Cobalt Complex with Thiazole-Based Ligand as New Pseudomonas aeruginosa Quorum Quencher, Biofilm Inhibitor and Virulence Attenuator
Abstract
1. Introduction
2. Results and Discussion
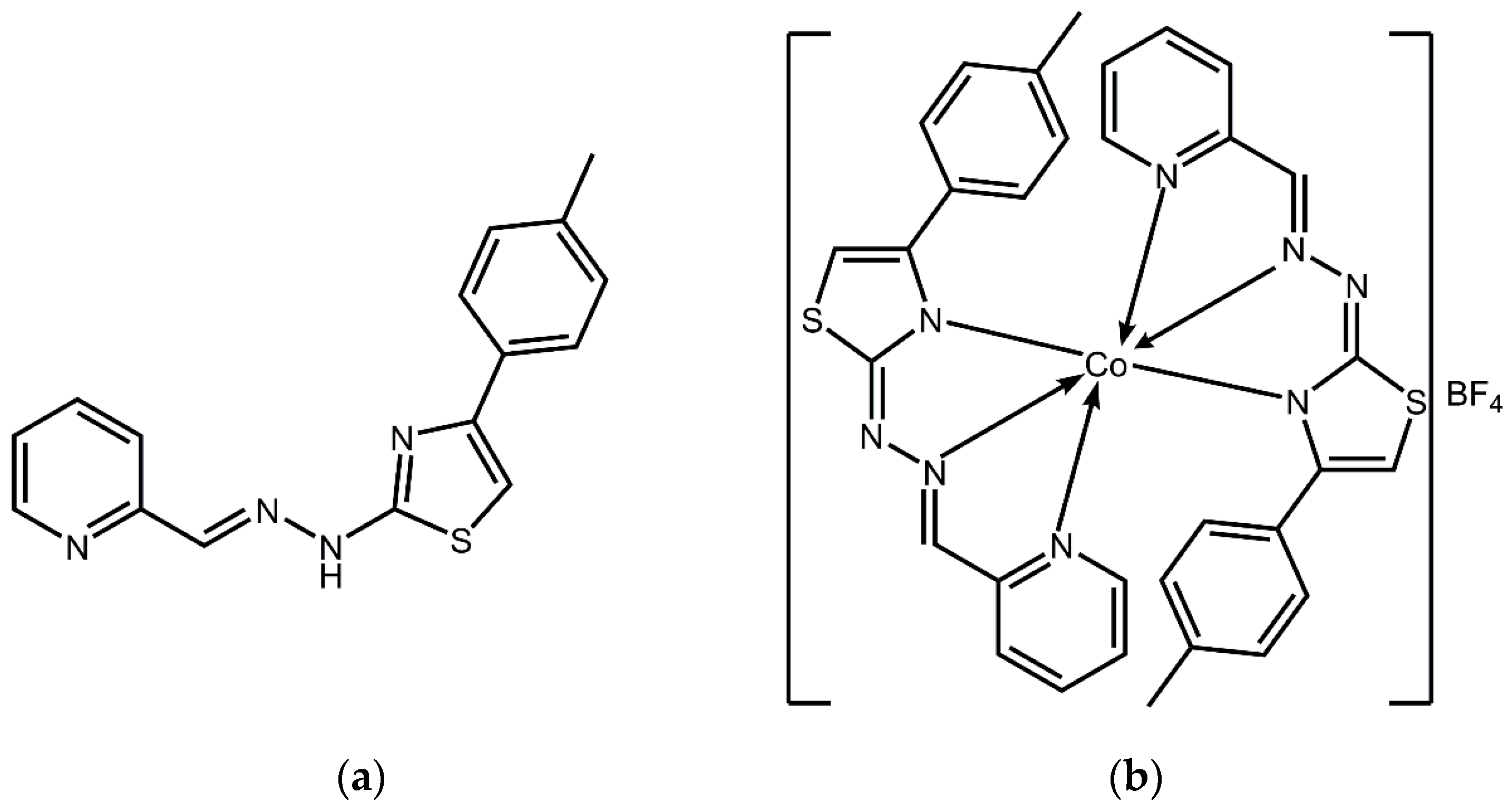
2.1. Cordination Complex Stability Analysis
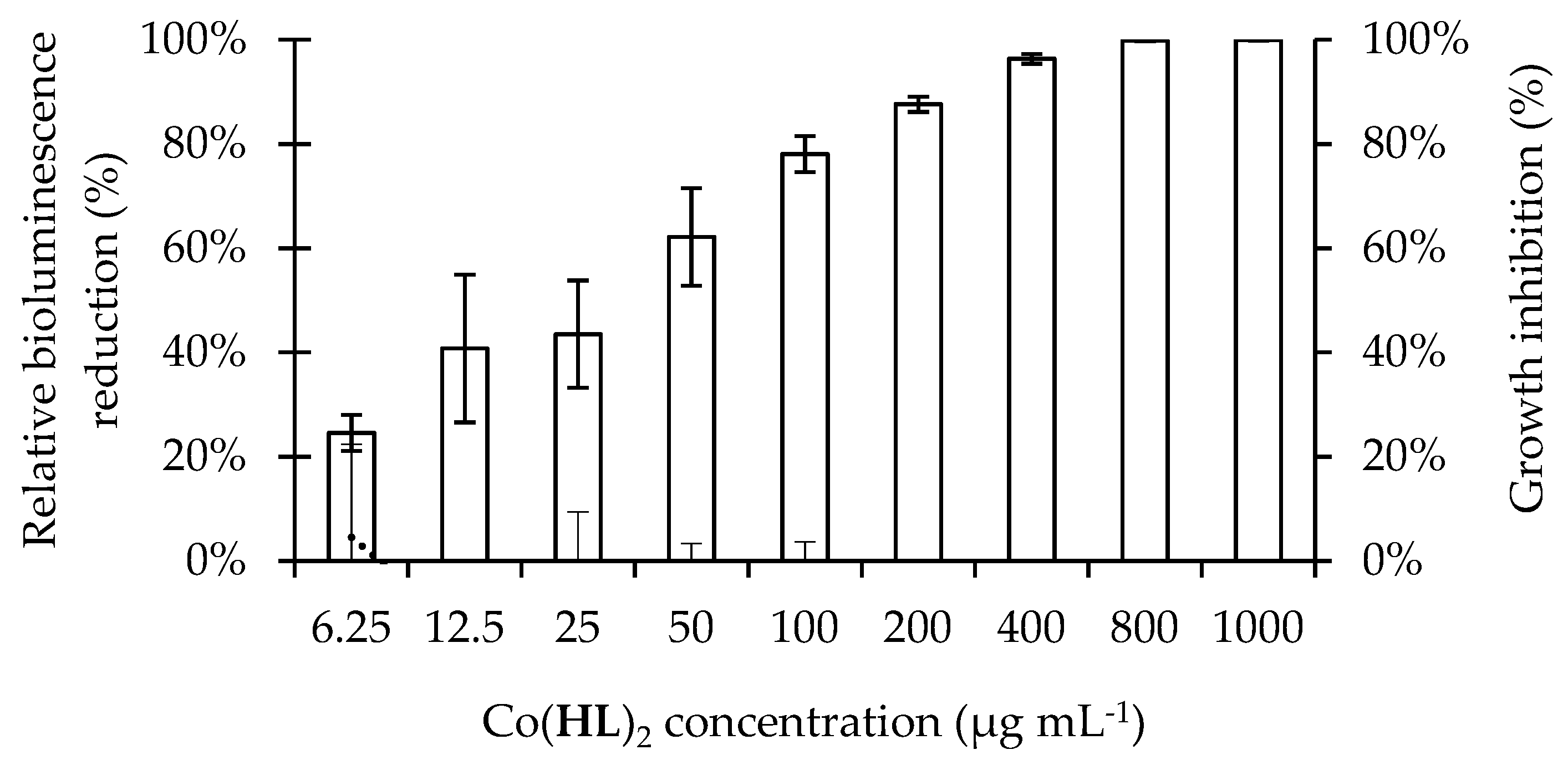
2.2. Antimicrobial Activity of Co(HL)2
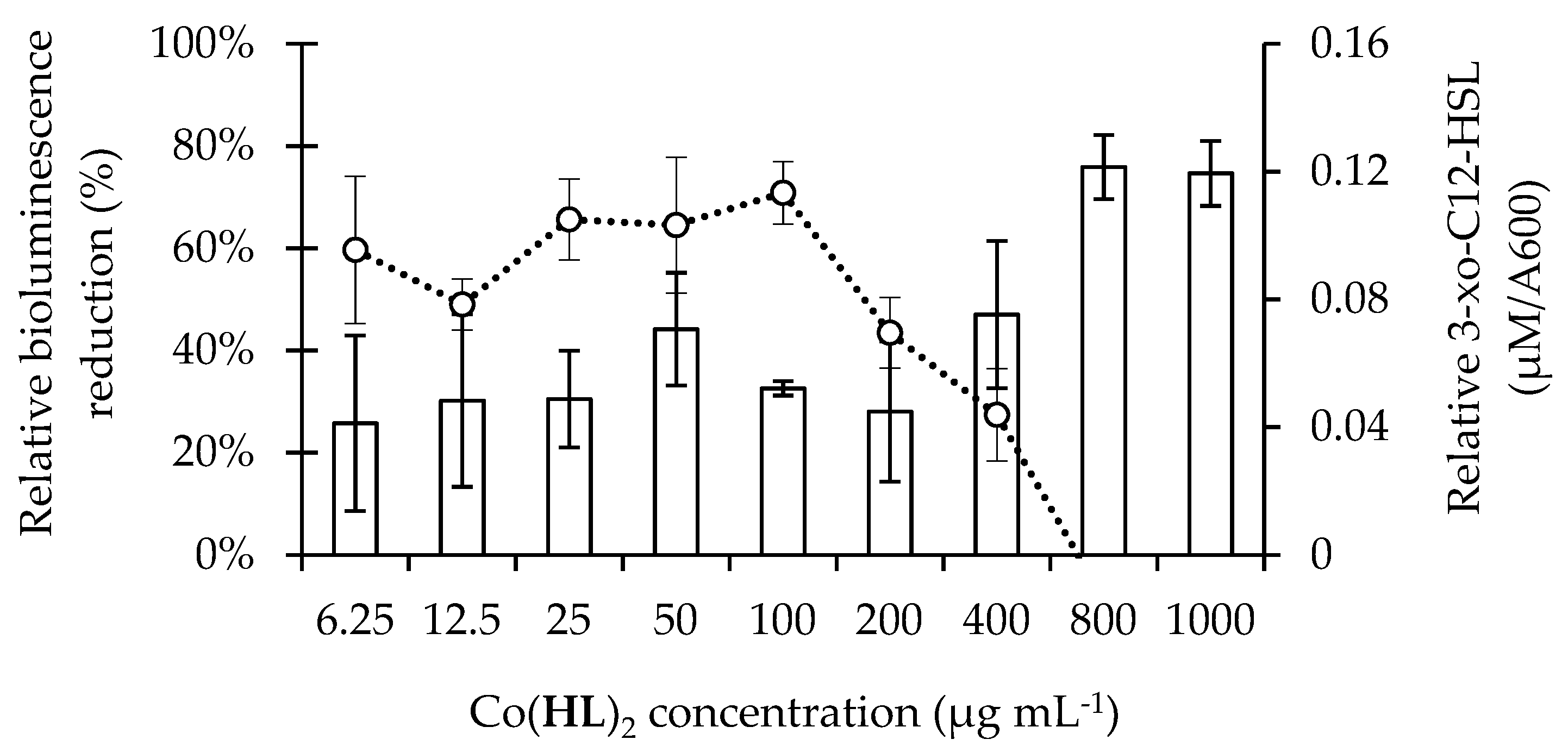
2.3. Co(HL)2 Mediated Inhibition of the 3-oxo-C12-HSL-Dependent QS System of P. aeruginosa
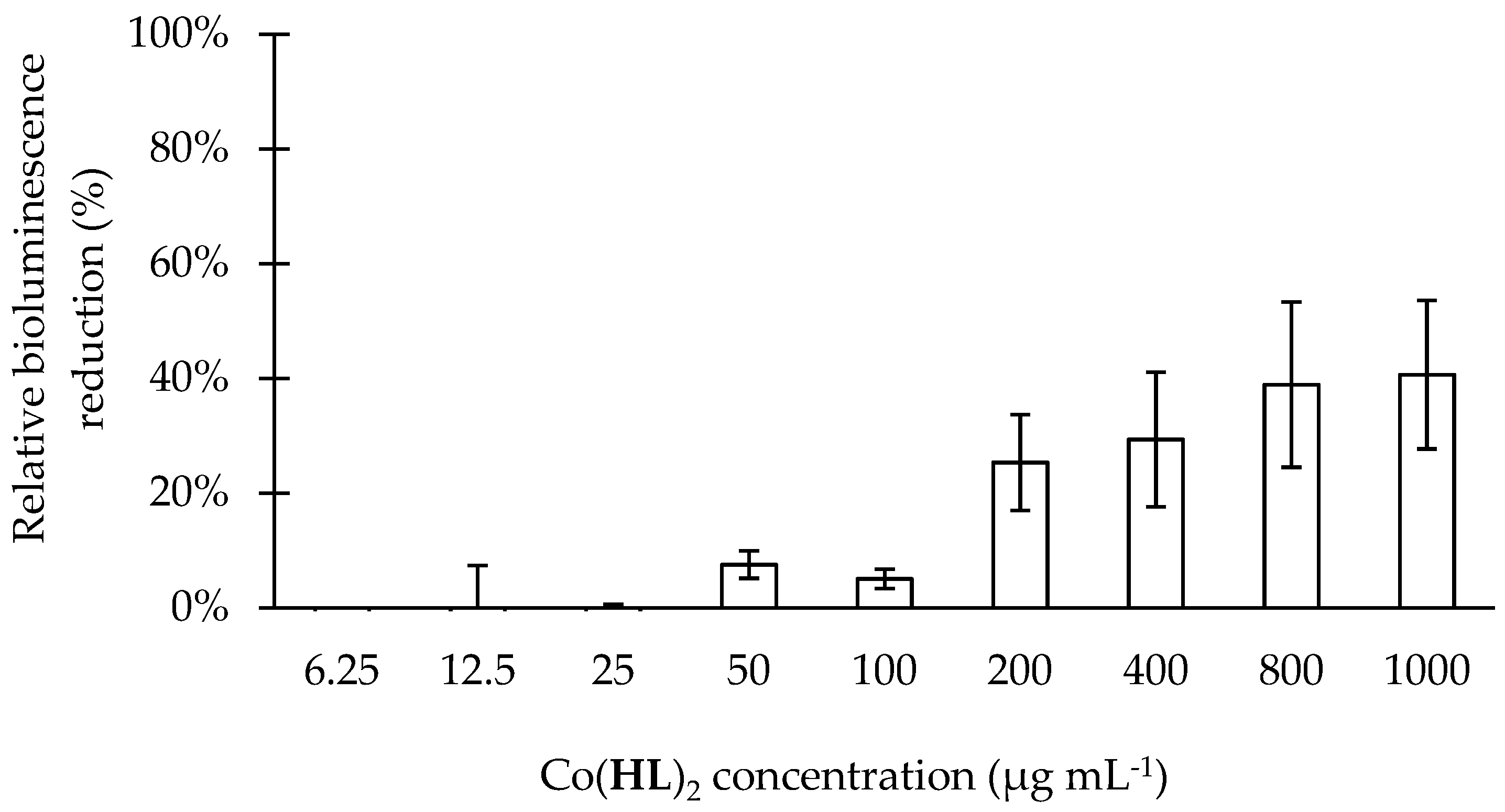
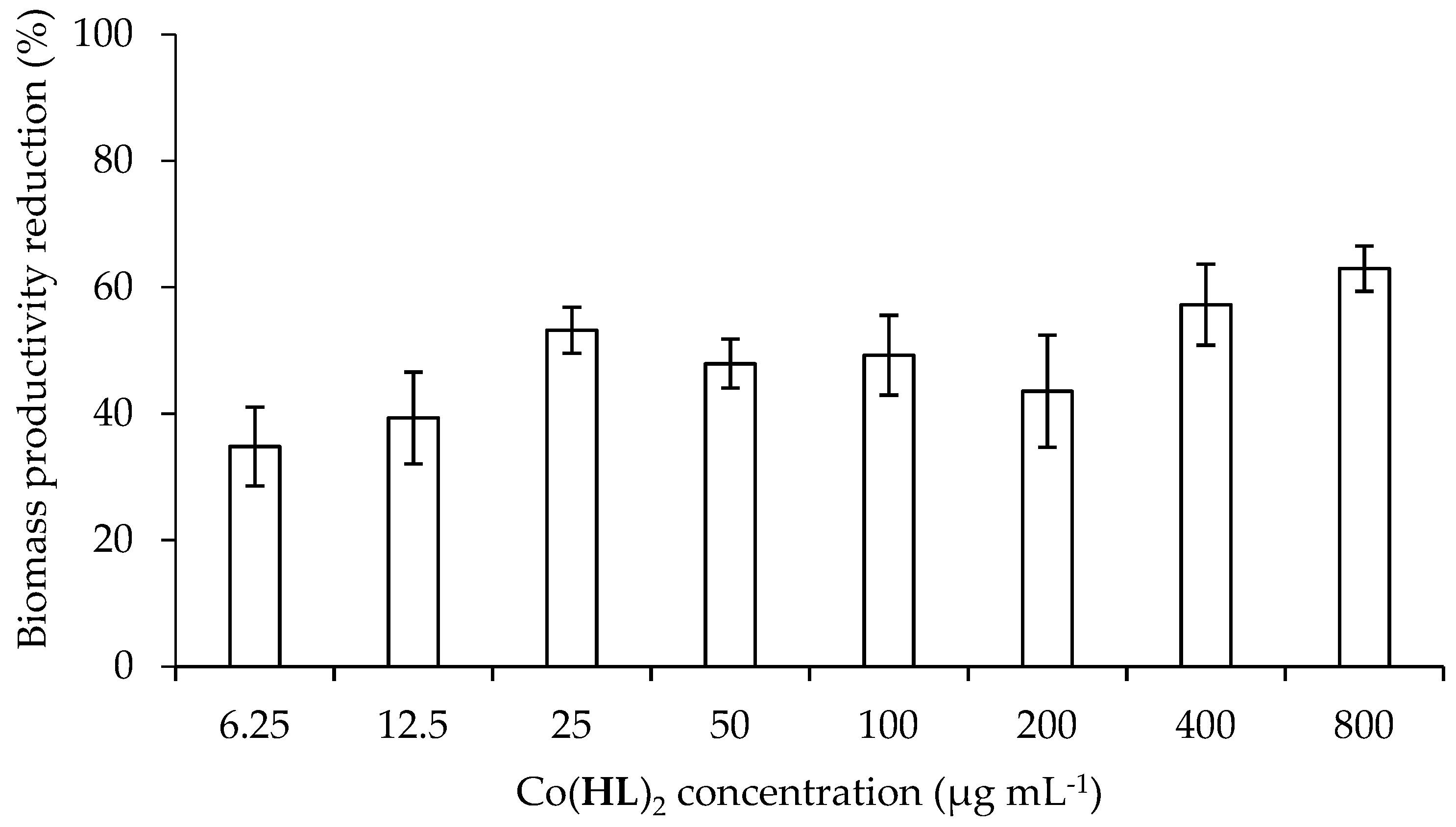
2.4. Effect of Co(HL)2 on the Prevention of Biofilm Formation
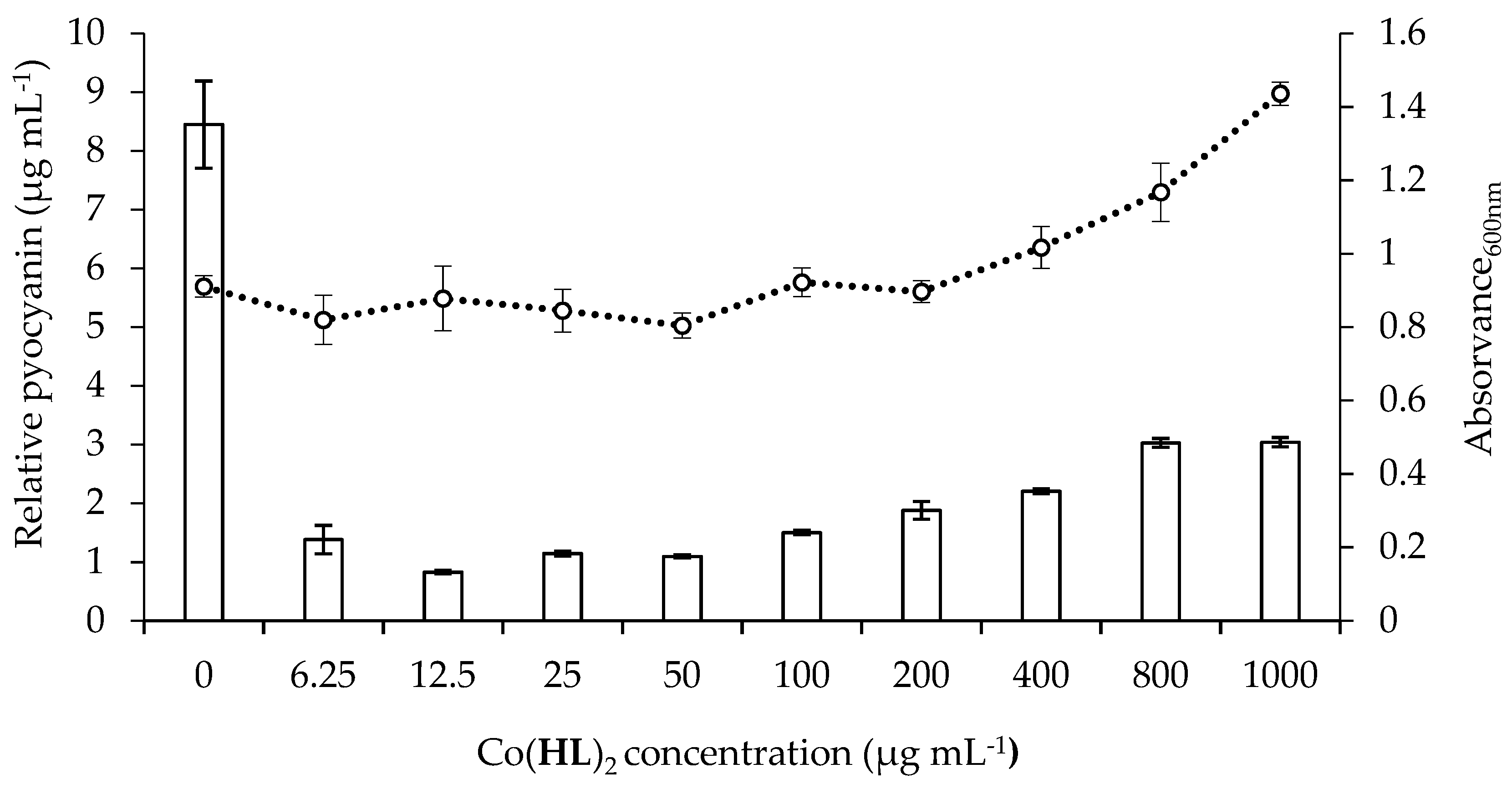
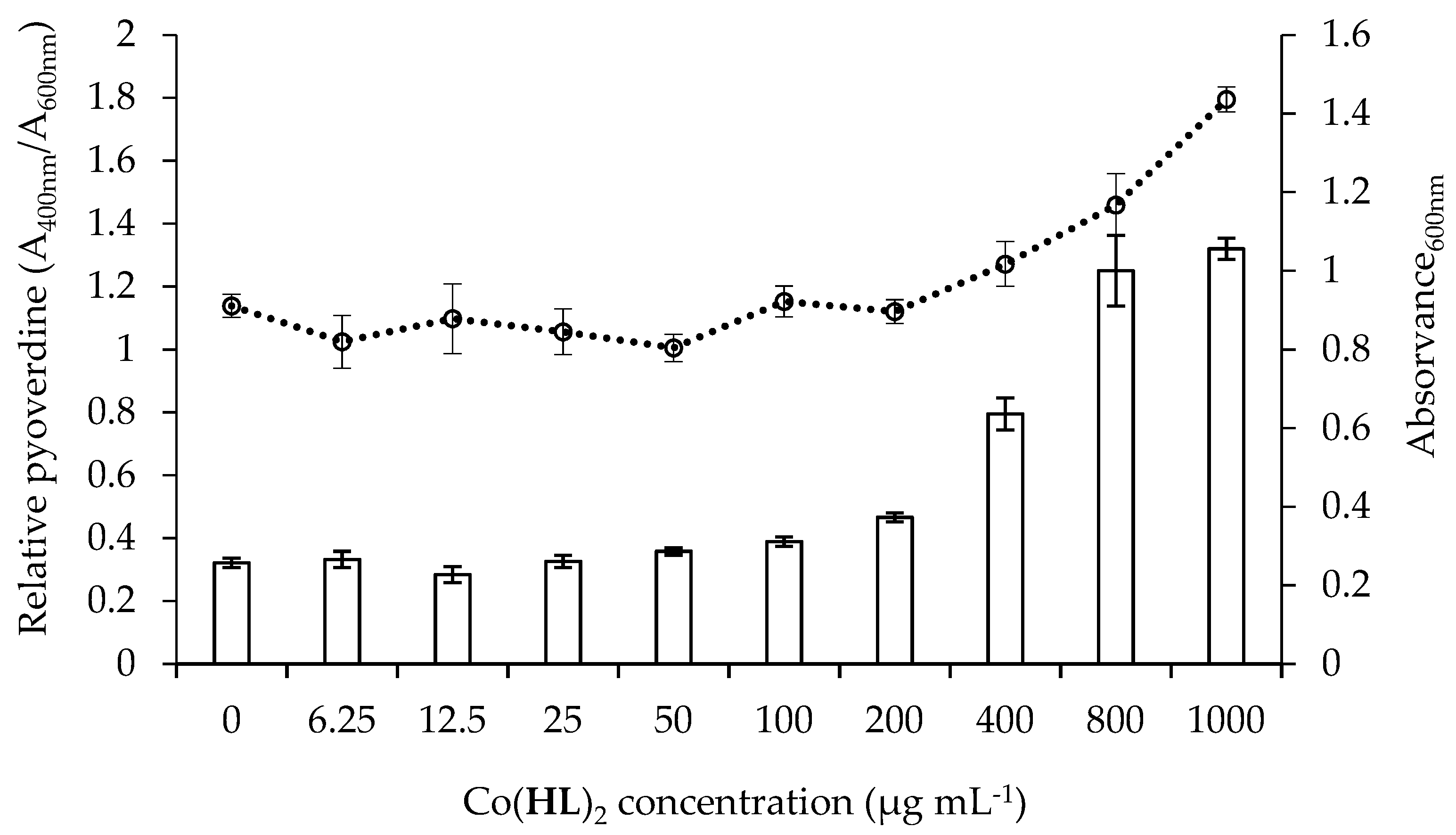
2.5. Effect of Co(HL)2 on the Production of Pyocyanin and Pyoverdine Virulence Factors
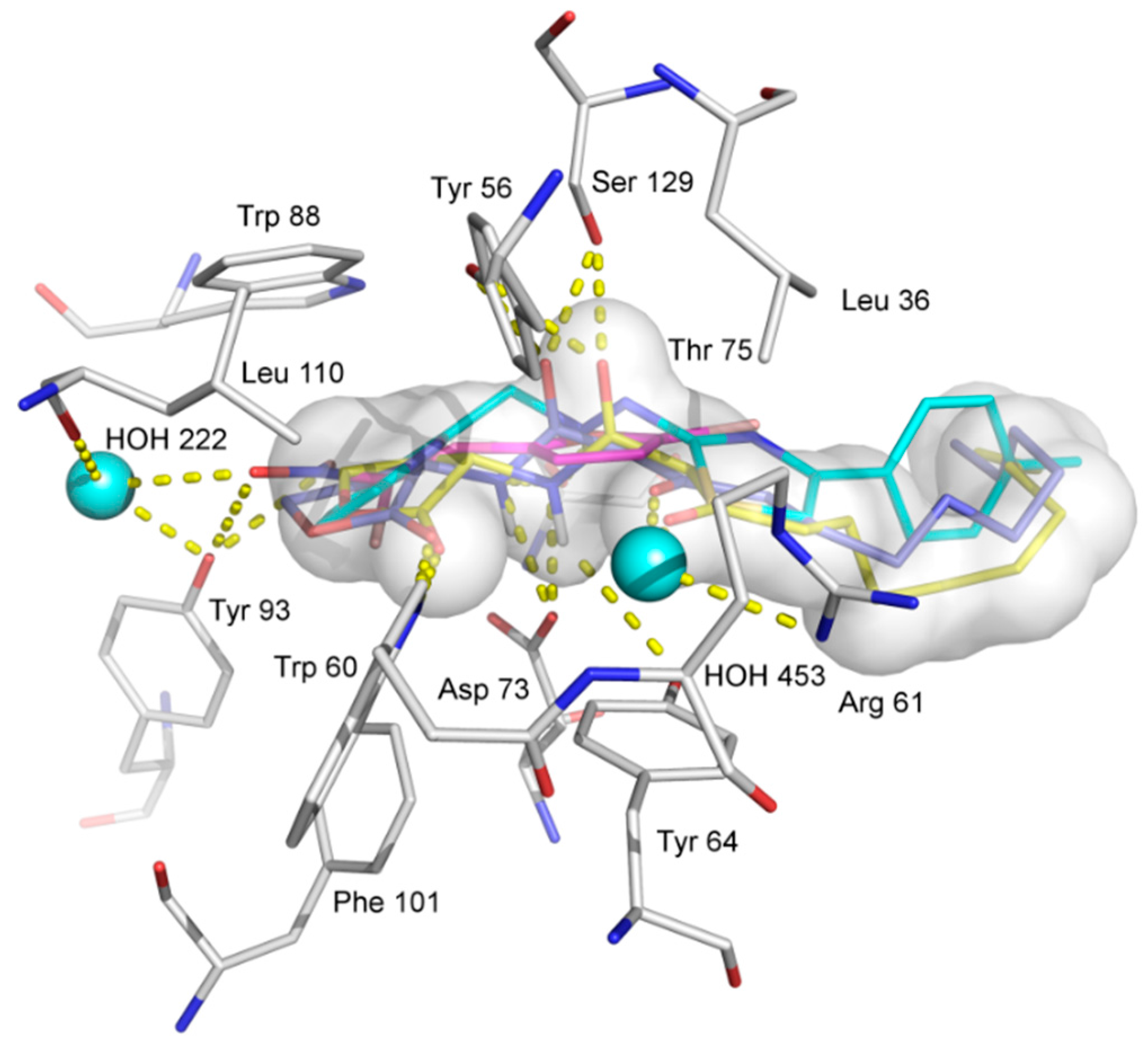
2.6. Molecular Docking Analysis
3. Materials and Methods
3.1. Reagents, Apparatus, and Synthesis
3.2. Bacterial Strains, Growth Conditions, and Solutions
3.3. Minimum Inhibitory and Bactericidal Concentrations (MIC and MBC)
3.4. High-Throughput QS Inhibition Screening
3.4.1. Autoinducer Synthesis Assay
3.4.2. Autoinducer Response Assay
3.5. Biofilm Prevention Assays
3.6. Virulence Factors Assays
3.6.1. Pyocyanin Production
3.6.2. Pyoverdine Production
3.7. Molecular Modeling of Co(HL)2 with LasR Receptor
3.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Kalia, V.C. Quorum sensing inhibitors: An overview. Biotechnol. Adv. 2013, 31, 224–245. [Google Scholar] [CrossRef] [PubMed]
- LaSarre, B.; Federle, M.J. Exploiting quorum sensing to confuse bacterial pathogens. Microbiol. Mol. Biol. Rev. 2013, 77, 73–111. [Google Scholar] [CrossRef] [PubMed]
