, M.; Perez-Perez, J.M. Quantitative analysis of adventitious root growth phenotypes in carnation stem cuttings. PLoS ONE 2015, 10, e0133123. [Google Scholar] [CrossRef] [PubMed]
- Harada, T.; Torii, Y.; Morita, S.; Masumura, T.; Satoh, S. Differential expression of genes identified by suppression subtractive hybridization in petals of opening carnation flowers. J. Exp. Bot. 2010, 61, 2345–2354. [Google Scholar] [CrossRef] [PubMed]
- Yagi, M.; Kosugi, S.; Hirakawa, H.; Ohmiya, A.; Tanase, K.; Harada, T.; Kishimoto, K.; Nakayama, M.; Ichimura, K.; Onozaki, T.; et al. Sequence analysis of the genome of carnation (Dianthus caryophyllus L.). DNA Res. 2014, 21, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Tanase, K.; Nishitani, C.; Hirakawa, H.; Isobe, S.; Tabata, S.; Ohmiya, A.; Onozaki, T. Transcriptome analysis of carnation (Dianthus caryophyllus L.) based on next-generation sequencing technology. BMC Genom. 2012, 13, 292. [Google Scholar] [CrossRef] [PubMed]
- Morita, S.; Sugiyama, S.; Tateishi, A.; Satoh, S. Identification and characterization of plasma membrane intrinsic protein (PIP) aquaporin genes in petals of opening carnation flowers. Horticult. J. 2017, 86, 78–86. [Google Scholar] [CrossRef]
- Deshmukh, R.K.; Ma, J.F.; Bélanger, R.R. Editorial: Role of silicon in plants. Front. Plant Sci. 2017, 8, 1858. [Google Scholar] [CrossRef] [PubMed]
- Anderberg, H.I.; Kjellbom, P.; Johanson, U. Annotation of Selaginella moellendorffii major intrinsic proteins and the evolution of the protein family in terrestrial plants. Front. Plant Sci. 2012, 3, 33. [Google Scholar] [CrossRef] [PubMed]
- De, R.S.; Sabaghian, E.; Li, Z.; Saeys, Y.; Van, Y.D.P. Coordinated functional divergence of genes after genome duplication in Arabidopsis thaliana. Plant Cell 2017, 29. [Google Scholar] [CrossRef]
- Wang, M.; Wang, P.; Min, L.; Ye, Z.; Li, G.; Tu, L.; Chao, S.; Li, J.; Yang, Q.; Zhang, X. Evolutionary dynamics of 3D genome architecture following polyploidization in cotton. Nat. Plants 2018, 4, 90. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Sankararamakrishnan, R. Genome-wide analysis of major intrinsic proteins in the tree plant Populus trichocarpa: Characterization of XIP subfamily of aquaporins from evolutionary perspective. BMC Plant Biol. 2009, 9, 134. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.Y.; Ali, Z.; Wang, C.B.; Xu, L.; Yi, J.X.; Xu, Z.L.; Liu, X.Q.; He, X.L.; Huang, Y.H.; Khan, I.A. Genome-wide sequence characterization and expression analysis of major intrinsic proteins in soybean (Glycine max L.). PLoS ONE 2013, 8, e56312. [Google Scholar] [CrossRef] [PubMed]
- Ren, R.; Wang, H.; Guo, C.; Zhang, N.; Zeng, L.; Chen, Y.; Ma, H.; Qi, J. Wide-spread whole genome duplications contribute to genome complexity and species diversity in Angiosperms. Mol. Plant 2018, 11, 357–358. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, J.; Yu, J.W.; Park, S.W. Genome-wide analysis and expression profiling of the Solanum tuberosum aquaporins. Plant Physiol. Biochem. 2013, 73, 392–404. [Google Scholar] [CrossRef] [PubMed]
