Characterization of the Chloroplast Genome Sequence of Acer miaotaiense: Comparative and Phylogenetic Analyses
Abstract
1. Introduction
2. Results and Discussion
2.1. Genomic Features
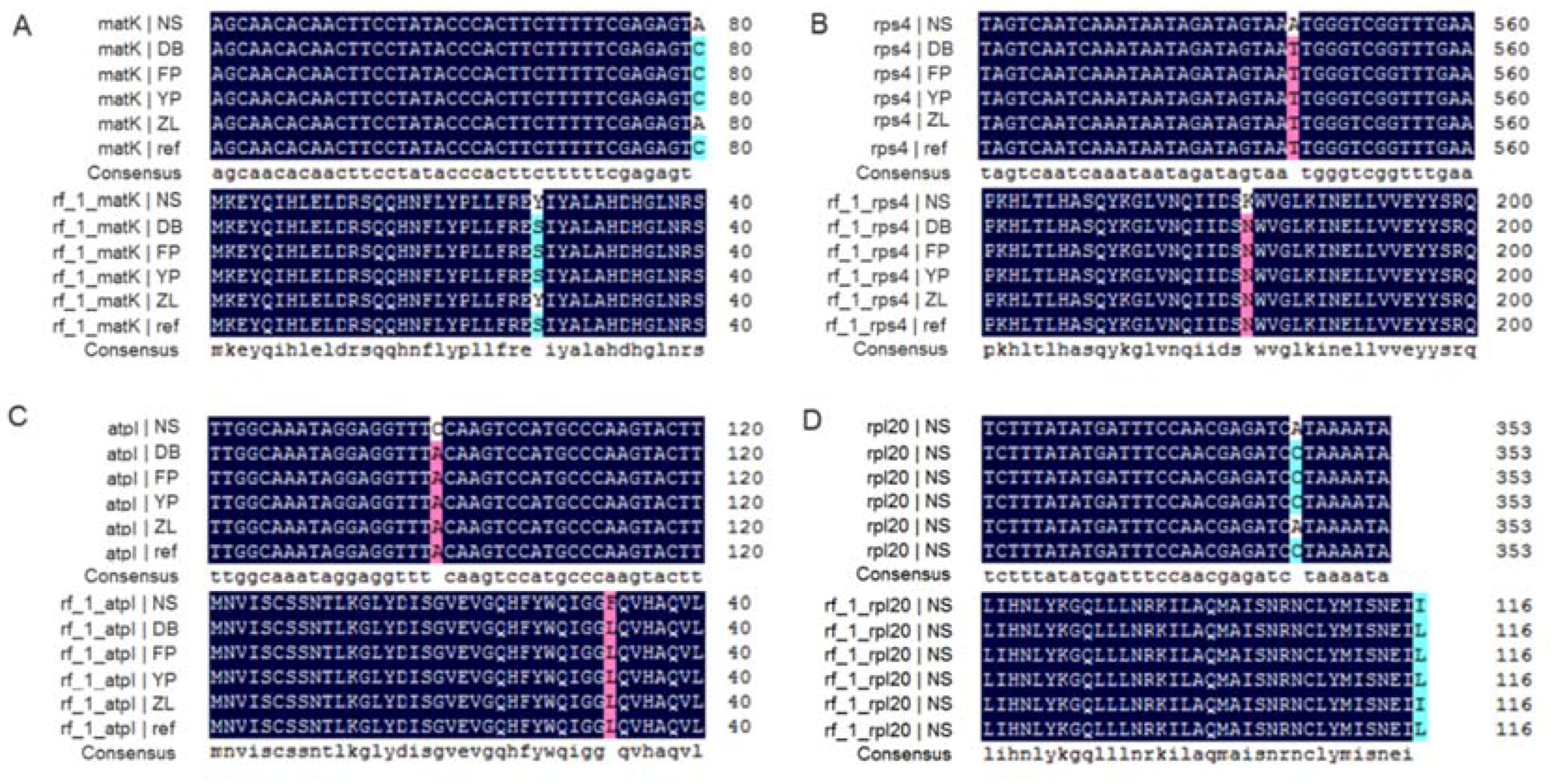