- Abreu, A.C.; McBain, A.J.; Simões, M. Plants as sources of new antimicrobials and resistance-modifying agents. Nat. Prod. Rep. 2012, 29, 1007–1021. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 2001, 45, 999–1007. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, P.; Allison, D.G.; McBain, A.J. Biofilms in vitro and in vivo: Do singular mechanisms imply cross-resistance? J. Appl. Microbiol. 2002, 92, 98S–110S. [Google Scholar] [CrossRef] [PubMed]
- Landini, P.; Antoniani, D.; Burgess, J.G.; Nijland, R. Molecular mechanisms of compounds affecting bacterial biofilm formation and dispersal. Appl. Microbiol. Biotechnol. 2010, 86, 813–823. [Google Scholar] [CrossRef] [PubMed]
- Sahner, J.H.; Empting, M.; Kamal, A.; Weidel, E.; Groh, M.; Börger, C.; Hartmann, R.W. Exploring the chemical space of ureidothiophene-2-carboxylic acids as inhibitors of the quorum sensing enzyme PqsD from Pseudomonas aeruginosa. Eur. J. Med. Chem. 2015, 96, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Rückert, C.; Birmes, F.S.; Müller, C.; Niewerth, H.; Winkler, A.; Fetzner, S.; Kalinowski, J. Complete genome sequence of Rhodococcus erythropolis BG43 (DSM 46869), a degrader of Pseudomonas aeruginosa quorum sensing signal molecules. J. Biotechnol. 2015, 211, 99–100. [Google Scholar] [CrossRef] [PubMed]
- Imperi, F.; Massai, F.; Ramachandran Pillai, C.; Longo, F.; Zennaro, E.; Rampioni, G.; Visca, P.; Leoni, L. New life for an old drug: The anthelmintic drug niclosamide inhibits Pseudomonas aeruginosa quorum sensing. Antimicrob. Agents Chemother. 2013, 57, 996–1005. