16S rDNA Profiling to Reveal the Influence of Seed-Applied Biostimulants on the Rhizosphere of Young Maize Plants
Abstract
1. Introduction
2. Results
2.1. Effects of Biostimulants on Plant Growth
2.2. Total Bacterial Community: Structure and Diversity
2.3. Effects of Biostimulants on the Rhizospheric Microbial Community of Maize
3. Discussion
4. Materials and Methods
4.1. Biostimulants Description
4.2. Evaluation of the Biostimulants’ Effects on Plant Growth
4.3. Site Description and Soil Sample Collection
4.4. DNA Extraction from Rhizospheric Soils
4.5. Rhizobiome Identification by 16S rRNA Gene Amplification and Sequencing, and Data Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- EBIC. What Are Biostimulants? 2012. Available online: http://www.biostimulants.eu/about/what-are-biostimulants (accessed on 1 January 2012).
- Trouvelot, S.; Héloir, M.; Poinssot, B.; Gauthier, A.; Paris, F.; Guillier, C. Carbohydrates in plant immunity and plant protection: Roles and potential application as foliar sprays. Front. Plant Sci. 2014, 5, 592. [Google Scholar] [CrossRef] [PubMed]
- Calvo, P.; Nelson, L.; Kloepper, J.W. Agricultural use of biostimulants. Plant Soil 2014, 383, 3–41. [Google Scholar] [CrossRef]
- Rose, M.T.; Patti, A.F.; Little, K.R.; Brown, A.L.; Jackson, W.R.; Cavagnaro, T.R. A meta-analysis and review of plant-growth response to humic substances: Practical implication for agriculture. In Advances in Agronomy; Sparks, D.L., Ed.; Academic Press: New York, NY, USA, 2014; Volume 124, pp. 37–89. [Google Scholar]
- Colla, G.; Rouphael, Y.; Canaguier, R.; Svecova, E.; Cardarelli, M. Biostimulant action of a plant-derived protein hydrolysate produced through enzymatic hydrolysis. Front. Plant Sci. 2014, 5, 448. [Google Scholar] [CrossRef] [PubMed]
- Popko, M.; Michalak, I.; Wilk, R.; Gramza, M.; Chojnacka, K.; Henryk Górecki, H. Effect of the New Plant Growth Biostimulants Based on Amino Acids on Yield and Grain Quality of Winter Wheat. Molecules 2018, 23, 470. [Google Scholar] [CrossRef] [PubMed]
- Brown, P.; Saa, S. Biostimulants in agriculture. Front. Plant Sci. 2015, 6, 671. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Montaño, F.; Alías-Villegas, C.; Bellogín, R.A.; del Cerro, P.; Espuny, M.R.; Jiménez-Guerrero, I.; López-Baena, F.J.; Ollero, F.J.; Cubo, T. Plant growth promotion in cereal and leguminous agricultural important plants: From microorganism capacities to crop production. Microbiol. Res. 2014, 169, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Timmusk, S.; Behers, L.; Muthoni, J.; Muraya, A.; Aronsson, A.-C. Perspectives and challenges of microbial application for crop improvement. Front. Plant Sci. 2017, 8, 49. [Google Scholar] [CrossRef] [PubMed]
- Vejan, P.; Abdullah, R.; Khadiran, T.; Salmah Ismail, S.; Boyce, A.M. Role of Plant Growth Promoting Rhizobacteria in Agricultural Sustainability—A Review. Molecules 2016, 21, 573. [Google Scholar] [CrossRef] [PubMed]
