Dicyanamide Bridged Cu(II)36-Metallacrown-6 Complex with 1,4,7-Triisopropyl-1,4,7-Triazacyclononane and Binding Properties with DNA
Abstract
:1. Introduction
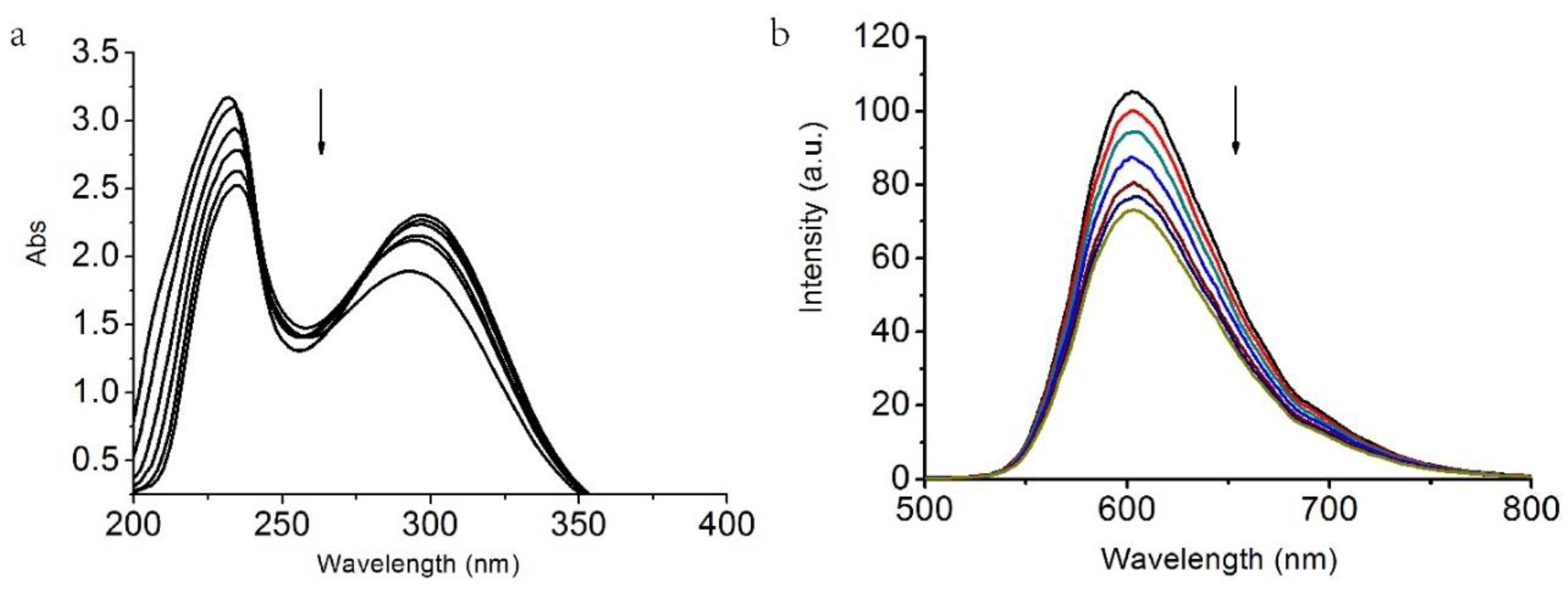
2. DNA Binding
3. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References and Notes
- Chow, C.Y.; Bolvin, H.; Campbell, V.E.; Guillot, R.; Kampf, J.W.; Wernsdorfer, W.; Gendron, F.; Autschbach, J.; Pecoraro, V.L.; Mallah, T. Assessing the exchange coupling in binuclear lanthanide(III) complexes and the slow relaxation of the magnetization in the antiferromagnetically coupled Dy2 derivative. Chem. Sci. 2015, 6, 4148–4159. [Google Scholar] [CrossRef] [PubMed]
- Boron, T.T.; Kampf, J.W.; Pecoraro, V.L. A mixed 3d−4f 14-metallacrown-5 complex that displays slow magnetic relaxation through geometric control of magnetoanisotropy. Inorg. Chem. 2010, 49, 9104–9106. [Google Scholar] [CrossRef] [PubMed]
- Deb, A.; Boron, T.T., III; Itou, M.; Sakurai, Y.; Mallah, T.; Pecoraro, V.L.; Penner-Hahn, J.E. Understanding spin structure in metallacrown single-molecule magnets using magnetic compton scattering. J. Am. Chem. Soc. 2014, 136, 4889–4892. [Google Scholar] [CrossRef] [PubMed]
- Chow, C.Y.; Guillot, R.; Riviere, E.; Kampf, J.W.; Mallah, T.; Pecoraro, V.L. Synthesis and magnetic characterization of Fe(III)-based 9-metallacrown-3 complexes which exhibit magnetorefrigerant properties. Inorg. Chem. 2016, 55, 10238–10247. [Google Scholar] [CrossRef] [PubMed]