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Juárez, I.; García-Contreras, R.; Velázquez-Guadarrama, N.; Soto-Hernández, M.; Martínez-Vázquez, M. Amphypterygium adstringens Anacardic Acid Mixture Inhibits Quorum Sensing-controlled Virulence Factors of Chromobacterium violaceum and Pseudomonas aeruginosa. Arch. Med. Res. 2013, 44, 488–494. [Google Scholar] [CrossRef] [PubMed]
- Busetti, A.; Shaw, G.; Megaw, J.; Gorman, S.; Maggs, C.; Gilmore, B. Marine-Derived Quorum-Sensing Inhibitory Activities Enhance the Antibacterial Efficacy of Tobramycin against Pseudomonas aeruginosa. Mar. Drugs 2015, 13, 1–28. [Google Scholar] [CrossRef] [PubMed]
- El-Mowafy, S.; Shaaban, M.; Abd El Galil, K.H. Sodium ascorbate as a quorum sensing inhibitor of Pseudomonas aeruginosa. J. Appl. Microbiol. 2014, 117, 1388–1399. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Zhang, L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Prot. Cell 2015, 6, 26–41. [Google Scholar] [CrossRef] [PubMed]
- Le Berre, R.; Nguyen, S.; Nowak, E.; Kipnis, E.; Pierre, M.; Ader, F.; Courcol, R.; Guery, B.; Faure, K. Quorum-sensing activity and related virulence factor expression in clinically pathogenic isolates of Pseudomonas aeruginosa. Clin. Microbiol. Infect. 2008, 14, 337–343. [Google Scholar] [CrossRef] [PubMed]
- De Kievit, T. Quorum sensing in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 2009, 11, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Ayati, A.; Emami, S.; Asadipour, A.; Shafiee, A.; Foroumadi, A. Recent applications of 1, 3-thiazole core structure in the identification of new lead compounds and drug discovery. Eur. J. Med. Chem. 2015, 97, 699–718. [Google Scholar] [CrossRef] [PubMed]
