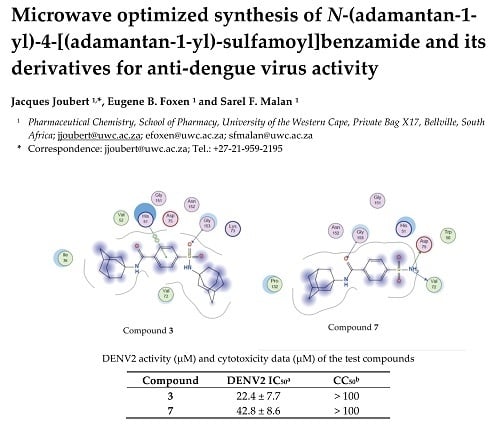
Microwave Optimized Synthesis of N-(adamantan-1-yl)-4-[(adamantan-1-yl)-sulfamoyl]benzamide and Its Derivatives for Anti-Dengue Virus Activity
Abstract
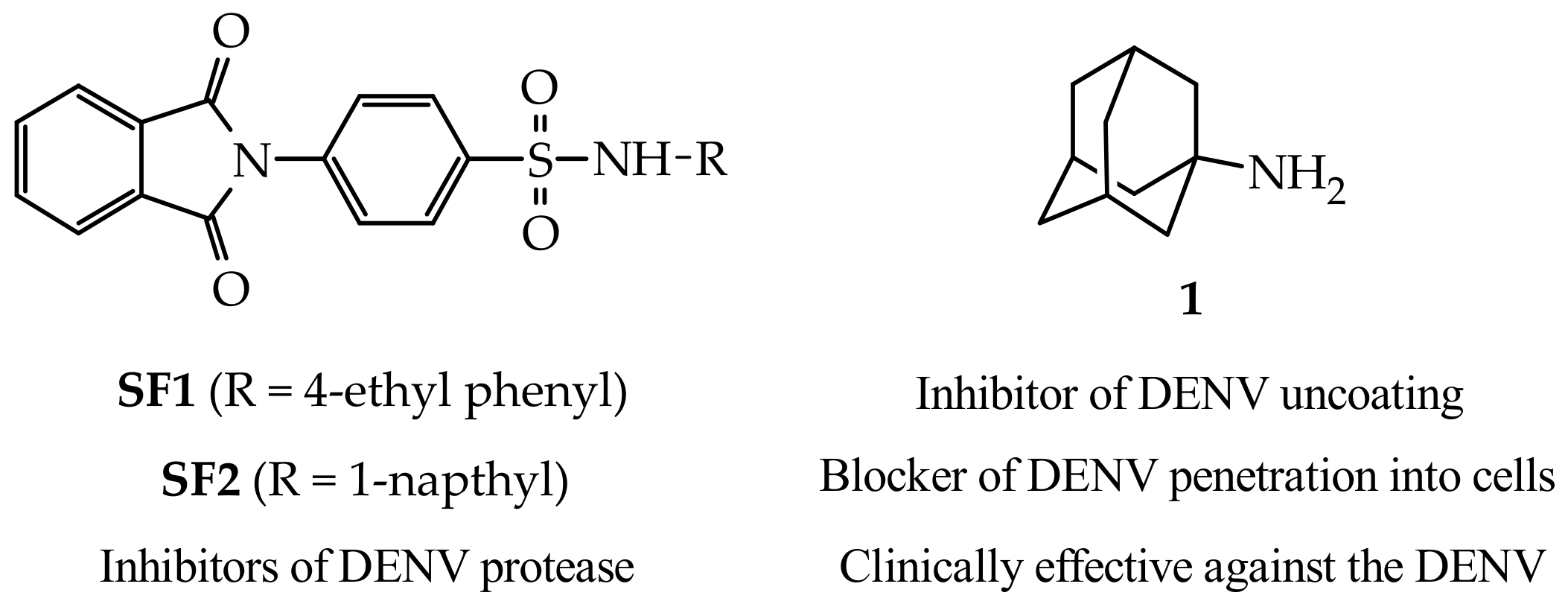
1. Introduction
2. Results and Discussion
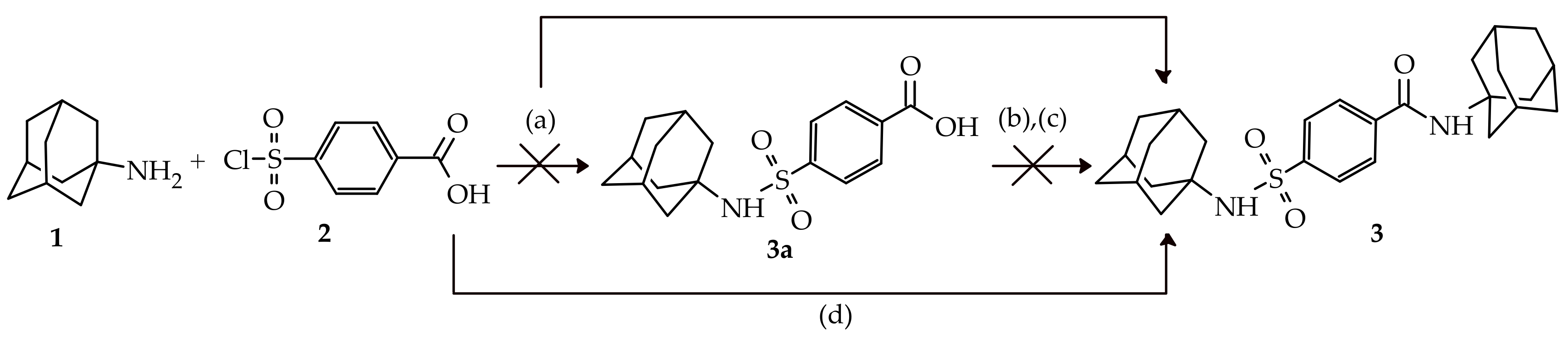
2.1. Chemistry
2.2. In-Vitro Anti-DENV2 Activity and Cytotoxicity Evaluations
2.3. Molecular Modelling
3. Materials and Methods
3.1. Chemistry
3.1.1. General Information
3.1.2. Synthesis of N-(adamantan-1-yl)-4-[(adamantan-1-yl)sulfamoyl]benzamide (3)
3.1.3. Synthesis of N-(adamantan-1-yl)benzamide (6)
3.1.4. Synthesis of N-(adamantan-1-yl)-4-sulfamoylbenzamide (7)
3.2. Anti-DENV2 Activity Assay
3.2.1. Cells and Viruses
3.2.2. Cell-Based Flavivirus Immunodetection (CFI) Assay
3.3. Cytotoxicity Assay
3.4. Molecular Modelling
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef] [PubMed]
- Henchal, E.A.; Putnak, J.R. The Dengue Viruses. Clin. Microbiol. Rev. 1990, 3, 376–396. [Google Scholar] [CrossRef] [PubMed]
- Poh, M.K.; Yip, A.; Zhang, S.; Priestle, J.P.; Ma, N.L.; Smit, J.M.; Wilschut, J.; Shi, P.; Wenk, M.R.; Schul, W. A small molecule fusion inhibitor of dengue virus. Antivir. Res. 2009, 84, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Holmes, E.C; Burch, S.S. The causes and consequences of genetic variation in dengue virus. Trends Microbiol. 2000, 8, 74–77. [Google Scholar] [CrossRef]
- Pugachev, K.V.; Guirakhoo, F.; Trent, D.W.; Monath, T.P. Traditional and novel approaches to flavivirus vaccines. Int. J. Parasitol. 2003, 33, 567–582. [Google Scholar] [CrossRef]
- Timiri, A.K.; Selvarasu, S.; Kesherwani, M.; Vijayan, V.; Sinha, B.N.; Devadasan, V.; Jayaprakash, V. Synthesis and molecular modelling studies of novel sulphonamide derivatives as dengue virus 2 protease inhibitors. Bioorgan. Chem. 2015, 62, 74–82. [Google Scholar] [CrossRef] [PubMed]