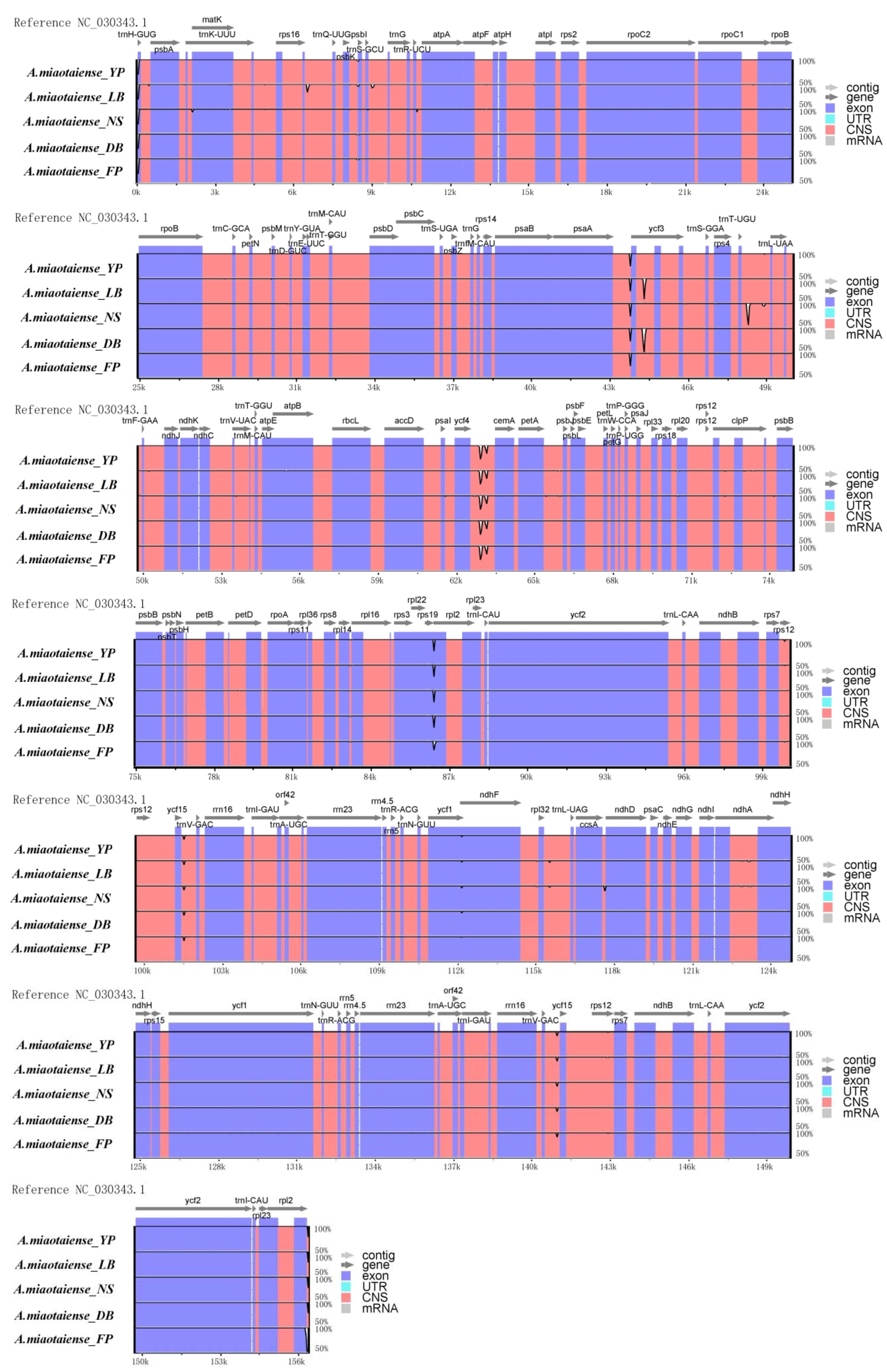
2.2. Comparative Genomic Analysis
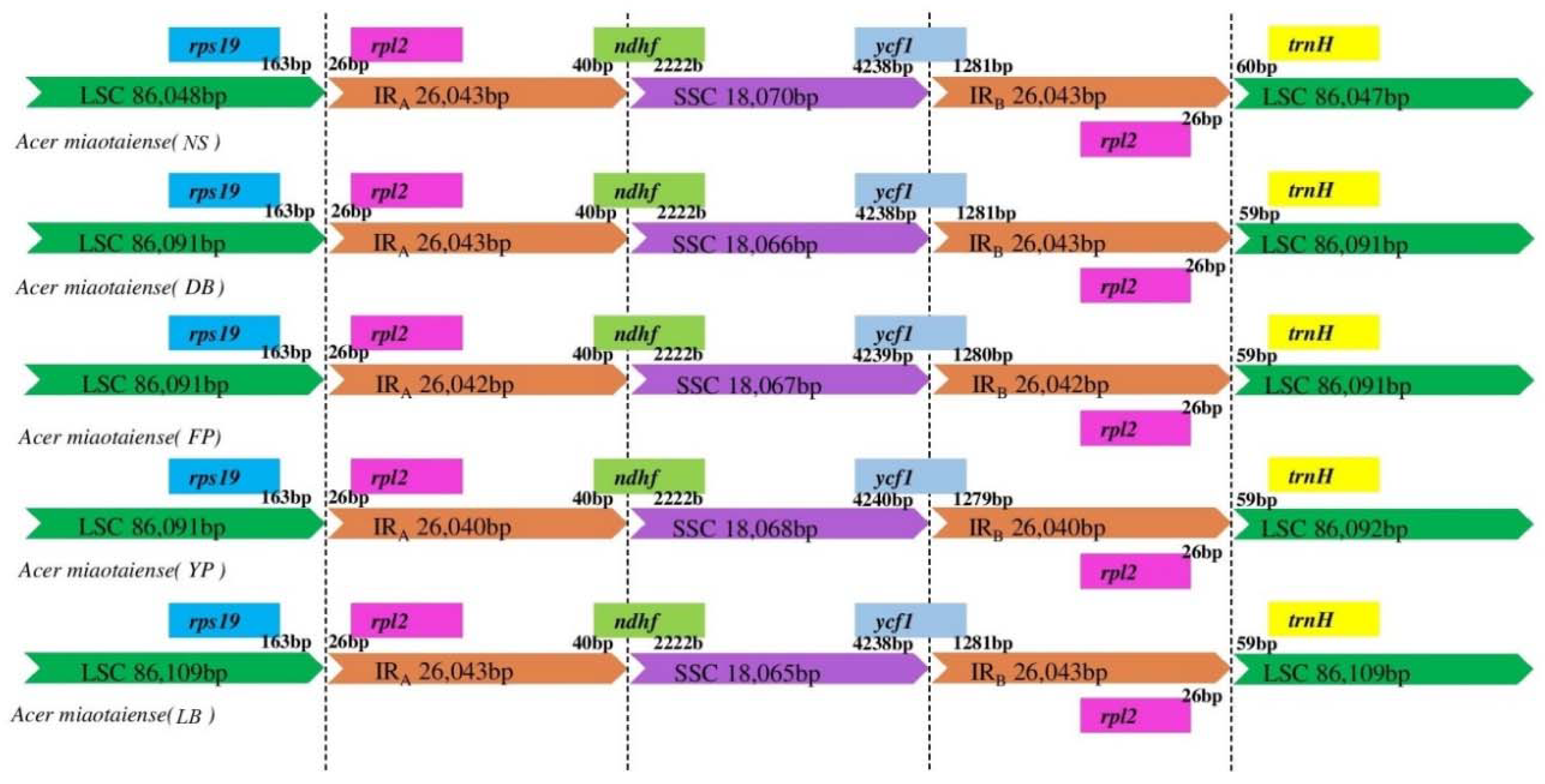
2.3. IR Expansion and Contraction
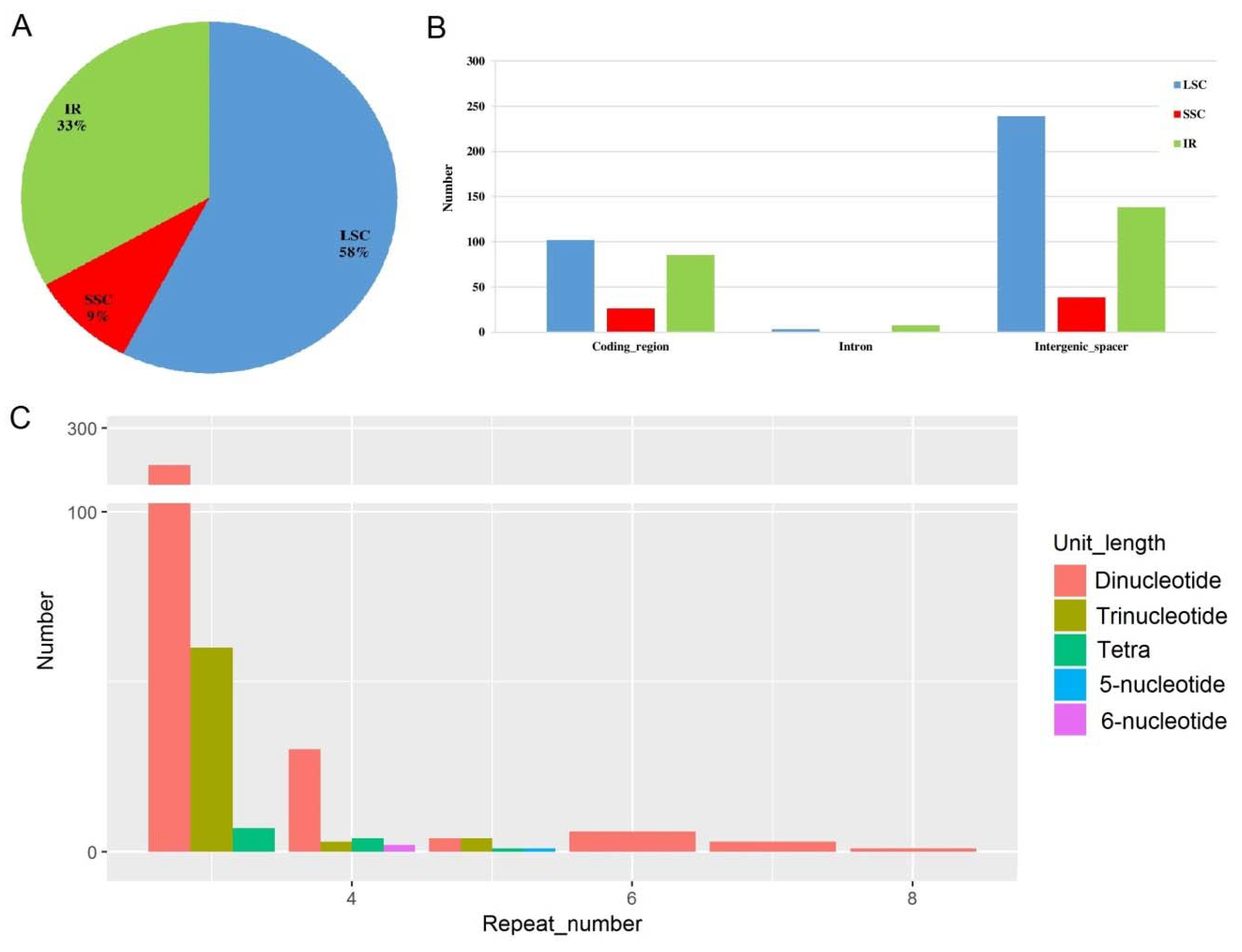
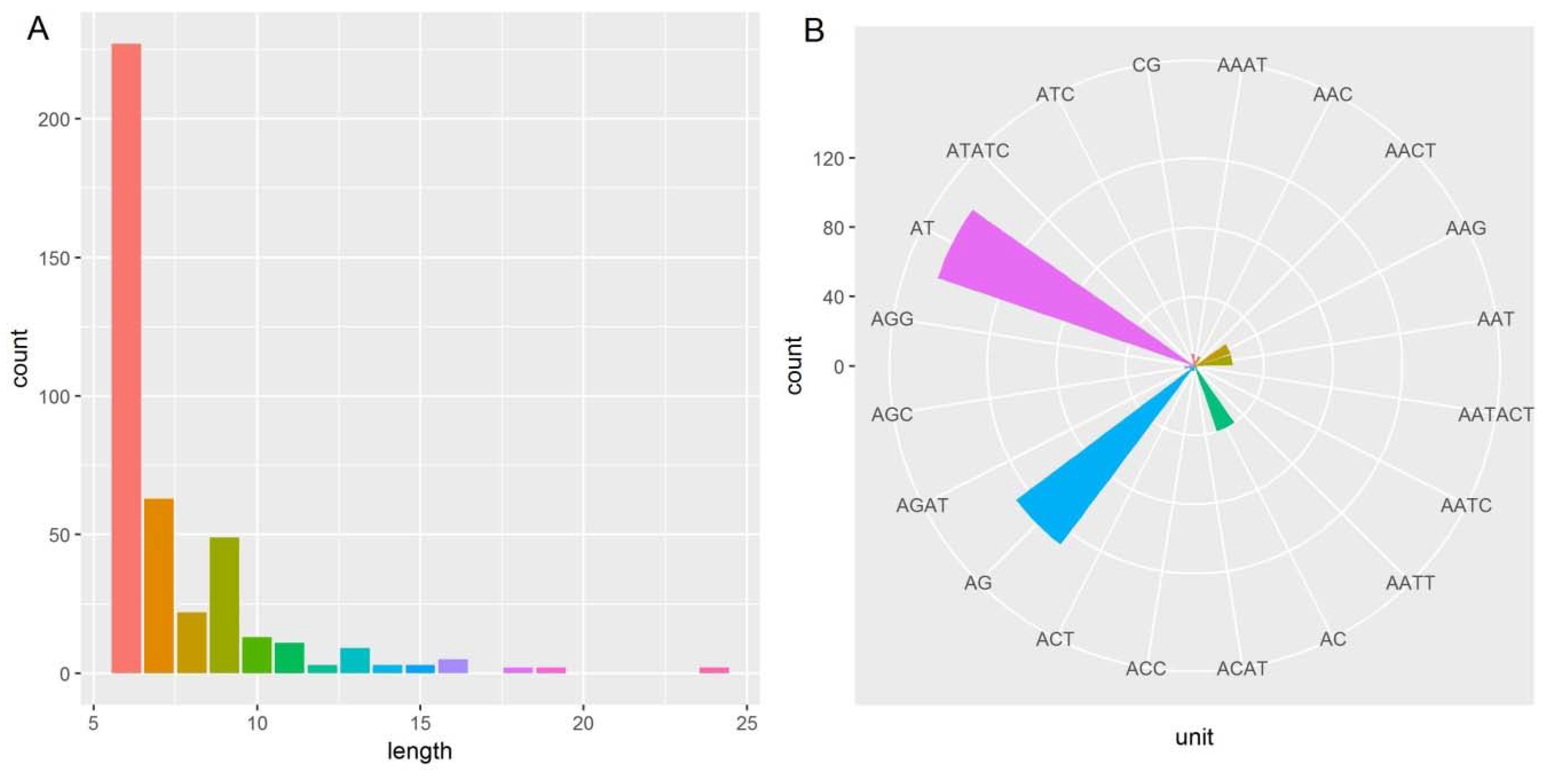
2.4. Microsatellite Detection Analysis