- Colla, G.; Hoagland, L.; Ruzzi, M.; Cardarelli, M.; Bonini, P.; Canaguier, R.; Rouphael, Y. Biostimulant Action of protein hydrolysates: Unraveling their effects on plant physiology and microbiome. Front. Plant Sci. 2017, 8, 2202. [Google Scholar] [CrossRef] [PubMed]
- Dal Cortivo, C.; Barion, G.; Visioli, G.; Mattarozzi, M.; Mosca, G.; Vamerali, T. Increased root growth and nitrogen accumulation followed by PGPR inoculation in common wheat: Assessment of plant-microbe interactions by ESEM. Agric. Ecosyst. Environ. 2017, 247, 396–408. [Google Scholar] [CrossRef]
- Tejada, M.; Benitez, C.; Gomez, I.; Parrado, J. Use of biostimulants on soil restoration: Effects on soil biochemical properties and microbial community. Appl. Soil Ecol. 2011, 49, 11–17. [Google Scholar] [CrossRef]
- Galeote-Correa, D.; Bedmar, E.J.; Fernández-González, A.J.; Fernández-López, M.; Arone, G.J. Bacterial communities in the rhizosphere of amilaceous maize (Zea mays L.) as assessed by pyrosequencing. Front. Plant Sci. 2016, 7, 1016. [Google Scholar] [CrossRef] [PubMed]
- Bakker, M.; Chaparro, J.; Manter, D.; Vivanco, J. Impacts of bulk soil microbial community structure on rhizosphere microbiomes of Zea mays. Plant Soil 2015, 392, 1–12. [Google Scholar] [CrossRef]
- Peiffer, J.A.; Lery, R.E. Exploring the maize rhizosphere microbiome in the field: A glimpse into highly complex system. Commun. Integr. Biol. 2013, 6, e25177. [Google Scholar] [CrossRef] [PubMed]
- Peiffer, J.A.; Spor, A.; Koren, O.; Jin, Z.; Tringe, S.G.; Dangl, J.L.; Buckler, E.S.; Ley, R.E. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. USA 2013, 110, 6548–6553. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Rui, J.; Xiong, J.; Li, J.; He, Z.; Zhou, J.; Yannarell, A.C.; Mackie, R.I. Functional potential of soil microbial communities in the maize rhizosphere. PLoS ONE 2014, 9, e112609. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Rui, J.; Mao, Y.; Yannarell, A.; Mackie, R. Dynamics of the bacterial community structure in the rhizosphere of a maize cultivar. Soil Biol. Biochem. 2014, 68, 392–401. [Google Scholar] [CrossRef]
- Carrera, L.M.; Buyer, J.S.; Vinyard, B.; Abdul-Baki, A.A.; Sikora, L.J.; Teasdale, J.R. Effects of cover crops, compost, and manure amendments on soil microbial community structure in tomato production systems. Appl. Soil Ecol. 2007, 37, 247–255. [Google Scholar] [CrossRef]
- Jack, A.L.H.; Rangarajan, A.; Culman, S.W.; Sooksa-Nguan, T.; Thies, J.E. Choice of organic amendments in tomato transplants has lasting effects on bacterial rhizosphere communities and crop performance in the field. Appl. Soil Ecol. 2011, 48, 94–101. [Google Scholar] [CrossRef]
- Van Passel, M.W.J.; Kant, R.; Palva, A.; Copeland, A.; Lucas, S.; Lapidus, A.; Glavina del Rio, T.; Pitluck, S.; Goltsman, E.; Clum, A.; et al. Genome sequence of the Verrucomicrobium Opitutus terrae PB90-1, an abundant inhabitant of rice paddy soil ecosystems. J. Bacteriol. 2011, 193, 2367–2368. [Google Scholar] [CrossRef] [PubMed]