- Dynowski, M.; Schaaf, G.; Loque, D.; Moran, O.; Ludewig, U. Plant plasma membrane water channels conduct the signalling molecule H2O2. Biochem. J. 2008, 414, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Uehlein, N.; Lovisolo, C.; Siefritz, F.; Kaldenhoff, R. The tobacco aquaporin NtAQP1 is a membrane CO2 pore with physiological functions. Nature 2003, 425, 734. [Google Scholar] [CrossRef] [PubMed]
- Uehlein, N.; Otto, B.; Eilingsfeld, A.; Itel, F.; Meier, W.; Kaldenhoff, R. Gas-tight triblock-copolymer membranes are converted to CO2 permeable by insertion of plant aquaporins. Sci. Rep. 2012, 2, 538. [Google Scholar] [CrossRef] [PubMed]
- Mitaniueno, N.; Yamaji, N.; Zhao, F.J.; Zhao, F.J.; Ma, J.F. The aromatic/arginine selectivity filter of NIP aquaporins plays a critical role in substrate selectivity for silicon, boron, and arsenic. J. Exp. Bot. 2011, 62, 4391–4398. [Google Scholar] [CrossRef] [PubMed]
- Meli, R.; Pirozzi, C.; Pelagalli, A. New perspectives on the potential role of aquaporins (AQPs) in the physiology of inflammation. Plant Physiol. 2018, 9, 101. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Yang, L.; Gong, J.; Mo, Y.; Wang, J.; Cao, J.; An, F.; Xie, G. Genome-wide identification of Jatropha curcas aquaporin genes and the comparative analysis provides insights into the gene family expansion and evolution in Hevea brasiliensis. Front. Plant Sci. 2016, 7, 395. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Tong, X.; Han, M.; Hu, H.; Dai, F. Genome-wide identification and characterization of WD40 protein genes in the silkworm, Bombyx mori. Int. J. Mol. Sci. 2018, 10, 597. [Google Scholar] [CrossRef] [PubMed]
- Abdul, M.K.; Nath, U.K.; Park, J.I.; Biswas, M.K.; Choi, E.K.; Song, J.Y.; Kim, H.T.; Nou, I.S. Genome-wide identification, characterization, and expression profiling of glutathione s-transferase (GST) family in pumpkin reveals likely role in cold-stress tolerance. Genes 2018, 9, 84. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhang, Z.; Li, W.; Zhu, W.; Ren, Z.; Wang, Z.; Li, L.; Jia, L.; Zhu, S.; Ma, Z. Genome-wide identification and comparative analysis of the 3-Hydroxy-3-methylglutaryl Coenzyme A Reductase (HMGR) gene family in Gossypium. Molecules 2018, 23, 193. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, R.; Bélanger, R.R. Molecular evolution of aquaporins and silicon influx in plants. Funct. Ecol. 2016, 30, 1277–1285. [Google Scholar] [CrossRef]
- Soundararajan, P.; Manivannan, A.; Cho, Y.S.; Jeong, B.R. Exogenous Supplementation of Silicon Improved the Recovery of Hyperhydric Shoots in Dianthus caryophyllus L. by Stabilizing the Physiology and Protein Expression. Front. Plant Sci. 2017, 8, 738. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.X.; Moon, S.; Jung, K.H. Genome-wide expression analysis of rice aquaporin genes and development of a functional gene network mediated by aquaporin expression in roots. Planta 2013, 238, 669–681. [Google Scholar] [CrossRef] [PubMed]
- Cohen, D.; Bogeat-Triboulot, M.B.; Vialet-Chabrand, S.; Merret, R.; Courty, P.E.; Moretti, S.; Bizet, F.; Guilliot, A.; Hummel, I. Developmental and environmental regulation of aquaporin gene expression across Populus species: Divergence or redundancy? PLoS ONE 2013, 8, e55506. [Google Scholar] [CrossRef] [PubMed]
- Kaldenhoff, R.; Kolling, A.; Richter, G. Regulation of the Arabidopsis thaliana aquaporin gene AthH2 (PIP1b). J. Photochem. Photobiol. B 1996, 36, 351–354. [Google Scholar] [CrossRef]