- Elshaflu, H.; Bjelogrlić, S.; Muller, C.D.; Todorović, T.R.; Rodić, M.; Marinković, A.; Filipović, N.R. Co (III) complex with (E)-2-(2-(pyridine-2-ylmethylene) hydrazinyl)-4-(4-tolyl)-1, 3-thiazole: Structure and activity against 2-D and 3-D cancer cell models. J. Coord. Chem. 2016, 69, 3354–3366. [Google Scholar] [CrossRef]
- Filipović, N.R.; Elshaflu, H.; Grubišić, S.; Jovanović, L.S.; Rodić, M.; Novaković, I.; Malešević, A.; Djordjević, I.S.; Li, H.; Šojić, N. Co (III) complexes of (1,3-selenazol-2-yl) hydrazones and their sulphur analogues. Dalton Trans. 2017, 46, 2910–2924. [Google Scholar] [CrossRef] [PubMed]
- Choo, J.H.; Rukayadi, Y.; Hwang, J.K. Inhibition of bacterial quorum sensing by vanilla extract. Lett. Appl. Microbiol. 2006, 42, 637–641. [Google Scholar] [CrossRef] [PubMed]
- Borges, A.; Abreu, A.; Dias, C.; Saavedra, M.; Borges, F.; Simões, M. New perspectives on the use of phytochemicals as an emergent strategy to control bacterial infections including biofilms. Molecules 2016, 21, 877. [Google Scholar] [CrossRef] [PubMed]
- Borges, A.; Saavedra, M.J.; Simões, M. Insights on antimicrobial resistance, biofilms and the use of phytochemicals as new antimicrobial agents. Curr. Med. Chem. 2015, 22, 2590–2614. [Google Scholar] [CrossRef] [PubMed]
- Kirisits, M.J.; Parsek, M.R. Does Pseudomonas aeruginosa use intercellular signalling to build biofilm communities? Cell. Microbiol. 2006, 8, 1841–1849. [Google Scholar] [CrossRef] [PubMed]
- Zegans, M.E.; Wozniak, D.; Griffin, E.; Toutain-Kidd, C.M.; Hammond, J.H.; Garfoot, A.; Lam, J.S. Pseudomonas aeruginosa exopolysaccharide Psl promotes resistance to the biofilm inhibitor polysorbate 80. Antimicrob. Agents Chemother. 2012, 56, 4112–41122. [Google Scholar] [CrossRef] [PubMed]