- Dabert, P.; Sialve, B.; Delgenès, J.P.; Moletta, R.; Godon, J.-J. Characterisation of the microbial 16S rDNA diversity of an aerobic phosphorus-removal ecosystem and monitoring of its transition to nitrate respiration. Appl. Microbiol. Biotechnol. 2001, 55, 500–509. [Google Scholar] [CrossRef] [PubMed]
- Taylor, W.J.; Draughon, F.A. Nannocystis exedens: A potential biocompetitive agent against Aspergillus flavus and Aspergillus parasiticus. J. Food Prot. 2001, 64, 1030–1034. [Google Scholar] [CrossRef] [PubMed]
- Ertani, A.; Cavani, L.; Pizzeghello, D.; Brandellero, E.; Altissimo, A.; Ciavatta, C.; Nardi, S. Biostimulant activity of two protein hydrolyzates in the growth and nitrogen metabolism of maize seedlings. J. Plant Nutr. Soil Sci. 2009, 172, 237–244. [Google Scholar] [CrossRef]
- Ertani, A.; Schiavon, M.; Muscolo, A.; Nardi, S. Alfalfa plant-derived biostimulant stimulate short-term growth of salt stressed Zea mays L. plants. Plant Soil 2013, 364, 145–158. [Google Scholar] [CrossRef]
- Francis, P.B.; Larry, D.; Earnest, L.D.; Bryant, K. Maize growth and yield response to a biostimulant amendment. J. Crop Improv. 2016, 30, 632–640. [Google Scholar] [CrossRef]
- Du Jardin, P. Plant biostimulants: Definition, concept, main categories and regulation. Sci. Hortic. 2015, 196, 3–14. [Google Scholar] [CrossRef]
- Fleet, C.M.; Sun, T.P. A DELLAcate balance: The role of gibberellin in plant morphogenesis. Curr. Opin. Plant Biol. 2005, 8, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Overvoorde, P.; Fukaki, H.; Beeckman, T. Auxin control of root development. Cold Spring Harb. Perspect. Biol. 2010, 2, a001537. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Subler, S.; Edwards, C.A. Effects of agricultural biostimulants on soil microbial activity and nitrogen dynamics. Appl. Ecol. 2002, 19, 249–259. [Google Scholar] [CrossRef]
- Owen, A.G.; Jones, D.L. Competition for amino acids between wheat roots and rhizosphere microorganisms and the role of amino acids in plant N acquisition. Soil Biol. Biochem. 2001, 33, 651–657. [Google Scholar] [CrossRef]
- Castellanos, T.; Dohrmann, A.B.; Imfeld, G.; Baumgarte, S.; Tebbe, C.C. Search of environmental descriptors to exaplain the variability of the bacterial diversity from maize rhizospheres across regional scale. Eur. J. Soil Biol. 2009, 45, 383–393. [Google Scholar] [CrossRef]
- Berg, G.; Smalla, K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 2009, 68, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Green, J.; Bohannan, B.J. Spatial scaling of microbial diversity. Trends Ecol. Evol. 2006, 21, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Q.; Wang, F.; Zhang, J.; Chen, Y.; Zhang, C.; Liu, G.; Zhang, H.; Ma, C.; Zhang, J. The Variation in the Rhizosphere Microbiome of Cotton with Soil Type, Genotype and Developmental Stage. Sci. Rep. 2017, 7, 3940. [Google Scholar] [CrossRef] [PubMed]