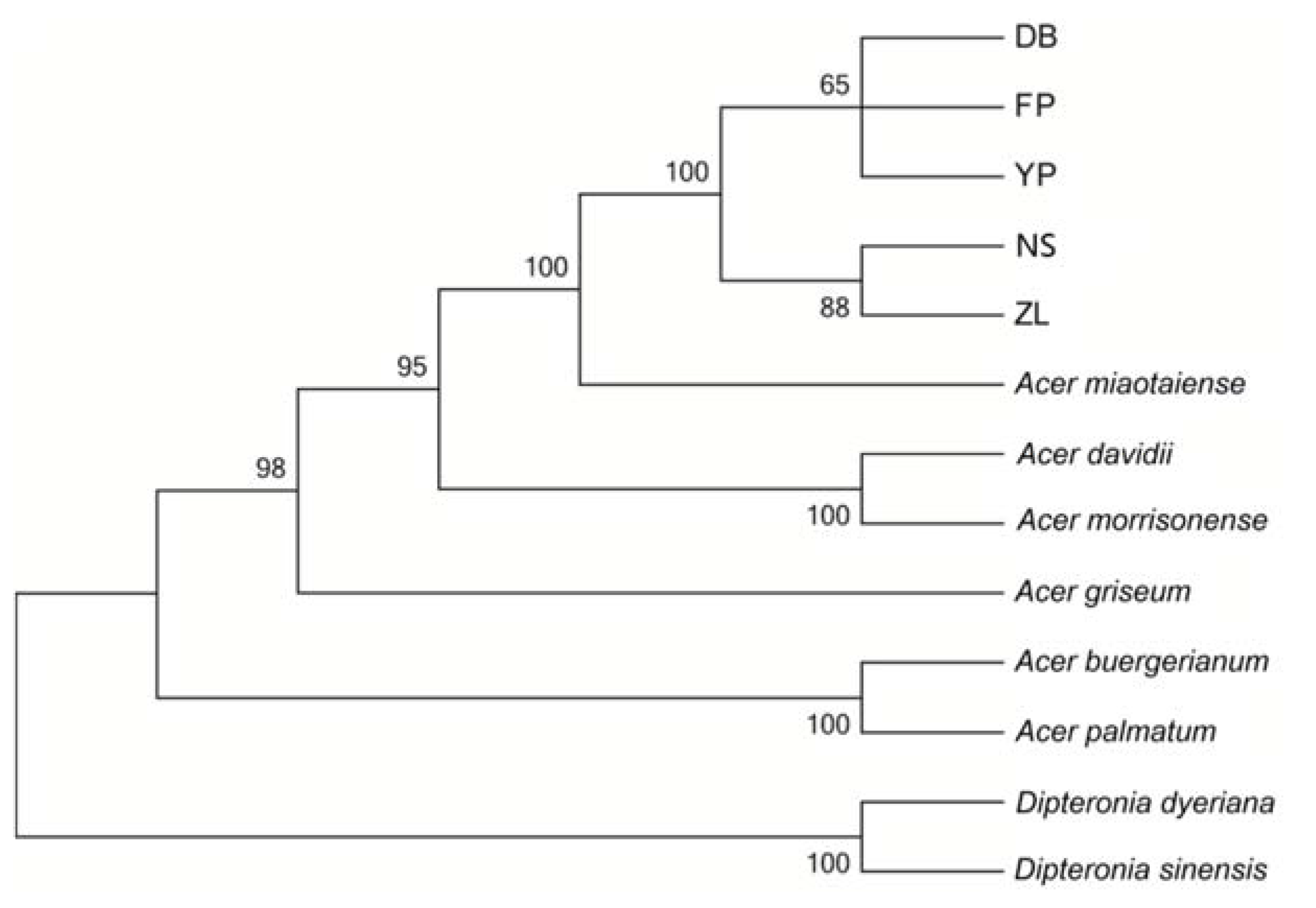
2.5. Phylogenetic Analysis
3. Materials and Methods
3.1. Plant Materials DNA Isolation
3.2. Chloroplast Genome Sequencing, Assembly, and Annotation
3.3. Comparative Genome Analysis
3.4. Microsatellite Detection Analysis
3.5. Phylogenetic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- IUCN. The IUCN Red List of Threatened Species. Available online: http://www.iucnredlist.org/details/46387/0 (accessed on 4 May 2018).