- De la Cruz, L.; Nguyen, T.H.D.; Ozawa, K.; Shin, J.; Graham, B.; Huber, T.; Otting, G. Binding of low molecular weight inhibitors promotes large conformational changes in the dengue virus NS2B–NS3 protease: fold analysis by pseudocontact shifts. J. Am. Chem. Soc. 2011, 133, 19205–19215. [Google Scholar] [CrossRef] [PubMed]
- Koff, W.C.; Elm., J.R.; Halstead, S.B. Inhibition of dengue virus replication by amantadine hydrochloride. Antimicrob. Agents Chemother. 1980, 18, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.C.; Chen, W.C. Treatment effectiveness of amantadine against dengue virus infection. Am. J. Case Rep. 2016, 17, 921–924. [Google Scholar] [CrossRef] [PubMed]
- Joubert, J.; Geldenhuys, W.J.; Van der Schyf, C.J.; Oliver, D.W.; Kruger, H.G.; Govender, T.; Malan, S.F. Polycyclic cage structures as lipophilic scaffolds for neuroactive drugs. ChemMedChem 2012, 7, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.C.; Cho, Y.H.; Lee, H.K.; Cho, S.H. A Facile One-Pot Transformation of Carboxylic Acids to Amides. Synth. Commun. 2007, 25, 2877–2881. [Google Scholar] [CrossRef]
- Joubert, J.; van Dyk, S.; Green, I.R.; Malan, S.F. Synthesis and evaluation of fluorescent heterocyclic aminoadamantanes as multifunctional neuroprotective agents. Bioorgan. Med. Chem. 2011, 19, 3935–3944. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Patel, S.J.; Vangrevelinghe, E.; Hao, Y.X.; Rao, R.; Jaber, D.; Schul, W.; Gu, F.; Heudi, O.; Ngai, L.M.; et al. A small-molecule dengue virus entry inhibitor Antimicrob. Agents Chemother. 2009, 53, 1823–1831. [Google Scholar] [CrossRef] [PubMed]
- Molecular Operating Environment MOE, Version 2018.01. Available online: http//www.chemcomp.com (accessed on 1 February 2018).
- Erbel, P.; Schiering, N.; D’Arcy, A.; Renatus, M.; Kroemer, M.; Lim, S.P.; Yin, Z.; Keller, T.H.; Vasudevan, S.G.; Hommel, U. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat. Struct. Mol. Biol. 2006, 13, 372–373. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of compounds 3, 6 and 7 are available from the authors upon request. |