- Escuer, A.; Mayans, J.; Font-Bardia, M. Linked nickel metallacrowns from a phosphonate/2-pyridyloximate blend of ligands: Structure and magnetic properties. Inorg. Chem. 2016, 55, 3161–3168. [Google Scholar] [CrossRef] [PubMed]
- Jankolovists, J.; Andolina, C.M.; Kampf, J.W.; Raymond, K.N.; Pecoraro, V.L. Assembly of near-infrared luminescent lanthanide host(host–guest) complexes with a metallacrown sandwich motif. Angew. Chem. Int. Ed. 2011, 50, 9660–9664. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, E.R.; Eliseeva, S.V.; Jankolovist, J.; Olmstead, M.M.; Petoud, S.; Pecoraro, V.L. Highly emitting near-infrared lanthanide “encapsulated sandwich” metallacrown complexes with excitation shifted toward lower energy. J. Am. Chem. Soc. 2014, 136, 1526–1534. [Google Scholar] [CrossRef] [PubMed]
- Chow, C.Y.; Eliseeva, S.V.; Trivedi, E.R.; Nguyen, T.N.; Kampf, J.W.; Petoud, S.; Pecoraro, V.L. Ga3+/Ln3+ metallacrowns: A promising family of highly luminescent lanthanide complexes that covers visible and near-infrared domains. J. Am. Chem. Soc. 2016, 138, 5100–5109. [Google Scholar] [CrossRef] [PubMed]
- Lah, M.S.; Pecoraro, V.L. Isolation and characterization of {MnII[MnIII(salicylhydroximate)]4(acetate)2(DMF)6}.2DMF: An inorganic analog of M2+(12-crown-4). J. Am. Chem. Soc. 1989, 111, 7258–7259. [Google Scholar] [CrossRef]
- Cutland, A.D.; Malkani, R.G.; Kampf, J.W.; Pecoraro, V.L. Lanthanide [15]metallacrown-5 complexes form nitrate-selective chiral cavities. Angew. Chem. Int. Ed. 2000, 39, 2689–2692. [Google Scholar] [CrossRef]