- Zhang, Y.; Li, B.; Chen, H.; Wang, Y. Characterization of the complete chloroplast genome of Acer miaotaiense (Sapindales: Aceraceae), a rare and vulnerable tree species endemic to China. Conserv. Genet. Resour. 2016, 8, 383–385. [Google Scholar] [CrossRef]
- Tang, W.; Wang, J.; Xu, J.; Wang, L.; Huang, J.; Chen, Y. Advances of Chemical Composition of Medicinal Plants in Aceraceae. North. Hortic. 2012, 36, 194–200. [Google Scholar]
- Cao, X.; Chen, K. Discussion on the endangering factors of Acer miaotaiense. Acta Bot. Boreali-Occident. Sin. 1996, 16, 12–15. [Google Scholar]
- Li, S.; Zheng, M. Discussion on the endangering factors of Acer miaotaiense. Shaanxi For. Technol. 2018, 46, 19–22. [Google Scholar]
- Qin, H.; Yang, Y.; Dong, S.; He, Q.; Jia, Y.; Zhao, L.; Yu, S.; Liu, H.; Liu, B.; Yan, Y.; et al. Threatened Species List of China’s Higher Plants. Biodivers. Sci. 2017, 25, 696–744. [Google Scholar] [CrossRef]
- Li, S.; Yan, G.; Zhao, G. Population genetic structure and genetic diversity of Acer miaotaiense. J. Northwest Univ. Nat. Sci. Ed. 2005, 35, 71–74. [Google Scholar] [CrossRef]
- Liu, J.; Qi, Z.C.; Zhao, Y.P.; Fu, C.X.; Xiang, Q.Y. Complete cpDNA genome sequence of Smilax china and phylogenetic placement of Liliales—Influences of gene partitions and taxon sampling. Mol. Phylogenet. Evol. 2012, 64, 545–562. [Google Scholar] [CrossRef] [PubMed]
- Brunkard, J.O.; Runkel, A.M.; Zambryski, P.C. Chloroplasts extend stromules independently and in response to internal redox signals. Proc. Natl. Acad. Sci. USA 2015, 112, 10044–10049. [Google Scholar] [CrossRef] [PubMed]
- Wicke, S.; Schneeweiss, G.M.; DePamphilis, C.W.; Müller, K.F.; Quandt, D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011, 76, 273–297. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.K.; Raubeson, L.A.; Boore, J.L.; DePamphilis, C.W.; Chumley, T.W.; Haberle, R.C.; Wyman, S.K.; Alverson, A.J.; Peery, R.; Herman, S.J.; et al. Methods for obtaining and analyzing whole chloroplast genome sequences. Methods Enzymol. 2005, 395, 348–384. [Google Scholar] [CrossRef] [PubMed]
- Daniell, H.; Lin, C.-S.; Yu, M.; Chang, W.-J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.Y.; Fritsch, P.W.; Ma, P.F.; Wang, H.; Lu, L.; Li, D.Z. Plastid phylogenomics and adaptive evolution of Gaultheria series Trichophyllae (Ericaceae), a clade from sky islands of the Himalaya-Hengduan Mountains. Mol. Phylogenet. Evol. 2017, 110, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Ma, P.F.; Li, H.T.; Li, D.Z. Complete plastid genome sequencing of four tilia species (Malvaceae): A comparative analysis and phylogenetic implications. PLoS ONE 2015, 10, e0142705. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.-H.; Wu, H.-B.; Gao, L.-Z. The complete chloroplast genome sequence of the threatened trident maple Acer buergerianum (Aceraceae). Mitochondrial DNA Part B 2017, 2, 273–274. [Google Scholar] [CrossRef]
- Li, Z.H.; Xie, Y.S.; Zhou, T.; Jia, Y.; He, Y.L.; Yang, J. The complete chloroplast genome sequence of Acer morrisonense (Aceraceae). Mitochondrial DNA Part A DNA Mapp. Seq. Anal. 2017, 28, 309–310. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Yang, J.; He, Y.L.; He, Y.; Niu, C.; Gong, L.L.; Li, Z.H. Characterization of the whole chloroplast genome sequence of Acer davidii Franch (Aceraceae). Conserv. Genet. Resour. 2016, 8, 141–143. [Google Scholar] [CrossRef]