- Hansen, M.R.; Jakobsen, T.H.; Bang, C.G.; Cohrt, A.E.; Hansen, C.L.; Clausen, J.W.; Le Quement, S.T.; Tolker-Nielsen, T.; Givskov, M.; Nielsen, T.E. Triazole-containing N-acyl homoserine lactones targeting the quorum sensing system in Pseudomonas aeruginosa. Bioorg. Med. Chem. 2015, 23, 1638–1650. [Google Scholar] [CrossRef] [PubMed]
- Miller, L.C.; O’Loughlin, C.T.; Zhang, Z.; Siryaporn, A.; Silpe, J.E.; Bassler, B.L.; Semmelhack, M.F. Development of potent inhibitors of pyocyanin production in Pseudomonas aeruginosa. J. Med. Chem. 2015, 58, 1298–1306. [Google Scholar] [CrossRef] [PubMed]
- Allen, L.; Dockrell, D.H.; Pattery, T.; Lee, D.G.; Cornelis, P.; Hellewell, P.G.; Whyte, M.K. Pyocyanin production by Pseudomonas aeruginosa induces neutrophil apoptosis and impairs neutrophil-mediated host defenses in vivo. J. Immunol. 2005, 174, 3643–3649. [Google Scholar] [CrossRef] [PubMed]
- Lamont, I.L.; Beare, P.A.; Ochsner, U.; Vasil, A.I.; Vasil, M.L. Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA 2002, 99, 7072–7077. [Google Scholar] [CrossRef] [PubMed]
- Kiratisin, P.; Tucker, K.; Passador, L. LasR, a transcriptional activator of Pseudomonas aeruginosa virulence genes, functions as a multimer. J. Bacteriol. 2002, 184, 4912–4919. [Google Scholar] [CrossRef] [PubMed]
- Hambley, T. Developing new metal-based therapeutics: Challenges and opportunities. Dalton Trans. 2007, 43, 4929–4937. [Google Scholar] [CrossRef] [PubMed]
- Gianferrara, T.; Bratsos, I.; Alessio, E. A categorization of metal anticancer compounds based on their mode of action. Dalton Trans. 2009, 37, 7588–7598. [Google Scholar] [CrossRef] [PubMed]