- Jian, F.F.; Jiao, K.; Li, Y.; Zhao, P.S.; Lu, L.D. [Ni6(SCH2CH2OH)12]: A double crown [12]metallacrown-6 nickel(II) cluster. Angew. Chem. Int. Ed. 2003, 42, 5722–5724. [Google Scholar] [CrossRef] [PubMed]
- Guillerm, V.; Kim, D.; Liu, X.; Adil, K.; Luebke, R.; Eubank, J.F.; Lah, M.S.; Eddaoudi, M. A supermolecular building approach for the design and construction of metal-organic frameworks. Chem. Soc. Rev. 2014, 43, 6141–6172. [Google Scholar] [CrossRef] [PubMed]
- Mezei, G.; Baran, P.; Raptis, R.G. Anion encapsulation by neutral supramolecular assemblies of cyclic CuII complexes: A series of five polymerization isomers, [{cis-CuII(μ-OH)(μ-pz)}n], n=6, 8, 9, 12, and 14. Angew. Chem. Int. Ed. 2004, 43, 574–577. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.L.M.; Pedroso, E.F.; Tumpf, H.O.S.; Novak, M.A.; Richard, L.; Garcia, R.R.; Riviere, E.; Journaux, Y. A CuIICoII metallacyclophane-based metamagnet with a corrugated brick-wall sheet architecture. Angew. Chem. Int. Ed. 2004, 43, 956–958. [Google Scholar] [CrossRef] [PubMed]
- Zaleski, C.M.; Depperman, E.C.; Dendrinou-Samara, C.; Alexiou, M.; Kampf, J.W.; Kessissoglou, D.P.; Kirk, M.L.; Pecoraro, V.L. Metallacryptate single-molecule magnets: Effect of lower molecular symmetry on blocking temperature. J. Am. Chem. Soc. 2005, 127, 12862–12872. [Google Scholar] [CrossRef] [PubMed]
- Zaleski, C.M.; Depperman, E.C.; Kampf, J.W.; Kirk, M.L.; Pecoraro, V.L. Using LnIII[15-MCCuII(N)(S)-pheHA−5]3+ complexes to construct chiral single-molecule magnets and chains of single-molecule magnets. Inorg. Chem. 2006, 45, 10022–10024. [Google Scholar] [CrossRef] [PubMed]