- Jansen, R.K.; Cai, Z.; Raubeson, L.A.; Daniell, H.; DePamphilis, C.W.; Leebens-Mack, J.; Muller, K.F.; Guisinger-Bellian, M.; Haberle, R.C.; Hansen, A.K.; et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. USA 2007, 104, 19369–19374. [Google Scholar] [CrossRef] [PubMed]
- Magee, A.M.; Aspinall, S.; Rice, D.W.; Cusack, B.P.; Sémon, M.; Perry, A.S.; Stefanović, S.; Milbourne, D.; Barth, S.; Palmer, J.D.; et al. Localized hypermutation and associated gene losses in legume chloroplast genomes. Genome Res. 2010, 20, 1700–1710. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhou, J.G.; Chen, X.L.; Cui, Y.X.; Xu, Z.C.; Li, Y.H.; Song, J.Y.; Duan, B.Z.; Yao, H. Gene losses and partial deletion of small single-copy regions of the chloroplast genomes of two hemiparasitic Taxillus species. Sci. Rep. 2017, 7, 12834. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.K.; Saski, C.; Lee, S.B.; Hansen, A.K.; Daniell, H. Complete plastid genome sequences of three rosids (Castanea, Prunus, Theobroma): Evidence for at least two independent transfers of rpl22 to the nucleus. Mol. Biol. Evol. 2011, 28, 835–847. [Google Scholar] [CrossRef] [PubMed]
- Sheppard, A.E.; Timmis, J.N. Instability of plastid DNA in the nuclear genome. PLoS Genet. 2009, 5, e1000323. [Google Scholar] [CrossRef] [PubMed]
- Lohan, A.J.; Wolfe, K.H. A subset of conserved tRNA genes in plastid DNA of nongreen plants. Genetics 1998, 150, 425–433. [Google Scholar] [PubMed]
- Krause, K. From chloroplasts to “cryptic” plastids: Evolution of plastid genomes in parasitic plants. Curr. Genet. 2008, 54, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Ravin, N.V.; Gruzdev, E.V.; Beletsky, A.V.; Mazur, A.M.; Prokhortchouk, E.B.; Filyushin, M.A.; Kochieva, E.Z.; Kadnikov, V.V.; Mardanov, A.V.; Skryabin, K.G. The loss of photosynthetic pathways in the plastid and nuclear genomes of the non-photosynthetic mycoheterotrophic eudicot Monotropa hypopitys. BMC Plant Biol. 2016, 16, 238. [Google Scholar] [CrossRef] [PubMed]
- McCoy, S.R.; Kuehl, J.V.; Boore, J.L.; Raubeson, L.A. The complete plastid genome sequence of Welwitschia mirabilis: An unusually compact plastome with accelerated divergence rates. BMC Evol. Biol. 2008, 8, 130. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.S.; Lai, Y.T.; Lin, C.P.; Wang, Y.N.; Chaw, S.M. Evolution of reduced and compact chloroplast genomes (cpDNAs) in gnetophytes: Selection toward a lower-cost strategy. Mol. Phylogenet. Evol. 2009, 52, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Liere, K.; Link, G. RNA-binding activity of the matK protein encodecd by the chloroplast trnk intron from mustard (Sinapis alba L.). Nucleic Acids Res. 1995, 23, 917–921. [Google Scholar] [CrossRef] [PubMed]
- Barthet, M.M.; Hilu, K.W. Expression of matK: Functional and evolutionary implications. Am. J. Bot. 2007, 94, 1402–1412. [Google Scholar] [CrossRef] [PubMed]
- Zoschke, R.; Nakamura, M.; Liere, K.; Sugiura, M.; Borner, T.; Schmitz-Linneweber, C. An organellar maturase associates with multiple group II introns. Proc. Natl. Acad. Sci. USA 2010, 107, 3245–3250. [Google Scholar] [CrossRef] [PubMed]
- Gay, N.J.; Walker, J.E. The atp operon: Nucleotide sequence of the promoter and the genes for the membrane proteins, and the δ subunit of Escherichia coli ATP-synthase. Nucleic Acids Res. 1981, 9, 3919–3926. [Google Scholar] [CrossRef] [PubMed]
- Schluenzen, F.; Tocilj, A.; Zarivach, R.; Harms, J.; Gluehmann, M.; Janell, D.; Bashan, A.; Bartels, H.; Agmon, I.; Franceschi, F.; et al. Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell 2000, 102, 615–623. [Google Scholar] [CrossRef]