- Heinen, R.B.; Ye, Q.; Chaumont, F. Role of aquaporins in leaf physiology. J. Exp. Bot. 2009, 60, 2971–2985. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Yang, F.; Gao, J. Isolation of Rh-TIP1; 1, an aquaporin gene and its expression in rose flowers in response to ethylene and water deficit. Postharvest Biol. Technol. 2009, 51, 407–413. [Google Scholar] [CrossRef]
- Fetter, K.; Van Wilder, V.; Moshelion, M.; Chaumont, F. Interactions between plasma membrane aquaporins modulate their water channel activity. Plant Cell 2004, 16, 215–228. [Google Scholar] [CrossRef] [PubMed]
- Zelazny, E.; Borst, J.W.; Muylaert, M.; Batoko, H.; Hemminga, M.A.; Chaumont, F. FRET imaging in living maize cells reveals that plasma membrane aquaporins interact to regulate their subcellular localization. Proc. Natl. Acad. Sci. USA 2007, 104, 12359–12364. [Google Scholar] [CrossRef] [PubMed]
- Yaneff, A.; Sigaut, L.; Marquez, M.; Alleva, K.; Pietrasanta, L.I.; Amodeo, G. Heteromerization of PIP aquaporins affects their intrinsic permeability. Proc. Natl. Acad. Sci. USA 2014, 111, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Zelazny, E.; Miecielica, U.; Borst, J.W.; Hemminga, M.A.; Chaumont, F. An N-terminal diacidic motif is required for the trafficking of maize aquaporins ZmPIP2; 4 and ZmPIP2; 5 to the plasma membrane. Plant J. 2009, 57, 346–355. [Google Scholar] [CrossRef] [PubMed]
- Daniels, M.J.; Mirkov, T.E.; Chrispeels, M.J. The plasma membrane of Arabidopsis thaliana contains a mercury-insensitive aquaporin that is a homolog of the tonoplast water channel protein TIP. Plant Physiol. 1994, 106, 1325–1333. [Google Scholar] [CrossRef] [PubMed]
- Yamada, S.; Katsuhara, M.; Kelly, W.B.; Michalowski, C.B.; Bohnert, H.J. A family of transcripts encoding water channel proteins: Tissue-specific expression in the common ice plant. Plant Cell 1995, 7, 1129–1142. [Google Scholar] [CrossRef] [PubMed]
- Chaumont, F.; Barrieu, F.; Jung, R.; Chrispeels, M.J. Plasma membrane intrinsic proteins from maize cluster in two sequence subgroups with differential aquaporin activity. Plant Physiol. 2000, 122, 1025–1034. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.D.; Herman, E.M.; Chrispeels, M.J. An abundant, highly conserved tonoplast protein in seeds. Plant Physiol. 1989, 91, 1006–1013. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Gong, J.; Huang, Q.; Mo, Y.; Yang, L.; Xie, G. Gene structures, evolution, classification and expression profiles of the aquaporin gene family in castor bean (Ricinus communis L.). PLoS ONE 2015, 10, e0141022. [Google Scholar] [CrossRef] [PubMed]
- Shivaraj, S.M.; Deshmukh, R.K.; Rai, R.; Bélanger, R.; Agrawal, P.K.; Dash, P.K. Genome-wide identification, characterization, and expression profile of aquaporin gene family in flax (Linum usitatissimum). Sci. Rep. 2017, 7, 46137. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Hou, X.; Huang, C.; Yan, Y.; Tie, W.; Ding, Z.; Wei, Y.; Liu, J.; Miao, H.; Lu, Z. Genome-wide identification and expression analyses of aquaporin gene family during development and abiotic stress in banana. Int. J. Mol. Sci. 2015, 16, 19728–19751. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Gong, J.; An, F.; Xie, G.; Wang, J.; Mo, Y.; Yang, L. Genome-wide identification of rubber tree (Hevea brasiliensis Muell. Arg.) aquaporin genes and their response to ethephon stimulation in the laticifer, a rubber-producing tissue. BMC Genom. 2015, 16, 1001. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef] [PubMed]