- Kamutando, C.N.; Vikram, S.; Kamgan-Nkuekam, G.; Makhalanyane, T.P.; Greve, M.; Le Roux, J.J.; Richardson, D.M.; Cowan, D.; Valverde, A. Soil nutritional status and biogeography influence rhizosphere microbial communities associated with the invasive tree Acacia dealbata. Sci. Rep. 2017, 7, 6472. [Google Scholar] [CrossRef] [PubMed]
- García-Salamanca, A.; Molina-Henares, M.A.; van Dillewijn, P.; Solano, J.; Pizarro- Tobías, P.; Roca, A.; Duque, E.; Ramos, J.L. Bacterial diversity in the rhizosphere of maize and the surrounding carbonate-rich bulk soil. Microb. Biotechnol. 2013, 6, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Johnston-Monje, D.; Lundberg, D.S.; Lazarovits, G.; Reis, V.M.; Raizada, M.N. Bacterial populations in juvenile maize rhizosphere originate from both seed and soil. Plant Soil 2016, 405, 337–355. [Google Scholar] [CrossRef]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef] [PubMed]
- Hocking, D.; Cook, E.D. Myxobacteria exert partial control of damping-off and root rot disease in container grown tree seedling. Can. J. Microbiol. 1972, 18, 1156–1157. [Google Scholar] [CrossRef]
- Rosenberg, E.; Varon, M. Antibiotics and lytic enzymes. In Myxobactera: Development and Cell Interactions; Rosenberg, E., Ed.; Springer: New York, NY, USA, 1984; pp. 109–125. [Google Scholar]
- Lancashire, P.D.; Bleiholder, H.; Van Den Boom, T.; Langeluddeke, P.; Stauss, R.; Weber, E.; Witzenberger, A. A uniform decimal code for growth stages of crops and weeds. Ann. Appl. Biol. 1991, 119, 561–601. [Google Scholar] [CrossRef]
- Milani, C.; Hevia, A.; Foroni, E.; Duranti, S.; Turroni, F.; Lugli, G.A.; Sanchez, B.; Martín, R.; Gueimonde, M.; van Sinderen, D.; et al. Assessing the fecal microbiota: An optimized ion torrent 16S rRNA gene-based analysis protocol. PLoS ONE 2013, 8, e68739. [Google Scholar] [CrossRef] [PubMed]
- Mancabelli, L.; Ferrario, C.; Milani, C.; Mangifesta, M.; Turroni, F.; Duranti, S.; Lugli, G.A.; Viappiani, A.; Ossiprandi, M.C.; van Sinderen, D.; et al. Insides into the biodiversity of the gut microbiota of broiler chickens. Environ. Microbiol. 2016, 18, 4727–4738. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Gonzalez Peña, A.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Meth. 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |

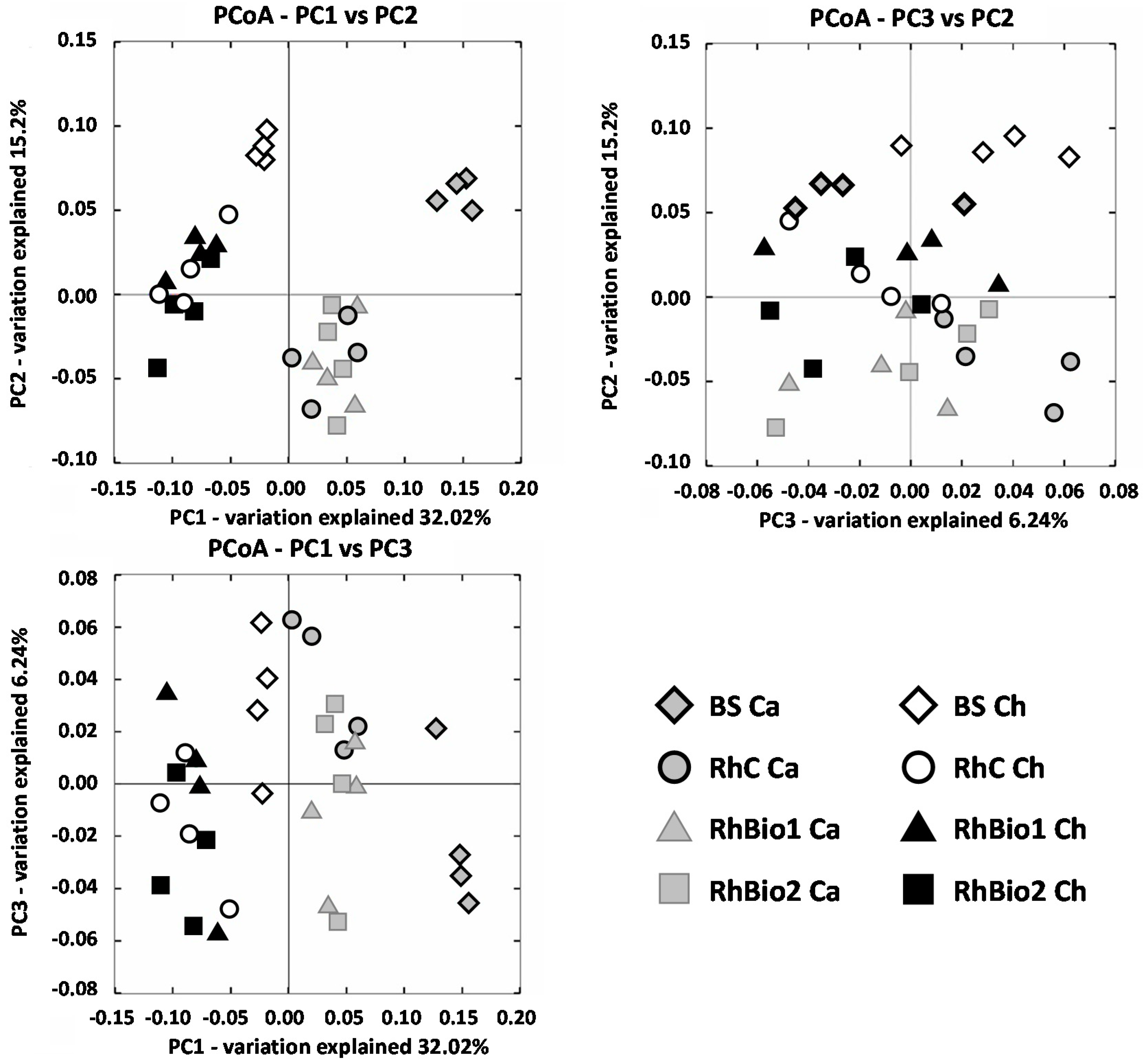
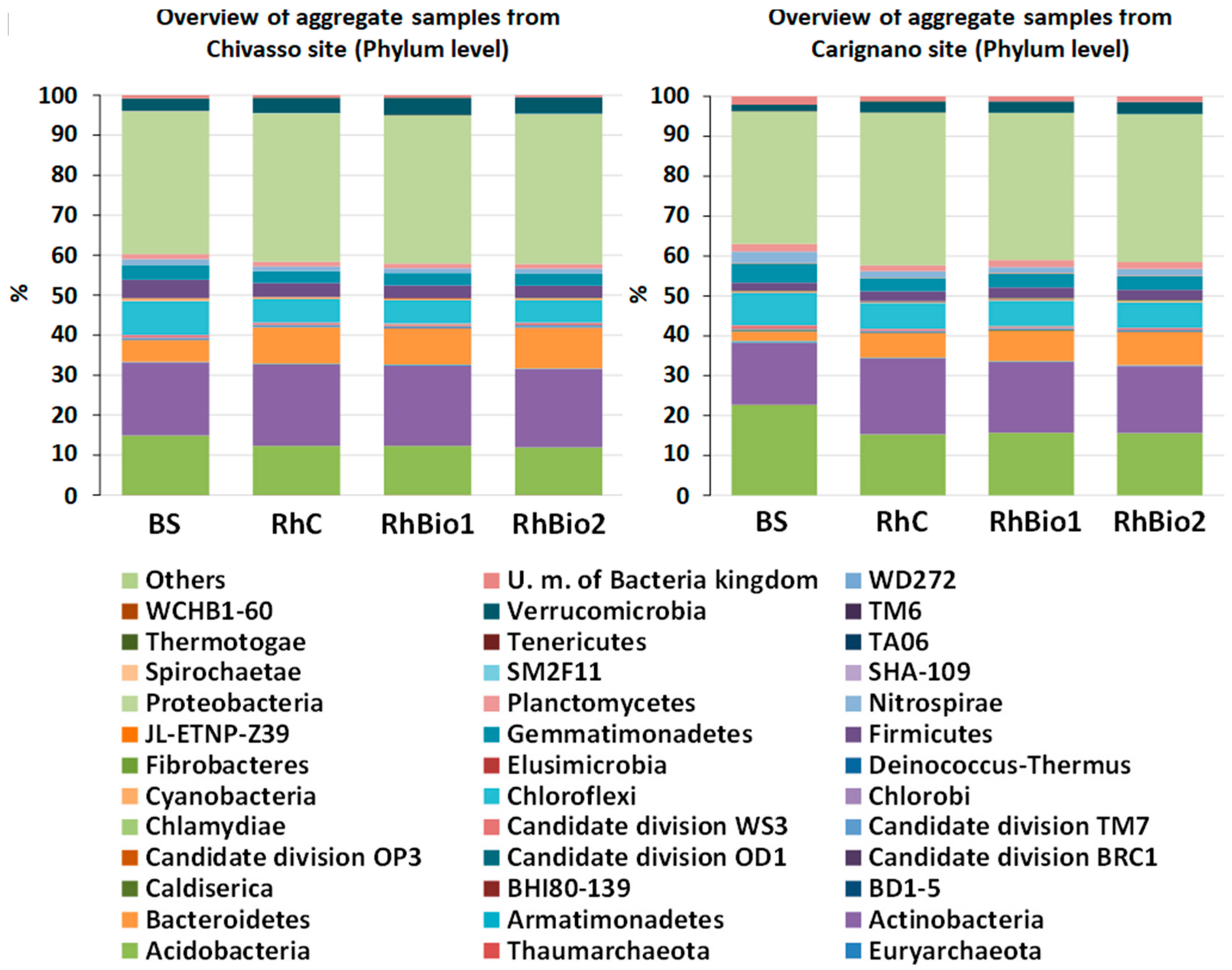
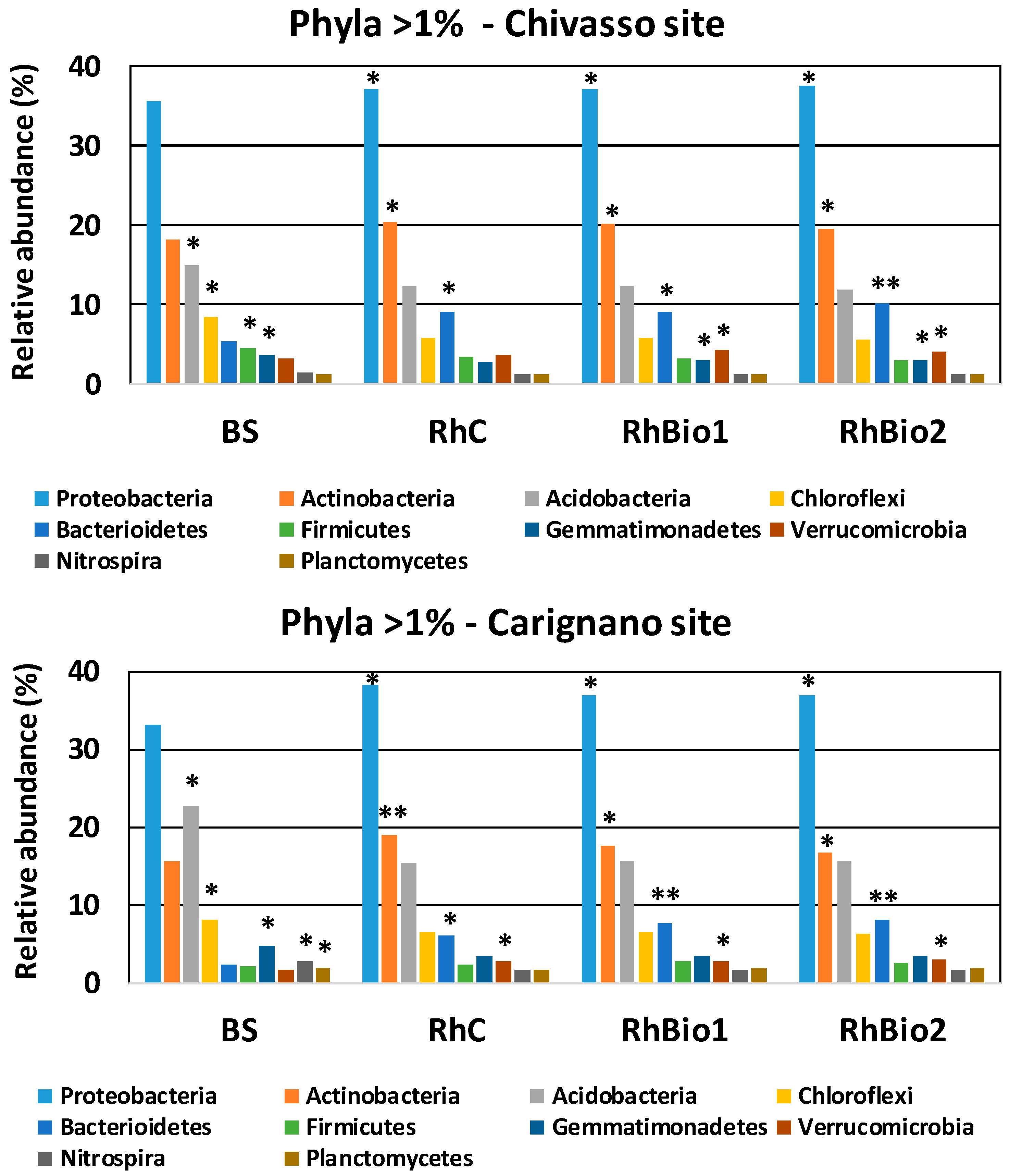
| Sites | Treatment | Number of OTUs | Chao1 Estimate of Richness | Shannon’s Diversity Index |
|---|---|---|---|---|
| Carignano | BS | 4973 ± 96a | 6135 ± 148a | 9.982 ± 0.021a |
| RhC | 4824 ± 122a | 6107 ± 65a | 9.813 ± 0.066a | |
| RhBio1 | 5035 ± 125a | 6114 ± 45a | 9.832 ± 0.115a | |
| RhBio2 | 4970 ± 258a | 6207 ± 123a | 9.883 ± 0.104a | |
| Chivasso | BS | 5522 ± 327a | 6397 ± 464a | 9.948 ± 0.244a |
| RhC | 5271 ± 132a | 6260 ± 182a | 10.005 ± 0.103a | |
| RhBio1 | 5203 ± 148a | 6242 ± 115a | 10.041 ± 0.117a | |
| RhBio2 | 5023 ± 622a | 6210 ± 130a | 10.067 ± 0.108a |