- Boron, T.T., III; Lutter, J.C.; Daly, C.I.; Chow, C.Y.; Davis, A.H.; Nimthong-Roldan, A.; Zeller, M.; Kampf, J.W.; Zaleski, C.M.; Pecoraro, V.L. The nature of the bridging anion controls the single-molecule magnetic properties of DyX4M 12-metallacrown-4 complexes. Inorg. Chem. 2016, 55, 10597–10607. [Google Scholar] [CrossRef] [PubMed]
- Moon, D.; Song, J.; Kim, B.J.; Suh, B.J.; Lah, M.S. Three-dimensional helical coordination networks of a hexanuclear manganese metallamacrocycle as a helical tecton. Inorg. Chem. 2004, 26, 8230–8232. [Google Scholar] [CrossRef] [PubMed]
- Samara, C.D.; Psomas, G.; Iordanidis, L.; Tangoulis, V.; Kessissoglou, D.P. Host–guest interaction of 12-MC-4, 15-MC-5, and fused 12-MC-4 metallacrowns with mononuclear and binuclear carboxylato complexes: Structure and magnetic behavior. Chem. Eur. J. 2001, 7, 5041–5051. [Google Scholar] [CrossRef]
- Gibney, B.G.; Stemmler, A.J.; Pilotek, S.; Kampf, J.W.; Pecoraro, V.L. Generalizing the metallacrown analogy: Ligand variation and solution stability of the VVO 9-metallacrown-3 structure type. Inorg. Chem. 1993, 32, 6008–6015. [Google Scholar] [CrossRef]
- Chen, G.J.; Wang, Z.G.; Yang, Y.S.; Tian, J.L.; Yan, S.P. A nickel(II) metallamacrocycle complex with antiferromagnetic properties. Z. Anorg. Allg. Chem. 2013, 639, 475–477. [Google Scholar] [CrossRef]