- Baranwal, V.K.; Negi, N.; Khurana, P. Auxin response factor genes repertoire in Mulberry: Identification, and structural, functional and evolutionary analyses. Genes 2017, 8, 202. [Google Scholar] [CrossRef] [PubMed]
- Yuan, D.; Li, W.; Hua, Y.; King, G.J.; Xu, F.; Shi, L. Genome-wide identification and characterization of the aquaporin gene family and transcriptional responses to boron deficiency in Brassica napus. Front. Plant Sci. 2017, 8, 1336. [Google Scholar] [CrossRef] [PubMed]
- Vivancos, J.; Deshmukh, R.; Grégoire, C.; Rémus-Borel, W.; Belzile, F.; Bélanger, R.R. Identification and characterization of silicon efflux transporters in horsetail (Equisetum arvense). J. Plant Physiol. 2016, 200, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Montpetit, J.; Vivancos, J.; Mitaniueno, N.; Yamaji, N.; Rémusborel, W.; Belzile, F.; Ma, J.F.; Bélanger, R.R. Cloning, functional characterization and heterologous expression of TaLsi1, a wheat silicon transporter gene. Plant Mol. Biol. 2012, 79, 35. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, R.K.; Vivancos, J.; Guerin, V.; Sonah, H.; Labbe, C.; Belzile, F.; Bélanger, R.R. Identification and functional characterization of silicon transporters in soybean using comparative genomics of major intrinsic proteins in Arabidopsis and rice. Plant Mol. Biol. 2013, 83, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Martins Cde, P.; Pedrosa, A.M.; Du, D.; Goncalves, L.P.; Yu, Q.; Gmitter, F.G., Jr.; Costa, M.G. Genome-wide characterization and expression analysis of major intrinsic proteins during abiotic and biotic stresses in sweet orange (Citrus sinensis L. Osb.). PLoS ONE 2015, 10, e0138786. [Google Scholar]
- Deokar, A.A.; Tar’an, B. Genome-wide analysis of the aquaporin gene family in Chickpea (Cicer arietinum L.). Front. Plant Sci. 2016, 7, 1802. [Google Scholar] [CrossRef] [PubMed]
- Verma, J.K.; Wardhan, V.; Singh, D.; Chakraborty, S.; Chakraborty, N. Genome-wide identification of the Alba gene family in plants and stress-responsive expression of the rice Alba genes. Genes 2018, 9, 183. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2014, 31, 1296. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Suyama, M.; Torrents, D.; Bork, P. PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006, 34, W609. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Yang, ZH. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- Tzfadia, O.; Diels, T.; De, M.S.; Vandepoele, K.; Aharoni, A.; Van, d.P.Y. CoExpNetViz: Comparative co-expression networks construction and visualization tool. Front. Plant Sci. 2016, 6, 1194. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.L.; Zhou, Q.; Wang, Y.Y.; Wang, W.E.; Bao, M.Z.; Zhang, J.W. Identification of heat-responsive genes in carnation (Dianthus caryophyllus L.) by RNA-seq. Front. Plant Sci. 2015, 6, 519. [Google Scholar] [CrossRef] [PubMed]