- Heffern, M.; Yamamoto, N.; Holbrook, R.; Eckermann, A.; Meade, T. Cobalt derivatives as promising therapeutic agents. Curr. Opin. Chem. Biol. 2013, 17, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Liu, L.; Chao, W.; Zhong, H.; Wang, M.; Chen, X.; Lu, J.; Li, R.; Ma, D.; Leung, C. Identification of an iridium(III) complex with anti-bacterial and anti-cancer activity. Sci. Rep. 2015, 5, 14544. [Google Scholar] [CrossRef] [PubMed]
- Massai, F.; Imperi, F.; Quattrucci, S.; Zennaro, E.; Visca, P.; Leoni, L. A multitask biosensor for micro-volumetric detection of N-3-oxo-dodecanoyl-homoserine lactone quorum sensing signal. Biosens. Bioelectron. 2011, 26, 3444–3449. [Google Scholar] [CrossRef] [PubMed]
- Leoni, L.; Landini, P. Microbiological methods for target-oriented screening of biofilm inhibitors. In Donelli G. (eds) Microbial Biofilms; Methods in Molecular Biology (Methods and Protocols); Springer Science: New York, NY, USA, 2014; pp. 175–186. [Google Scholar]
- Ferreira, C.; Pereira, A.M.; Pereira, M.C.; Melo, L.F.; Simões, M. Physiological changes induced by the quaternary ammonium compound benzyldimethyldodecylammonium chloride on Pseudomonas fluorescens. J. Antimicrob. Chemother. 2011, 66, 1036–1043. [Google Scholar] [CrossRef] [PubMed]
- Borges, A.; Saavedra, M.J.; Simões, M. The activity of ferulic and gallic acids in biofilm prevention and control of pathogenic bacteria. Biofouling 2012, 28, 755–767. [Google Scholar] [CrossRef] [PubMed]
- Simões, L.C.; Simões, M.; Vieira, M.J. Influence of the diversity of bacterial isolates from drinking water on resistance of biofilms to disinfection. Appl. Environ. Microbiol. 2010, 76, 6673–6679. [Google Scholar] [CrossRef] [PubMed]