- Synthesis of complexes 1: 0.0578 g (0.15 mmol) of LCuCl2 was dissolved in 20 mL of methanol, 0.0134 g (0.15 mmol) of NaN(CN)2 was added as solid to the nickel solution, stirred, after all the solid was dissolved, 0.0276 g (0.15 mmol) of KPF6 was added. Single crystals of 1 was obtained by slow evaporation of a methanol solution of the power containing a very small amount of water. Anal. Calcd (%) for 1, C102H199Cu6F36N36O0.5P6: C, 38.95; H, 6.73; N, 15.97. Found: C, 38.88; H, 6.33; N, 16.00. IR for 1 (KBr, cm−1): 3446s, 2972s, 2333s, 2259s, 2206vs, 2178vs, 1383s, 1146m, 942m, 840vs, 551s. Crystal data: 1, Mr = 3189.06, Rhombohedral, , a = 28.417(3) Å, b = 28.417(3) Å, c = 15.6781(19) Å, α = 90°, β = 90°, γ = 120°, V = 0964(2) Å3, Z = 3, ρcalcd = 1.449 g cm−3, 2θmax = 25.02° (−33 ≤ h ≤ 17, −33 ≤ k ≤ 33, −18 ≤ l ≤ 18), T = 294(2) K, F(000) = 4965. GOF = 1.066. R1 (wR2) = 0.0421 (0.1133). 18801 reflections were collected, 4320 were unique (Rint = 0.0511). CCDC-626300.
- Crystal data: [LNi(N(CN)2)]2(BPh4)2, Mr = 1398.83, Triclinic, , a = 10.2350(17) Å, b = 10.8420(19) Å, c = 18.055(3) Å, α = 74.422(3)°, β = 88.842(3)°, γ = 78.520(3)°, V = 1890.2(6) Å3, Z = 1, ρcalcd = 1.229 g cm−3, 2θmax = 26.40° (−12 ≤ h ≤ 12, −5 ≤ k ≤ 13, −22 ≤ l ≤ 22), T = 294(2) K, F(000) = 748. R1 (wR2) = 0.0442 (0.0860). GOF = 1.006. 10744 reflections were collected, 7607 were unique [Rint = 0.0255]. CCDC-629545.