- Hima Kumari, P.; Anil Kumar, S.; Ramesh, K.; Sudhakar Reddy, P.; Nagaraju, M.; Bhanu Prakash, A.; Shah, T.; Henderson, A.; Srivastava, R.K.; Rajasheker, G.; et al. Genome-wide identification and analysis of Arabidopsis sodium proton antiporter (NHX) and human sodium proton exchanger (NHE) homologs in sorghum bicolor. Genes 2018, 9, 236. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protocols 2012, 7, 562. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; Hu, R.; Bao, M.; Fu, X. Cloning and expression analysis of the aquaporin genes in carnation. Acta Horticult. Sin. 2017, 44, 515–527. [Google Scholar]
- Cao, Y.; Han, Y.; Li, D.; Yi, L.; Cai, Y. MYB transcription factors in chinese pear (Pyrus bretschneideri Rehd.): Genome-wide identification, classification, and expression profiling during fruit development. Front. Plant Sci. 2016, 7, 577. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |
| Subfamily | Gene | Accession ID | AA Length | MW (kDa) | pI | TMD | Subcellular Localization | Putative Allele ORF |
|---|---|---|---|---|---|---|---|---|
| PIP | DcaPIP1;1 | Dca52692.1 | 285 | 30.4083 | 8.86 | 6 | plas | |
| DcaPIP1;2 | Dca27473.1 | 284 | 30.3473 | 8.84 | 6 | plas | Dca29035.1 | |
| * DcaPIP1;3 | Dca31714.1 | 289 | 30.857 | 9.14 | 6 | plas | ||
| * DcaPIP2;1 | Dca59969.1 | 284 | 30.4886 | 9.23 | 6 | plas | Dca47122.1 | |
| * DcaPIP2;2 | Dca36456.1 | 291 | 31.2254 | 9.2 | 6 | plas | ||
| * DcaPIP2;3 | Dca37662.1 | 278 | 29.6184 | 8.58 | 6 | plas | Dca51181.1 | |
| * DcaPIP2;4 | Dca7764.1 | 286 | 30.7608 | 8.58 | 6 | plas | Dca23336.1 | |
| * DcaPIP2;5 | Dca12710.1 | 339 | 36.7057 | 9.57 | 6 | plas | ||
| * DcaPIP2;6 | Dca21274.1 | 295 | 31.586 | 9.21 | 5 | plas | ||
| TIP | DcaTIP1;1 | Dca5708.1 | 251 | 25.7188 | 5.36 | 6 | vacu | |
| DcaTIP1;2 | Dca9296.1 | 281 | 30.0387 | 5.11 | 7 | vacu | ||
| * DcaTIP1;3 | Dca44001.1 | 252 | 25.9331 | 5.43 | 5 | vacu | Dca51356.1 | |
| * DcaTIP1;4 | Dca50626.1 | 253 | 25.9171 | 5.04 | 6 | vacu | ||
| * DcaTIP2;1 | Dca15468.1 | 250 | 25.3706 | 5.33 | 6 | vacu | ||
| * DcaTIP2;2 | Dca20683.1 | 247 | 25.1731 | 5.76 | 6 | vacu | ||
| DcaTIP3;1 | Dca4416.1 | 258 | 27.2657 | 6.7 | 6 | vacu | Dca20060.1 | |
| DcaTIP3;2 | Dca4417.1 | 258 | 27.2918 | 7.15 | 6 | vacu | ||
| DcaTIP3;3 | Dca20061.1 | 290 | 30.7138 | 7.15 | 7 | plas | ||
| DcaTIP4;1 | Dca14656.1 | 248 | 25.8422 | 5.34 | 7 | vacu | ||
| DcaTIP5;1 | Dca14871.1 | 252 | 26.2813 | 6.39 | 6 | vacu | ||
| NIP | * DcaNIP4;1 | Dca15023.1 | 250 | 26.7095 | 9.35 | 6 | plas | |
| DcaNIP5;1 | Dca40630.1 | 304 | 31.5687 | 8.38 | 6 | plas | ||
| DcaNIP5;2 | Dca29915.1 | 308 | 31.7267 | 8.75 | 6 | plas | ||
| DcaNIP6;1 | Dca29994.1 | 307 | 31.8083 | 6.42 | 6 | plas | ||
| DcaNIP6;2 | Dca42214.1 | 247 | 25.8832 | 8.76 | 6 | plas | ||
| DcaNIP6;3 | Dca25595.1 | 254 | 26.2389 | 6.05 | 6 | plas | ||
| SIP | DcaSIP1;1 | Dca30655.1 | 251 | 26.3213 | 9.62 | 6 | plas | |
| * DcaSIP2;1 | Dca8588.1 | 271 | 26.0338 | 8.41 | 6 | vacu/plas | ||
| DcaSIP2;2 | Dca13275.1 | 237 | 24.448 | 9.54 | 5 | plas | ||
| XIP | DcaXIP1;1 | Dca56078.1 | 220 | 28.9014 | 9.05 | 7 | plas |
| Gene | NPA | Ar/R Selectivity Filter | Froger’s Position | Hove et al. | Azad et al. | |
|---|---|---|---|---|---|---|
| LB | LE | H2,H5,LE1,LE2 | P1,P2,P3,P4,P5 | |||
| DcaPIP1;1 | SGGHINPAVT | GTGINPARSLG | F,H,T,R | Q,S,A,F,W | Boron,CO2,H2O2,Urea | CO2,H2O2 |
| DcaPIP1;2 | SGGHINPAVT | GTGINPARSLG | F,H,T,R | Q,S,A,F,W | Boron,CO2,H2O2,Urea | CO2,H2O2 |