| Taxonomy | RhC | RhBio1 | Relative Difference a | p-Value | RhBio2 | Relative Difference a | p-Value |
|---|---|---|---|---|---|---|---|
| U.M. of Subgroup 5 order | 0.01% | 0.004% | 143% | 0.038 * | 0.007% | 35% | 0.297 |
| U.M. of Holophagae class | 0.024% | 0.033% | −29% | 0.289 | 0.042% | −44% | 0.049 * |
| Smaragdicoccus | 0.002% | 0.002% | −22% | 0.717 | 0.000% | 399% | 0.008 ** |
| Quadrisphaera | 0.004% | 0.003% | 46% | 0.512 | 0.002% | 174% | 0.043 * |
| Frigoribacterium | 0.000% | 0.002% | −81% | 0.021 * | 0.000% | 22% | 0.895 |
| Pseudoclavibacter | 0.01% | 0.004% | 148% | 0.003 * | 0.008% | 19% | 0.469 |
| Microlunatus | 0.378% | 0.311% | 21% | 0.072 | 0.267% | 41% | 0.026 * |
| Crossiella | 0.005% | 0.002% | 249% | 0.016 * | 0.003% | 60% | 0.301 |
| Actinocorallia | 0.024% | 0.046% | −47% | 0.012 * | 0.032% | −24% | 0.222 |
| Euzebya | 0.000% | 0.002% | absent in RhC | 0.024 * | 0.001% | absent in RhC | 0.356 |
| Chryseolinea | 0.233% | 0.333% | −30% | 0.083 | 0.351% | −34% | 0.046 * |
| U.M. of Cytophagaceae family | 0.529% | 0.825% | −36% | 0.047 * | 0.807% | −34% | 0.191 |
| U.M. of AKYH767 family | 0.091% | 0.136% | −33% | 0.021 * | 0.1% | −9% | 0.584 |
| Niastella | 0.208% | 0.224% | −7% | 0.865 | 0.317% | −34% | 0.0 *** |
| Terrimonas | 0.284% | 0.328% | −13% | 0.181 | 0.355% | −20% | 0.027 * |
| U.M. of PHOS-HE51 family | 0.057% | 0.096% | −41% | 0.027 * | 0.089% | −36% | 0.148 |
| U.M. of 08D2Z23 family | 0.001% | 0.005% | −89% | 0.030 * | 0.001% | −64% | 0.443 |
| U.M. of Lineage Iia order (Elusimicrobia class) | 0.041% | 0.059% | −30% | 0.031 * | 0.039% | 4% | 0.747 |
| U.M. of Fibrobacteraceae family | 0.069% | 0.106% | −34% | 0.290 | 0.097% | −29% | 0.023 * |
| U.M. of Bacillaceae family | 0.08% | 0.06% | 34% | 0.680 | 0.045% | 78% | 0.011 * |
| Brevibacillus | 0.013% | 0.009% | 51% | 0.184 | 0.006% | 123% | 0.025 * |
| U.M. of Paenibacillaceae family | 0.045% | 0.027% | 67% | 0.013 * | 0.028% | 62% | 0.019 * |
| Lutispora | 0.008% | 0.002% | 203% | 0.035 * | 0.006% | 34% | 0.500 |
| Blautia | 0.006% | 0.003% | 94% | 0.045 * | 0.003% | 124% | 0.049 * |
| Pseudobutyrivibrio | 0.006% | 0.001% | 293% | 0.044 * | 0.004% | 45% | 0.482 |
| Desulfitobacterium | 0.003% | 0.002% | 46% | 0.385 | 0.001% | 211% | 0.046 * |
| Thermincola | 0.004% | 0.001% | 320% | 0.036 * | 0.003% | 67% | 0.370 |
| U.M. of Peptococcaceae family | 0.002% | 0.001% | 101% | 0.459 | 0.000% | absent in RhBio2 | 0.044 * |
| Ruminococcus | 0.003% | 0.003% | 15% | 0.787 | 0.001% | 562% | 0.042 * |
| Phascolarctobacterium | 0.002% | 0.003% | −38% | 0.658 | 0.0% | 399% | 0.008 ** |
| U.M. of OPB54 class | 0.046% | 0.028% | 67% | 0.007 ** | 0.03% | 55% | 0.033 * |
| Hyphomicrobium | 0.29% | 0.254% | 14% | 0.124 | 0.226% | 28% | 0.009 ** |
| U.M. of JG35-K1-AG5 family | 0.194% | 0.183% | 6% | 0.568 | 0.153% | 27% | 0.046 * |
| Rhodobium | 0.564% | 0.474% | 19% | 0.095 | 0.432% | 30% | 0.015 * |
| U.M. of Xanthobacteraceae family | 2.5% | 2.4% | 3% | 0.767 | 1.9% | 26% | 0.028 * |