- Meyer, A.; Gleizes, A.; Girerd, J.J.; Verdaguer, M.; Kahn, O. Crystal structures, magnetic anisotropy properties and orbital interactions in catena-(.mu.-nitrito)-bis(ethylenediamine)nickel(II) perchlorate and triiodide. Inorg. Chem. 1982, 21, 1729–1739. [Google Scholar] [CrossRef]
- Tjioe, L.; Meininger, A.; Joshi, T.; Spiccia, L.; Graham, B. Efficient plasmid DNA cleavage by copper(II) complexes of 1,4,7-triazacyclononane ligands featuring xylyl-linked guanidinium groups. Inorg. Chem. 2011, 50, 4327–4339. [Google Scholar] [CrossRef] [PubMed]
- Reichmann, M.E.; Rice, S.A.; Thomas, C.A.; Doty, P. A further examination of the molecular weight and size of desoxypentose nucleic acid. J. Am. Chem. Soc. 1954, 76, 3047–3053. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds 1 and 2 are available from the authors. |
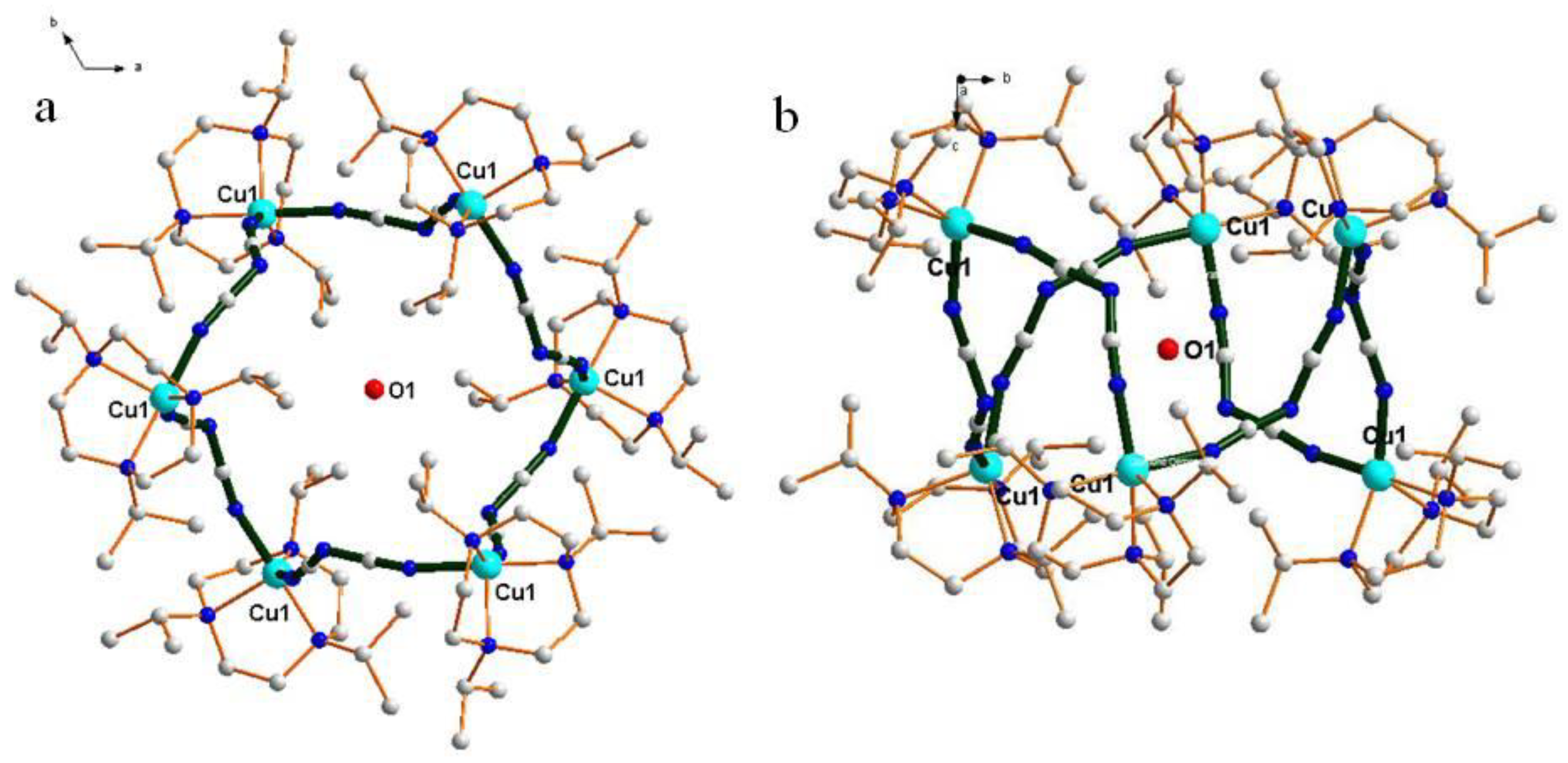
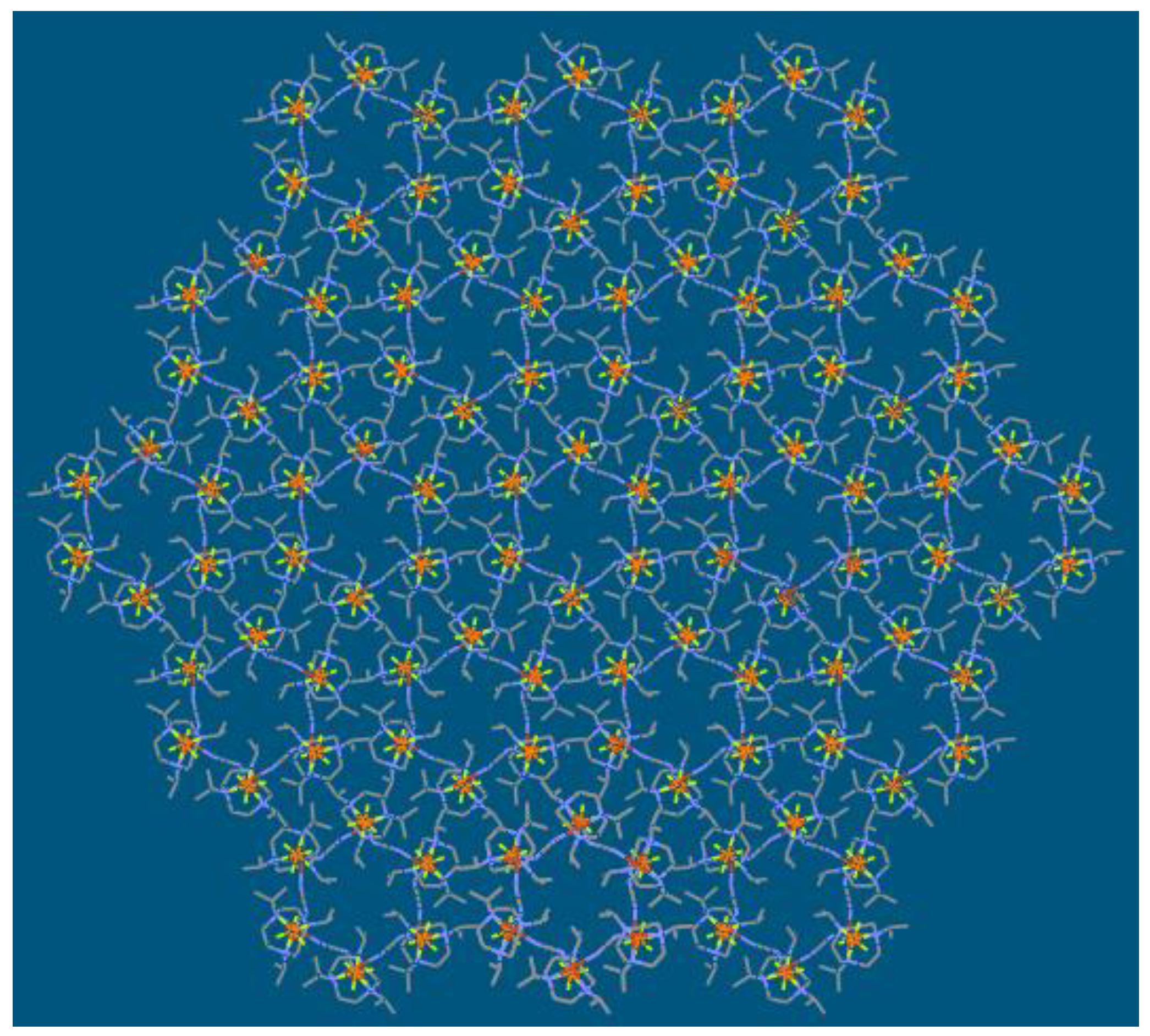
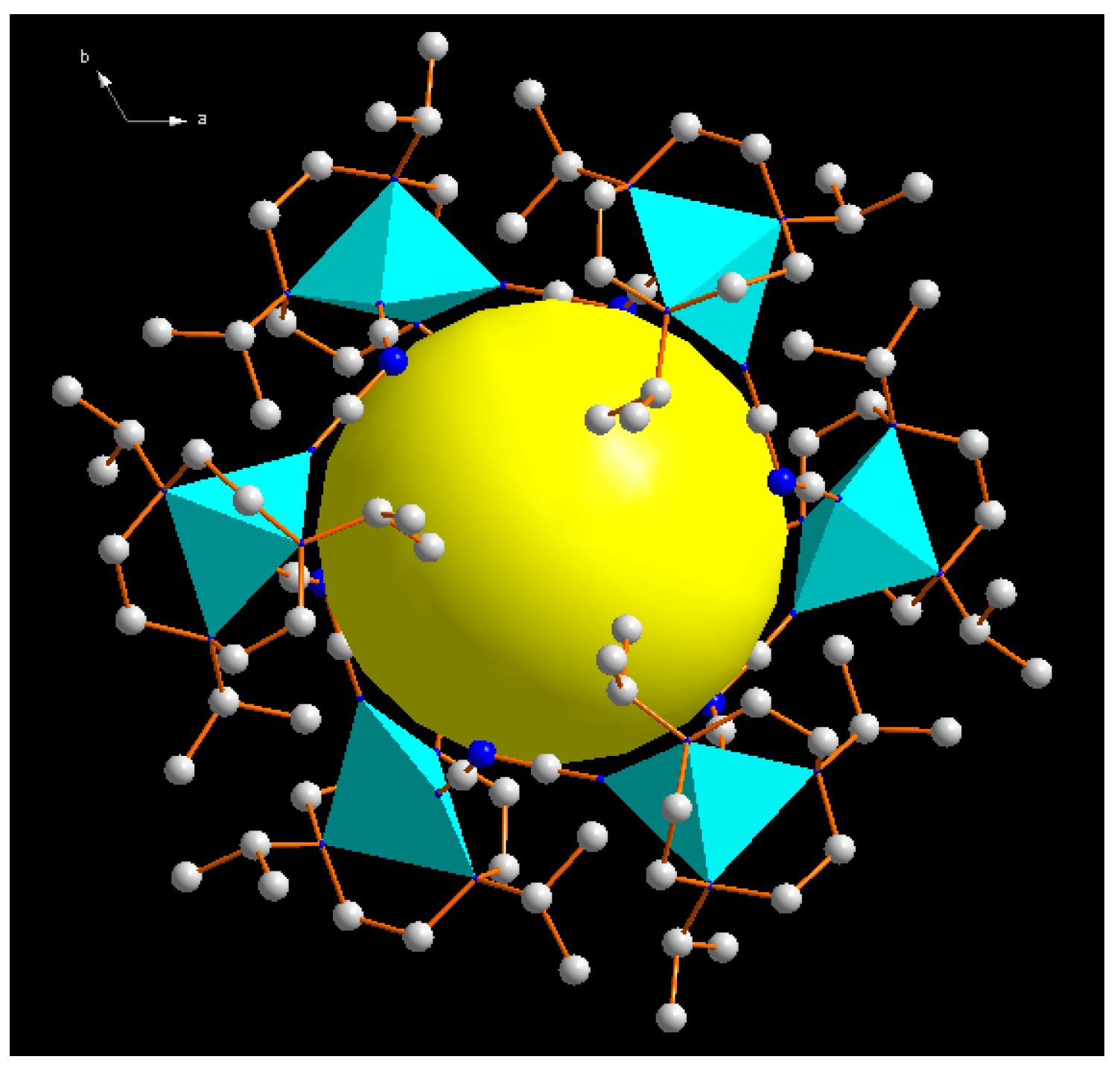
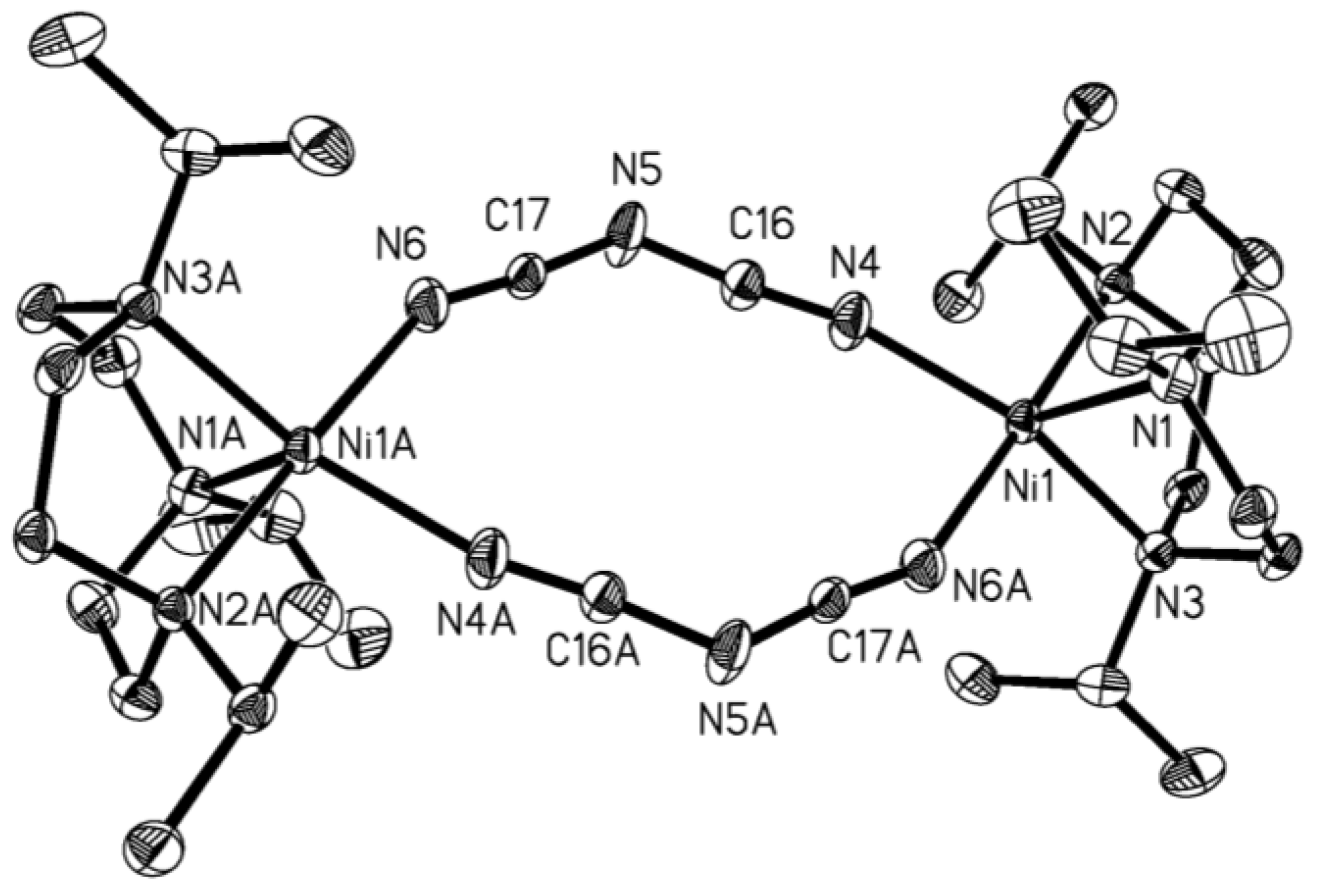
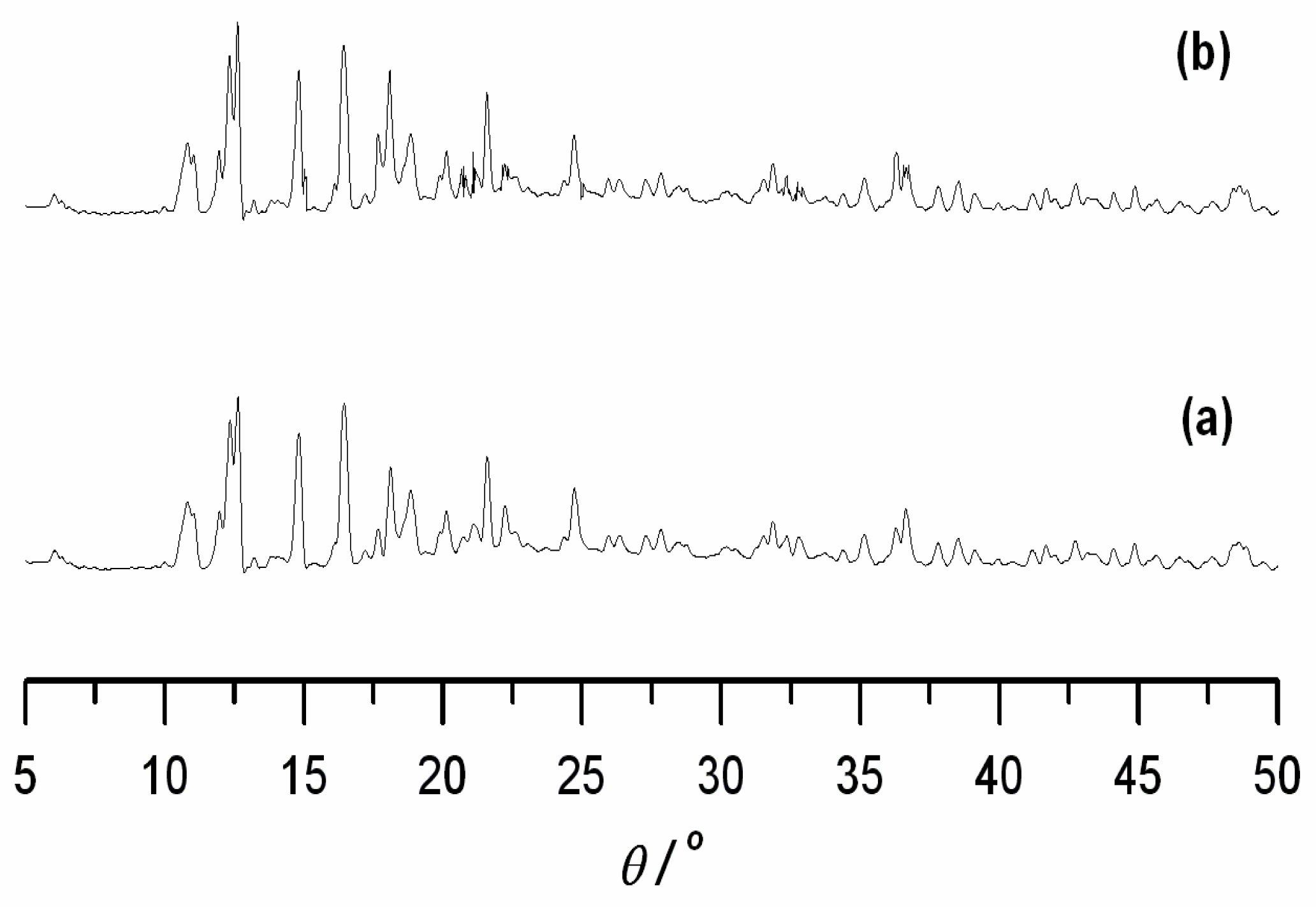
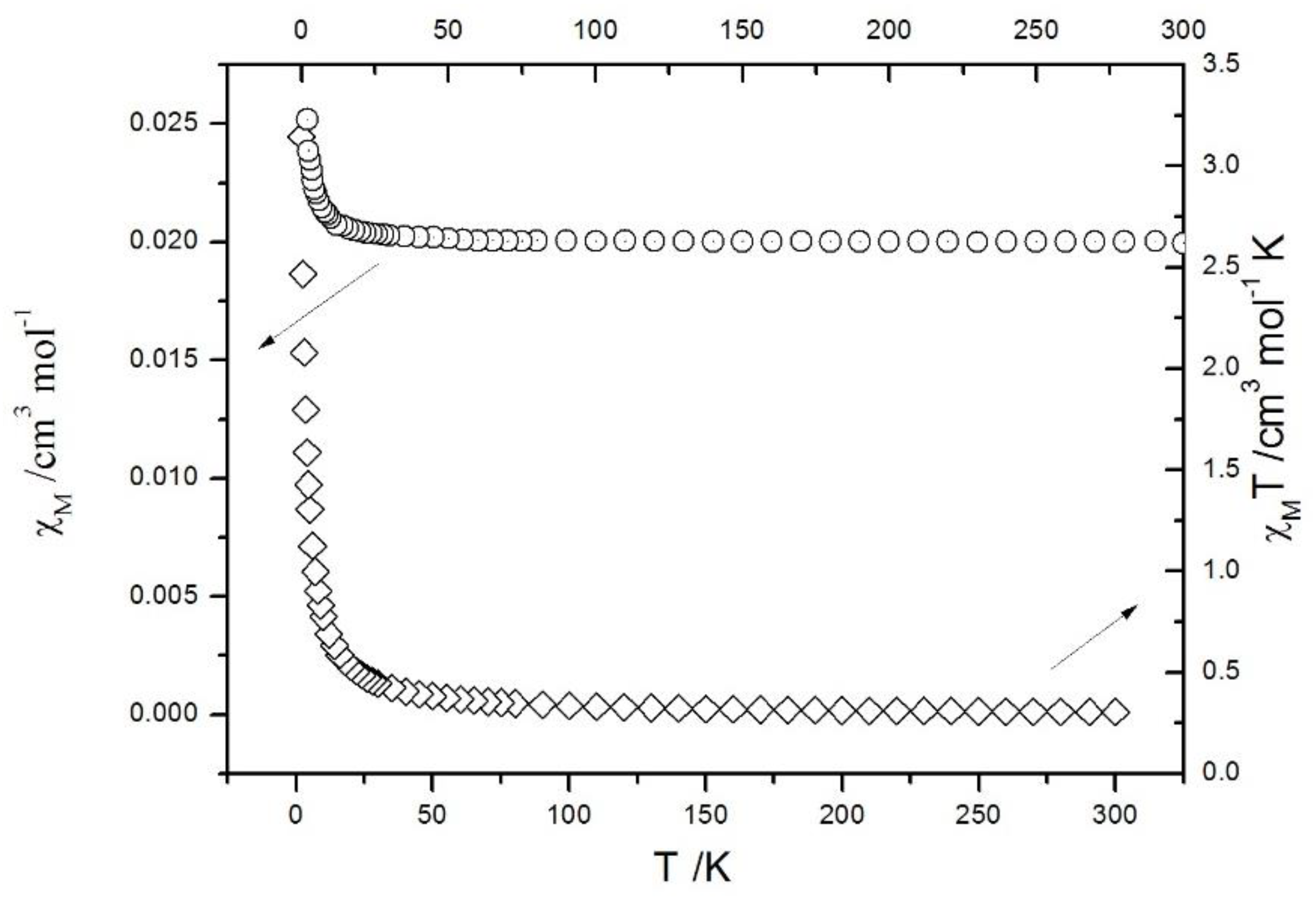
| Compound | 1 | 2 |
|---|---|---|
| Empirical formula | C17H33.17CuF6N6O0.08P | C82H106Ni2B2N12 |
| Formula weight | 531.51 | 1398.83 |
| Temperature | 294(2) K | 293(2) K |
| Wavelength | 0.71073 Å | 0.71073 Å |
| Crystal system, space group | Rhombohedral, R–3 | Triclinic, P–1 |
| Unit cell dimensions | a = 28.417(3) Å α = 90° | a = 10.2350(2) Å α = 74.4° |
| b = 28.417(3) Å β = 90° | b = 10.8420(2) Å β = 88.8° | |
| c = 15.6781(19) Å γ = 120° | c = 18.055(3) Å γ = 78.5° | |
| Volume | 10964(2) Å−3 | 1890.2(6) Å−3 |
| Z, Calculated density | 18, 1.449 Mg m−3 | 1, 1.229 Mg m−3 |