| DcaPIP1;3 | SGGHINPAVT | GTGINPARSLG | F,H,T,R | Q,S,A,F,W | Boron,CO2,H2O2,Urea | CO2,H2O2 |
| DcaPIP2;1 | SGGHINPAVT | GTGINPARSFG | F,H,T,R | M,S,A,F,W | Ammonia,Urea | Ammonia |
| DcaPIP2;2 | SGGHINPAVT | GTGINPARSFG | F,H,T,R | M,S,A,F,W | Ammonia,Urea | Ammonia |
| DcaPIP2;3 | SGGHINPAVT | GTGINPARSFG | F,H,T,R | Q,S,A,F,W | H2O2,Urea | H2O2 |
| DcaPIP2;4 | SGGHINPAVT | GTGINPARSFG | F,H,T,R | Q,S,A,F,W | H2O2,Urea | H2O2 |
| DcaPIP2;5 | SGGHINPAVT | GTGINPARSFG | F,H,T,R | Q,S,A,F,W | H2O2,Urea | H2O2 |
| DcaPIP2;6 | SGGHINPAVT | GTGINPARSLG | F,H,T,R | Q,S,A,F,W | Boron,CO2,H2O2,Urea | CO2 |
| DcaTIP1;1 | SGGHVNPAIT | GASMNPAVSFG | H,I,A,V | T,S,A,Y,W | ||
| DcaTIP1;2 | SGGHVNPAVT | GASMNPAVTFG | H,I,A,V | T,T,A,Y,W | Urea | |
| DcaTIP1;3 | SGGHVNPAIT | GASMNPAVSFG | H,I,A,V | T,S,A,Y,W | ||
| DcaTIP1;4 | SGGHVNPAVT | GASMNPAVSFG | H,I,A,V | T,S,A,Y,W | H2O2,Urea | H2O2,Urea |
| DcaTIP2;1 | SGGHLNPAVT | GGSMNPARSFG | H,I,G,R | T,S,A,Y,W | H2O2,Urea | H2O2,Urea |
| DcaTIP2;2 | SGGHVNPAVT | GGSMNPARSFG | H,I,G,R | T,S,A,Y,W | H2O2,Urea | H2O2,Urea |
| DcaTIP3;1 | SGGHVNPAVT | GASMNPARAFG | H,I,A,R | T,A,A,Y,W | H2O2,Urea | |
| DcaTIP3;2 | SGGHVNPAVT | GASMNPARAFG | H,I,A,R | T,A,A,Y,W | H2O2,Urea | |
| DcaTIP3;3 | SGGHVNPAVT | GASMNPARAFG | H,I,A,R | T,A,A,Y,W | H2O2,Urea | |
| DcaTIP4;1 | SGGHLNPAVT | AASMNPARSFG | H,I,A,R | T,S,A,Y,W | ||
| DcaTIP5;1 | SGGHVNPAVT | GGSMNPAYSFG | N,V,G,Y | T,S,A,Y,W | ||
| DcaNIP4;1 | SGAHFNPAVT | GASMNPARSIG | W,F,A,R | F,S,A,Y,I | ||
| DcaNIP5;1 | SGAHLNPSLT | GGSMNPVRTLG | A,I,G,R | F,T,A,Y,L | Boron | Boron |
| DcaNIP5;2 | SGAHLNPSLT | GGSMNPVRTLG | A,I,G,R | F,T,A,Y,L | Boron | Boron |
| DcaNIP6;1 | SGAHLNPALT | GASMNPVRTLG | A,I,A,R | F,T,A,Y,L | Boron | |
| DcaNIP6;2 | SGAHINLAVS | GGSMNPARTVG | S,I,G,R | Y,T,A,Y,I | ||
| DcaNIP6;3 | SKAHFNPAVT | GASMNPARTLG | S,I,A,R | Y,T,A,Y,M | ||
| DcaSIP1;1 | GGASFNPTGT | GFSMNPANAFG | I,V,P,N | M,A,A,Y,W | ||
| DcaSIP2;1 | KAGNYNPLTL | GGCMNPASVMG | S,Q,G,S | F,V,A,Y,W | ||
| DcaSIP2;2 | NGAAYNPLTV | GGCMNPASVMG | S,H,G,S | F,V,A,Y,W | ||
| DcaXIP1;1 | SGGHINPSVT | GAGMNPARCVG | I,T,A,R | V,C,A,Y,W | ||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kong, W.; Bendahmane, M.; Fu, X. Genome-Wide Identification and Characterization of Aquaporins and Their Role in the Flower Opening Processes in Carnation (Dianthus caryophyllus). Molecules 2018, 23, 1895. https://doi.org/10.3390/molecules23081895
Kong W, Bendahmane M, Fu X. Genome-Wide Identification and Characterization of Aquaporins and Their Role in the Flower Opening Processes in Carnation (Dianthus caryophyllus). Molecules. 2018; 23(8):1895. https://doi.org/10.3390/molecules23081895
Chicago/Turabian StyleKong, Weilong, Mohammed Bendahmane, and Xiaopeng Fu. 2018. "Genome-Wide Identification and Characterization of Aquaporins and Their Role in the Flower Opening Processes in Carnation (Dianthus caryophyllus)" Molecules 23, no. 8: 1895. https://doi.org/10.3390/molecules23081895
APA StyleKong, W., Bendahmane, M., & Fu, X. (2018). Genome-Wide Identification and Characterization of Aquaporins and Their Role in the Flower Opening Processes in Carnation (Dianthus caryophyllus). Molecules, 23(8), 1895. https://doi.org/10.3390/molecules23081895