| U.M. of Rhizobiales order | 0.78% | 0.748% | 4% | 0.455 | 0.656% | 19% | 0.017 * |
| Rhodovastum | 0.004% | 0.005% | −23% | 0.436 | 0.008% | −52% | 0.046 * |
| U.M. of I-10 family | 0.051% | 0.034% | 49% | 0.055 | 0.037% | 37% | 0.004 ** |
| Dongia | 0.007% | 0.001% | 362% | 0.048 * | 0.002% | 226% | 0.099 |
| U.M. of SM2D12 family | 0.003% | 0.004% | −6% | 0.901 | 0.009% | −61% | 0.023 * |
| U.M. of ARKDMS-49 class | 0.000% | 0.002% | absent in RhC | 0.041 * | 0.000% | absent in RhC and RhBio2 | |
| Rivibacter | 0.006% | 0.006% | −3% | 0.913 | 0.003% | 128% | 0.037 * |
| Nannocystis | 0.036% | 0.045% | −20% | 0.015 * | 0.052% | −31% | 0.001 *** |
| Aquicella | 0.016% | 0.007% | 138% | 0.019 * | 0.015% | 4% | 0.915 |
| U.M. of Solimonadaceae family | 0.161% | 0.233% | −31% | 0.149 | 0.264% | −39% | 0.037 * |
| Silanimonas | 0.006% | 0.002% | 298% | 0.048 * | 0.004% | 54% | 0.293 |
| Stenotrophomonas | 0.033% | 0.031% | 4% | 0.923 | 0.114% | −71% | 0.003 ** |
| U.M. of SC3-20 class | 0.002% | 0.004% | −52% | 0.302 | 0.005% | −64% | 0.035 * |
| Opitutus | 0.359% | 0.556% | −35% | 0.016 * | 0.515% | −30% | 0.045 * |
| Taxonomy | RhC | RhBio1 | Relative Difference a | p-Value | RhBio2 | Relative Difference a | p-Value |
|---|---|---|---|---|---|---|---|
| U.M. of Subgroup 19 order | 0.001% | 0.002% | −43% | 0.428 | 0.004% | −70% | 0.049 * |
| Uncultured bacterium gp7 | 0.009% | 0.019% | −54% | 0.009 * | 0.007% | 32% | 0.546 |
| U.M. of uncultured Thermaerobacter | 0.004% | 0.001% | 304% | 0.042 * | 0.006% | −37% | 0.438 |
| Geodermatophilus | 0.613% | 0.629% | −3% | 0.735 | 0.523% | 17% | 0.032 * |
| Quadrisphaera | 0.005% | 0.007% | −31% | 0.154 | 0.009% | −44% | 0.011 * |
| U.M. of Cellulomonadaceae family | 0.002% | 0.001% | 140% | 0.261 | 0.0% | absent in RhBio2 | 0.048 * |
| U.M. of Microbacteriaceae family | 0.228% | 0.164% | 39% | 0.031 * | 0.152% | 49% | 0.008 ** |
| Nocardioides | 0.71% | 0.656% | 8% | 0.446 | 0.53% | 34% | 0.006 ** |
| Actinoallomurus | 0.000% | 0.002% | absent in RhC | 0.047 * | 0.001% | absent in RhC | 0.356 |
| Gaiella | 1.7% | 1.3% | 18% | 0.071 | 1.3% | 23% | 0.012 * |
| U.M. of FCPU744 family | 0.006% | 0.001% | 506% | 0.023 * | 0.004% | 58% | 0.320 |
| U.M. of Thermoleophilia class | 0.005% | 0.002% | 157% | 0.028 * | 0.002% | 193% | 0.068 |
| Prevotella | 0.002% | 0.011% | −80% | 0.199 | 0.027% | −92% | 0.017 * |
| Hymenobacter | 0.012% | 0.011% | 10% | 0.817 | 0.004% | 224% | 0.026 * |
| Ohtaekwangia | 0.167% | 0.158% | 6% | 0.659 | 0.22% | −24% | 0.039 * |
| U.M. of AKYH767 family | 0.014% | 0.007% | 105% | 0.027 * | 0.007% | 90% | 0.020 * |
| U.M. of Chitinophagaceae family | 0.466% | 0.521% | −11% | 0.030 * | 0.511% | −9% | 0.386 |
| U.M. of Sphingobacteriaceae family | 0.109% | 0.131% | −17% | 0.478 | 0.078% | 40% | 0.013 * |
| U.M. of Anaerolineaceae family | 1.06% | 1.1% | −8% | 0.263 | 1.2% | −11% | 0.036 * |
| U.M. of Chloroflexia class | 0.001% | 0.005% | −76% | 0.019 * | 0.004% | −72% | 0.198 |