| Absorption coefficient | 1.024 mm−1 | 0.550 mm−1 |
| F(000) | 4965 | 748 |
| Crystal size | 0.26 × 0.20 × 0.16 mm3 | 0.24 × 0.20 × 0.14 mm3 |
| θ range for data collection | 1.43–25.02° | 1.99–26.40° |
| Limiting indices | −33 ≤ h ≤ 17, −33 ≤ k ≤ 33, −18 ≤ l ≤ 18 | −12 ≤ h ≤ 12, −5 ≤ k ≤ 13, −22 ≤ l ≤ 22 |
| Reflections collected/unique | 18,801/4320 [R(int) = 0.0511] | 10,744/7607 [R(int) = 0.0255] |
| Max. and min. transmission | 1.000000 and 0.712842 | 1.000000 and 0.818876 |
| Refinement method | Full-matrix least-squares on F2 | Full-matrix least-squares on F2 |
| Data/restraints/parameters | 4320/54/287 | 7607/0/448 |
| Goodness-of-fit on F2 | 1.066 | 1.006 |
| Final R indices [I > 2θ(I)] | R1 = 0.0421, wR2 = 0.1133 | R1 = 0.0442, wR2 = 0.0860 |
| R indices (all data) | R1 = 0.0691, wR2 = 0.1320 | R1 = 0.0774, wR2 = 0.0990 |
| Largest diff. peak and hole | 0.558 and −0.362 e Å−3 | 0.326 and −0.290 e Å−3 |
| 1 | |||
| Cu(1)-N(4) | 1.992(3) | Cu(1)-N(2) | 2.077(3) |
| Cu(1)-N(6) | 2.001(4) | Cu(1)-N(3) | 2.201(3) |
| Cu(1)-N(1) | 2.070(3) | ||
| N(4)-Cu(1)-N(6) | 86.05(14) | N(1)-Cu(1)-N(2) | 85.62(12) |
| N(4)-Cu(1)-N(1) | 95.30(13) | N(4)-Cu(1)-N(3) | 118.49(15) |
| N(6)-Cu(1)-N(1) | 178.58(14) | N(6)-Cu(1)-N(3) | 93.39(16) |
| N(4)-Cu(1)-N(2) | 154.96(15) | N(1)-Cu(1)-N(3) | 85.58(14) |
| N(6)-Cu(1)-N(2) | 93.35(14) | N(2)-Cu(1)-N(3) | 86.55(13) |
| 2 | |||
| Ni(1)-N(4) | 2.022(2) | Ni(1)-N(1) | 2.065(2) |
| Ni(1)-N(6) | 2.086(2) | Ni(1)-N(3) | 2.0897(19) |