| Compound | Time | % Yields | ||
|---|---|---|---|---|
| Conventional | MWAM | Conventional | MWAM | |
| 3 | 48 h 1 | 30 min 2 | 25 3 | 78 3 |
| 6 | 2 h | 10 min | 25 4 | 64 4 |
| 7 | 2 h | 10 min | 40 4 | 82 4 |
| Compound | DENV2 IC50 a | CC50 b |
|---|---|---|
| 1 | >100 | >100 |
| 3 | 22.4 ± 7.7 | >100 |
| 6 | >100 | >100 |
| 7 | 42.8 ± 8.6 | >100 |
| Compound | Binding Affinity | Residues Interacting with Ligand |
|---|---|---|
| SF1 | −7.835 | His51, Tyr151 |
| SF2 | −8.011 | His51, Leu128 |
| 1 | −4.234 | None |
| 3 | −7.413 | His51, Gly153 |
| 6 | −6.124 | None |
| 7 | −7.123 | Val72, Asp75, Gly153 |
| Bz-Nle-Lys-Arg-Arg-H | −11.323 | His51, Val72, Asp75, Asn152 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Joubert, J.; Foxen, E.B.; Malan, S.F. Microwave Optimized Synthesis of N-(adamantan-1-yl)-4-[(adamantan-1-yl)-sulfamoyl]benzamide and Its Derivatives for Anti-Dengue Virus Activity. Molecules 2018, 23, 1678. https://doi.org/10.3390/molecules23071678
Joubert J, Foxen EB, Malan SF. Microwave Optimized Synthesis of N-(adamantan-1-yl)-4-[(adamantan-1-yl)-sulfamoyl]benzamide and Its Derivatives for Anti-Dengue Virus Activity. Molecules. 2018; 23(7):1678. https://doi.org/10.3390/molecules23071678
Chicago/Turabian StyleJoubert, Jacques, Eugene B. Foxen, and Sarel F. Malan. 2018. "Microwave Optimized Synthesis of N-(adamantan-1-yl)-4-[(adamantan-1-yl)-sulfamoyl]benzamide and Its Derivatives for Anti-Dengue Virus Activity" Molecules 23, no. 7: 1678. https://doi.org/10.3390/molecules23071678
APA StyleJoubert, J., Foxen, E. B., & Malan, S. F. (2018). Microwave Optimized Synthesis of N-(adamantan-1-yl)-4-[(adamantan-1-yl)-sulfamoyl]benzamide and Its Derivatives for Anti-Dengue Virus Activity. Molecules, 23(7), 1678. https://doi.org/10.3390/molecules23071678