| Nitrolancea | 0.006% | 0.005% | 27% | 0.603 | 0.003% | 130% | 0.032 * |
| U.M. of LineageIIa order | 0.002% | 0.007% | −75% | 0.012 * | 0.006% | −70% | 0.023 * |
| Anaerobacillus | 0.004% | 0.008% | −53% | 0.038 | 0.004% | 7% | 0.908 |
| U.M. of Bacillaceae family | 0.021% | 0.028% | −23% | 0.455 | 0.031% | −30% | 0.018 * |
| Brevibacillus | 0.006% | 0.002% | 175% | 0.046 * | 0.004% | 82% | 0.118 |
| Cohnella | 0.082% | 0.114% | −28% | 0.021 * | 0.102% | −19% | 0.361 |
| U.M. of Thermoactinomycetaceae family | 0.001% | 0.001% | −42% | 0.632 | 0.004% | −85% | 0.010 ** |
| U.M. of Clostridiaceae 1 family | 0.015% | 0.006% | 141% | 0.003 ** | 0.01% | 41% | 0.066 |
| Lutispora | 0.002% | 0.000% | absent in RhBio1 | 0.024 * | 0.002% | 4% | 0.943 |
| U.M. of C47 class | 0.013% | 0.018% | −29% | 0.032 * | 0.016% | −20% | 0.512 |
| U.M. of Phycisphaeraceae family | 0.000% | 0.001% | absent in RhC | 0.135 | 0.002% | absent in RhC | 0.027 * |
| Planctomycete LX80 | 0.006% | 0.015% | −56% | 0.011 * | 0.014% | −53% | 0.164 |
| U.M. of Pla3 lineage class | 0.000% | 0.002% | absent in RhC | 0.003 ** | 0.004% | absent in RhC | 0.179 |
| Isosphaera | 0.000% | 0.002% | absent in RhC | 0.043 * | 0.002% | absent in RhC | 0.146 |
| Pir4 lineage (Planctomycetaceae family) | 0.142% | 0.18% | −21% | 0.100 | 0.17% | −16% | 0.043 * |
| Bosea | 0.08% | 0.06% | 35% | 0.121 | 0.052% | 55% | 0.021 * |
| U.M. of Hyphomicrobiaceae family | 0.057% | 0.044% | 28% | 0.034 * | 0.031% | 81% | 0.010 ** |
| Microvirga | 0.178% | 0.167% | 6% | 0.582 | 0.137% | 30% | 0.039 * |
| Rhizomicrobium | 0.051% | 0.041% | 24% | 0.535 | 0.031% | 64% | 0.032 * |
| Labrys | 0.004% | 0.005% | −8% | 0.862 | 0.021% | −79% | 0.037 * |
| U.M. of MNC12 family | 0.03% | 0.02% | 50% | 0.001 *** | 0.019% | 55% | 0.036 * |
| U.M. of MND8 family | 0.129% | 0.138% | −7% | 0.571 | 0.166% | −23% | 0.026 * |
| Ottowia | 0.006% | 0.011% | −48% | 0.105 | 0.001% | 442% | 0.023 * |
| Pseudorhodoferax | 0.000% | 0.0% | absent in RhBio1 | 0.000 | 0.001% | absent in RhC | 0.024 * |
| Noviherbaspirillum | 0.016% | 0.009% | 77% | 0.204 | 0.003% | 359% | 0.048 * |
| Pseudoduganella | 0.001% | 0.0% | 28% | 0.870 | 0.003% | −78% | 0.033 * |
| U.M. of AKYG597 family | 0.000% | 0.000% | absent in RhC | 0.356 | 0.002% | absent in RhC | 0.000 *** |
| U.M. of FFCH16767 family | 0.007% | 0.006% | 3% | 0.931 | 0.005% | 27% | 0.012 * |
| Nannocystis | 0.029% | 0.039% | −26% | 0.036 * | 0.027% | 8% | 0.421 |
| Sorangium | 0.216% | 0.159% | 36% | 0.050 * | 0.198% | 9% | 0.405 |
| Anaeromyxobacter dehalogenans | 0.008% | 0.016% | −53% | 0.001 *** | 0.016% | −52% | 0.042 * |
| U.M. of Deltaproteobacteria class | 0.105% | 0.125% | −16% | 0.045 * | 0.127% | −17% | 0.057 |
| U.M. of CHAB-XI-27 family | 0.002% | 0.002% | −13% | 0.868 | 0.0% | absent in RhBio2 | 0.024 * |
| Pseudoxanthomonas | 0.066% | 0.051% | 29% | 0.229 | 0.033% | 102% | 0.020 * |
| Thermomonas | 0.043% | 0.049% | −11% | 0.492 | 0.064% | −33% | 0.019 * |