- Borges, A.; Lopez-Romero, J.; Oliveira, D.; Giaouris, E.; Simões, M. Prevention, removal and inactivation of Escherichia coli and Staphylococcus aureus biofilms using selected monoterpenes of essential oils. J. Appl. Microbiol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Essar, D.W.; Eberly, L.; Hadero, A.; Crawford, I.P. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: Interchangeability of the two anthranilate synthase and evolutionary implications. J. Bacteriol. 1990, 172, 884–900. [Google Scholar] [CrossRef] [PubMed]
- Höfte, M.; Buysens, S.; Koedam, N.; Cornelis, P. Zinc affects siderophore-mediated high affinity iron uptake systems in the rhizosphere Pseudomonas aeruginosa 7NSK2. Biometals 1993, 6, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Rybtke, M.T.; Jakobsen, T.H.; Hentzer, M.; Bjarnsholt, T.; Givskov, M.; Tolker-Nielsen, T. Computer-aided identification of recognized drugs as Pseudomonas aeruginosa quorum-sensing inhibitors. Antimicrob. Agents Chemother. 2009, 53, 2432–2443. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.; Huey, R.; Lindstrom, W.; Sanner, M.; Belew, R.; Goodsell, D.; Olson, A. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A. Software News and Update AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [PubMed]
- García-Sosa, A.T.; Sild, S.; Maran, U. Docking and virtual screening using distributed grid technology. QSAR Comb. Sci. 2009, 28, 815–821. [Google Scholar] [CrossRef]
- Glisic, S.; Sencanski, M.; Perovic, V.; Stevanovic, S.; García-Sosa, A.T. Arginase flavonoid anti-Leishmanial in silico inhibitors flagged against anti-targets. Molecules 2016, 21, 589. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compound Co(HL)2 are available from the authors. |