- Moore, P.B.; Steitz, T.A. The Structural Basis of Large Ribosomal Subunit Function. Annu. Rev. Biochem. 2003, 72, 813–850. [Google Scholar] [CrossRef] [PubMed]
- Bobik, K.; Burch-Smith, T.M. Chloroplast signaling within, between and beyond cells. Front. Plant Sci. 2015, 6, 781. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.T.; Kim, K.J. Chloroplast genome differences between Asian and American Equisetum arvense (Equisetaceae) and the origin of the hypervariable trnY-trnE intergenic spacer. PLoS ONE 2014, 9, e103898. [Google Scholar] [CrossRef] [PubMed]
- Mu, X.; Wang, P.; Du, J.; Gao, Y.G.; Zhang, J. The chloroplast genome of Cerasus humilis: Genomic characterization and phylogenetic analysis. PLoS ONE 2018, 13, e0196473. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, Y.; Yang, H.; Zhou, B. Chloroplast Genome of the Folk Medicine and Vegetable Plant Talinum paniculatum (Jacq.) Gaertn.: Gene Organization, Comparative and Phylogenetic Analysis. Molecules 2018, 23, 857. [Google Scholar] [CrossRef]
- Gu, C.; Dong, B.; Xu, L.; Tembrock, L.R.; Zheng, S.; Wu, Z. The complete chloroplast genome of Heimia myrtifolia and comparative analysis within myrtales. Molecules 2018, 23, 846. [Google Scholar] [CrossRef] [PubMed]
- Jian, H.Y.; Zhang, Y.H.; Yan, H.J.; Qiu, X.Q.; Wang, Q.G.; Li, S.B.; Zhang, S.D. The complete chloroplast genome of a key ancestor of modern roses, Rosa chinensis var. spontanea, and a comparison with congeneric species. Molecules 2018, 23, 389. [Google Scholar] [CrossRef]
- Raman, G.; Park, V.; Kwak, M.; Lee, B.; Park, S.J. Characterization of the complete chloroplast genome of Arabis stellari and comparisons with related species. PLoS ONE 2017, 12, e0183197. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Guo, L.; Zhao, W.; Xu, J.; Li, Y.; Zhang, X.; Shen, X.; Wu, M.; Hou, X. Complete chloroplast genome sequence and phylogenetic analysis of Paeonia ostii. Molecules 2018, 23, 246. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Xu, C.; Li, D.; Jin, X.; Li, R.; Lu, Q.; Suo, Z. Comparative analysis of the complete chloroplast genome sequences in psammophytic Haloxylon species (Amaranthaceae). PeerJ 2016, 4, e2699. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Xu, C.; Cheng, T.; Lin, K.; Zhou, S. Sequencing angiosperm plastid genomes made easy: A complete set of universal primers and a case study on the phylogeny of saxifragales. Genome Biol. Evol. 2013, 5, 989–997. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Chen, C.; Wei, Y.; Chang, Y.; Bai, G.; Li, Z.; Kanwal, N.; Zhao, G. Comparative Transcriptome and Chloroplast Genome Analyses of Two Related Dipteronia Species. Front. Plant Sci. 2016, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liu, B.; Cheng, F.; Ramchiary, N.; Choi, S.R.; Lim, Y.P.; Wang, X.-W. Sequencing of Chloroplast Genome Using Whole Cellular DNA and Solexa Sequencing Technology. Front. Plant Sci. 2012, 3, 243. [Google Scholar] [CrossRef] [PubMed]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2016, 45, e18. [Google Scholar] [CrossRef]
- Lohse, M.; Drechsel, O.; Bock, R. OrganellarGenomeDRAW (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007, 52, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Mayor, C.; Brudno, M.; Schwartz, J.R.; Poliakov, A.; Rubin, E.M.; Frazer, K.A.; Pachter, L.S.; Dubchak, I. Vista: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 2000, 16, 1046–1047. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are available by contacting the corresponding author J.Z. (yyzhj@nwsuaf.edu.cn). |