| Turneriella | 0.003% | 0.004% | −28% | 0.650 | 0.008% | −63% | 0.034 * |
| U.M. of OPB35 soil group class | 0.507% | 0.528% | −4% | 0.480 | 0.616% | −18% | 0.014 * |
| Others | 0.003% | 0.009% | −61% | 0.002 ** | 0.006% | −44% | 0.292 |
| Putative Bacterial Functional Groups | Bacteria Taxa | % 1 |
|---|---|---|
| N-cycling (diazotrophic bacteria) | Opitutus genus; Cytophagaceae family; Chryseolinea genus; Terrimonas genus; U.M. PHOS-HE51 family; Rhodovasum genus; Cohnella genus; Pseudoduganella genus; Anaeromyxobacter dehalogenans species; Anaerobacillus genus; Planctomicetes phylum | 40 |
| Antimicrobial activity | Solimonadaceae family; Nannocystis genus; Ohtaekwangia genus; Quadrisphaera genus; Anaerolinaceae family; Turneriella genus; Thermoactinomycetaceae family; Actinoallomurus genus | 33 |
| Production of plant growth hormones, micronutrient bioavailability, S-cycling | Niastella genus; Chitinophagaceae family; Labrys genus; Chloroflexia genus; Isophaera family; Deltaproteobacteria class; Thermomonas genus | 25 |
| Parameters † | Measurement | Loam | Silty-Loam |
|---|---|---|---|
| Location | Chivasso | Carignano | |
| Geographical coordinates | 45°12′43″ N; 7°55′45″ E | 44°53′21″ N; 7°40′56″ E | |
| Altitude | m a.s.l. | 183 | 245 |
| Soil (USDA classification) | Typic Hapludalfs | Typic Udifluvents | |
| Sand (2–0.05 mm) | % | 42.9 | 28.7 |
| Silt (0.05–0.002 mm) | % | 49.3 | 64.6 |
| Clay (<0.002 mm) | % | 7.8 | 6.7 |
| pH | 6.7 | 8.0 | |
| Organic matter | % | 1.53 | 1.45 |
| Total Nitrogen | % | 0.081 | 0.110 |
| C/N | 11.0 | 7.7 | |
| Cation Exchange Capacity (C.E.C.) | cmolc/kg | 10.6 | 12.2 |
| Exchangeable Potassium | mg/kg | 94 | 49 |
| Available Phosphorus | mg/kg | 36 | 7 |
| Exchangeable Calcium | mg/kg | 875 | 2002 |
| Exchangeable Magnesium | mg/kg | 109 | 46 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Visioli, G.; Sanangelantoni, A.M.; Vamerali, T.; Dal Cortivo, C.; Blandino, M. 16S rDNA Profiling to Reveal the Influence of Seed-Applied Biostimulants on the Rhizosphere of Young Maize Plants. Molecules 2018, 23, 1461. https://doi.org/10.3390/molecules23061461
Visioli G, Sanangelantoni AM, Vamerali T, Dal Cortivo C, Blandino M. 16S rDNA Profiling to Reveal the Influence of Seed-Applied Biostimulants on the Rhizosphere of Young Maize Plants. Molecules. 2018; 23(6):1461. https://doi.org/10.3390/molecules23061461
Chicago/Turabian StyleVisioli, Giovanna, Anna Maria Sanangelantoni, Teofilo Vamerali, Cristian Dal Cortivo, and Massimo Blandino. 2018. "16S rDNA Profiling to Reveal the Influence of Seed-Applied Biostimulants on the Rhizosphere of Young Maize Plants" Molecules 23, no. 6: 1461. https://doi.org/10.3390/molecules23061461
APA StyleVisioli, G., Sanangelantoni, A. M., Vamerali, T., Dal Cortivo, C., & Blandino, M. (2018). 16S rDNA Profiling to Reveal the Influence of Seed-Applied Biostimulants on the Rhizosphere of Young Maize Plants. Molecules, 23(6), 1461. https://doi.org/10.3390/molecules23061461