| Ni(1)-N(2) | 2.0992(18) | ||
| N(4)-Ni(1)-N(1) | 117.56(9) | N(4)-Ni(1)-N(6) | 84.72(9) |
| N(1)-Ni(1)-N(6) | 93.79(9) | N(4)-Ni(1)-N(3) | 154.51(9) |
| N(1)-Ni(1)-N(3) | 87.87(8) | N(6)-Ni(1)-N(3) | 95.90(8) |
| N(4)-Ni(1)-N(2) | 93.74(8) | N(1)-Ni(1)-N(2) | 87.51(7) |
| N(6)-Ni(1)-N(2) | 178.33(8) | N(3)-Ni(1)-N(2) | 85.19(7) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.-S.; Liu, L.-J.; Ju, H.-Y.; Liu, X.-Y.; Li, Y.-G.; Yan, S.-P. Dicyanamide Bridged Cu(II)36-Metallacrown-6 Complex with 1,4,7-Triisopropyl-1,4,7-Triazacyclononane and Binding Properties with DNA. Molecules 2018, 23, 1269. https://doi.org/10.3390/molecules23061269
Yang Y-S, Liu L-J, Ju H-Y, Liu X-Y, Li Y-G, Yan S-P. Dicyanamide Bridged Cu(II)36-Metallacrown-6 Complex with 1,4,7-Triisopropyl-1,4,7-Triazacyclononane and Binding Properties with DNA. Molecules. 2018; 23(6):1269. https://doi.org/10.3390/molecules23061269
Chicago/Turabian StyleYang, Yong-Sheng, Li-Jun Liu, Hai-Yan Ju, Xiu-Ying Liu, Yu-Guang Li, and Shi-Ping Yan. 2018. "Dicyanamide Bridged Cu(II)36-Metallacrown-6 Complex with 1,4,7-Triisopropyl-1,4,7-Triazacyclononane and Binding Properties with DNA" Molecules 23, no. 6: 1269. https://doi.org/10.3390/molecules23061269
APA StyleYang, Y.-S., Liu, L.-J., Ju, H.-Y., Liu, X.-Y., Li, Y.-G., & Yan, S.-P. (2018). Dicyanamide Bridged Cu(II)36-Metallacrown-6 Complex with 1,4,7-Triisopropyl-1,4,7-Triazacyclononane and Binding Properties with DNA. Molecules, 23(6), 1269. https://doi.org/10.3390/molecules23061269