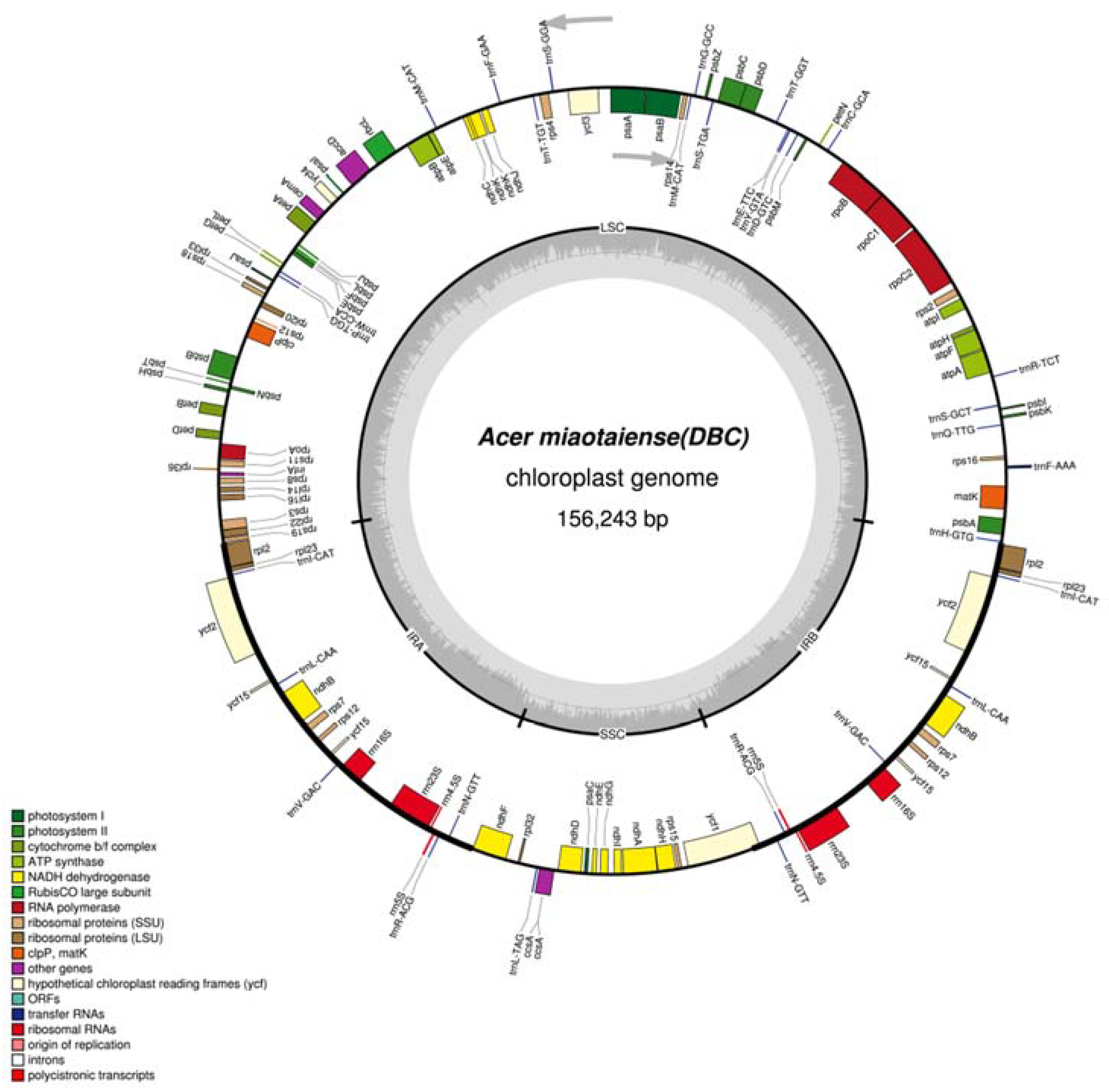
| Type | DB | YG | ZL | NS | FP | Average |
|---|---|---|---|---|---|---|
| Size (bp) | 156,243 | 156,239 | 156,260 | 156,204 | 156,242 | 156,238 |
| LSC length (bp) | 86,091 | 86,093 | 86,109 | 86,048 | 86,136 | 86,095 |
| SSC length (bp) | 18,068 | 18,068 | 18,067 | 18,072 | 18,068 | 18,069 |
| IR length (bp) | 52,084 | 52,078 | 52,084 | 52,084 | 52,038 | 52,074 |
| Number of gene | 130 | 130 | 130 | 130 | 130 | 130 |
| Protein-coding genes | 92 | 92 | 92 | 92 | 92 | 92 |
| tRNA genes | 30 | 30 | 30 | 30 | 30 | 30 |
| rRNA genes | 8 | 8 | 8 | 8 | 8 | 8 |
| Gene Functions | Gene Family | Gene Names |
|---|---|---|
| Photosynthesis | Subunits of ATP synthase | atpA, atpB, atpE, atpF, atpH, atpI |
| Subunits of NADH dehydrogenase | ndhA, ndhB, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Subunits of cytochrome | petA, petB, petD, petG, petL, petN | |
| Subunits of photosystem I | psaA, psaB, psaC, psaI, psaJ | |
| Subunits of photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| Subunit of rubisco | rbcL | |
| Other genes | Subunit of Acetyl-CoA-carboxylase | accD |
| c-type cytochrome synthesis gene | ccsA | |
| Envelop membrane protein | cemA | |
| Protease | clpP | |
| Translational initiation | infA | |
| Maturase | matK | |
| Self-replication | Large subunit of ribosome | rpl2, rpl14, rpl16, rpl20, rpl22, rpl23, rp32, rpl33, rpl36 |
| DNA dependent RNA polymerase | rpoA, rpoB, rpoC1, rpoC2 | |
| Small subunit of ribosome | rps2, rps3, rps4, rps7, rps8, rps11, rps12, rps14, rps15, rps16, rps18, rps19 | |
| rRNA Genes | rrn4.5S, rrn5S, rrn16S, rrn23S | |
| tRNA Genes | trnC-GCA, trnD-GTC, trnE-TTC, trnF-AAA, trnF-GAA, trnG-GCC, trnH-GTG, trnI-CAT, trnL-CAA, trnL-TAG, trnM-CAT, trnN-GTT, trnP-TGG, trnQ-TTG, trnR-ACG, trnR-TCT, trnS-GCT, trnS-GGA, trnS-TGA, trnT-GGT, trnT-TGT, trnV-GAC, trnW-CCA, trnY-GTA | |
| Unknown function | Conserved open reading frames | ycf1, ycf2, ycf3, ycf4, ycf15 |
| Code | Location | Longitude | Latitude | Altitude (m) |
|---|---|---|---|---|
| DB | Dianbingchang, Meixian country | E107°41′54″ | N34°4′19″ | 1427 |
| YP | Yangpigou, Taibai country | E107°40′54″ | N34°1′56″ | 1506 |
| ZL | Zhangliangmiao, Liuba country | E106°50′10″ | N33°40′58″ | 1224 |
| NS | Xunyangba, Ningshan country | E108°30′49″ | N33°33′40″ | 1224 |
| FP | Shiziba, Foping country | E107°52′31″ | N33°29′14″ | 945 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, J.; Xu, Y.; Xi, L.; Yang, J.; Chen, H.; Zhang, J. Characterization of the Chloroplast Genome Sequence of Acer miaotaiense: Comparative and Phylogenetic Analyses. Molecules 2018, 23, 1740. https://doi.org/10.3390/molecules23071740
Zhao J, Xu Y, Xi L, Yang J, Chen H, Zhang J. Characterization of the Chloroplast Genome Sequence of Acer miaotaiense: Comparative and Phylogenetic Analyses. Molecules. 2018; 23(7):1740. https://doi.org/10.3390/molecules23071740
Chicago/Turabian StyleZhao, Jiantao, Yao Xu, Linjie Xi, Junwei Yang, Hongwu Chen, and Jing Zhang. 2018. "Characterization of the Chloroplast Genome Sequence of Acer miaotaiense: Comparative and Phylogenetic Analyses" Molecules 23, no. 7: 1740. https://doi.org/10.3390/molecules23071740
APA StyleZhao, J., Xu, Y., Xi, L., Yang, J., Chen, H., & Zhang, J. (2018). Characterization of the Chloroplast Genome Sequence of Acer miaotaiense: Comparative and Phylogenetic Analyses. Molecules, 23(7), 1740. https://doi.org/10.3390/molecules23071740