| Compound | P. aeruginosa PA14 | P. aeruginosa PA14-R3 | ||
|---|---|---|---|---|
| MIC (µg mL−1) | MBC (µg mL−1) | MIC (µg mL−1) | MBC (µg mL−1) | |
| Co(HL)2 | 800 | >1000 1 | 800 | >1000 1 |
| Compound | QPLD | Autodock4 | Vina |
|---|---|---|---|
| Co(HL)2 | - | +51.94 | −4.0 |
| HL | −8.86 | −8.41 | −10.1 |
| Furvina | −4.71 | −5.74 | −7.0 |
| 3-oxo-C12-HSL | −8.60 | −7.39 | −8.7 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borges, A.; Simões, M.; Todorović, T.R.; Filipović, N.R.; García-Sosa, A.T. Cobalt Complex with Thiazole-Based Ligand as New Pseudomonas aeruginosa Quorum Quencher, Biofilm Inhibitor and Virulence Attenuator. Molecules 2018, 23, 1385. https://doi.org/10.3390/molecules23061385
Borges A, Simões M, Todorović TR, Filipović NR, García-Sosa AT. Cobalt Complex with Thiazole-Based Ligand as New Pseudomonas aeruginosa Quorum Quencher, Biofilm Inhibitor and Virulence Attenuator. Molecules. 2018; 23(6):1385. https://doi.org/10.3390/molecules23061385
Chicago/Turabian StyleBorges, Anabela, Manuel Simões, Tamara R. Todorović, Nenad R. Filipović, and Alfonso T. García-Sosa. 2018. "Cobalt Complex with Thiazole-Based Ligand as New Pseudomonas aeruginosa Quorum Quencher, Biofilm Inhibitor and Virulence Attenuator" Molecules 23, no. 6: 1385. https://doi.org/10.3390/molecules23061385
APA StyleBorges, A., Simões, M., Todorović, T. R., Filipović, N. R., & García-Sosa, A. T. (2018). Cobalt Complex with Thiazole-Based Ligand as New Pseudomonas aeruginosa Quorum Quencher, Biofilm Inhibitor and Virulence Attenuator. Molecules, 23(6), 1385. https://doi.org/10.3390/molecules23061385









