Discovery of Pyrazolo[4,3-c]quinolines Derivatives as Potential Anti-Inflammatory Agents through Inhibiting of NO Production
Abstract
:1. Introduction
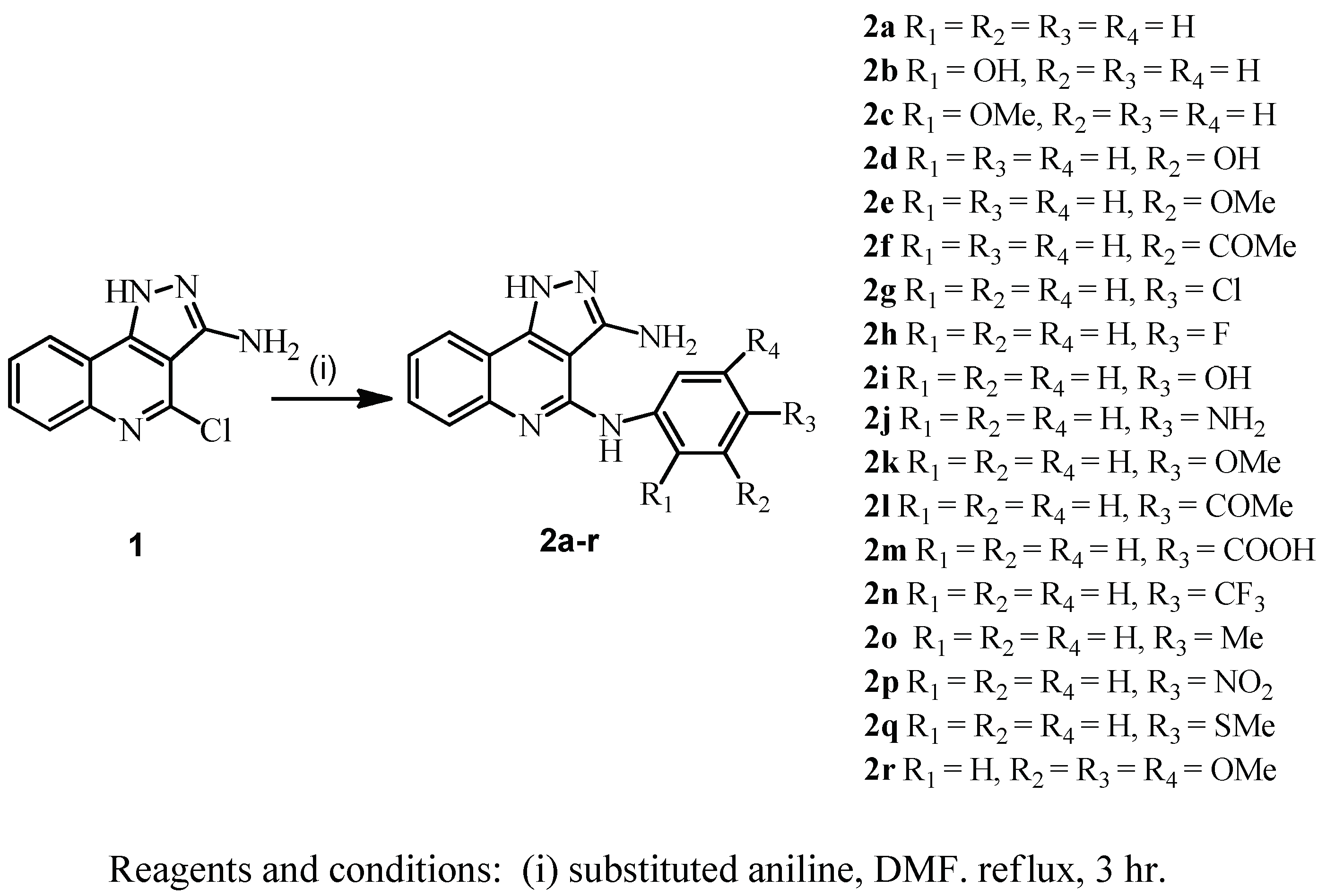
2. Chemistry
3. Results and Discussion
3.1. Lipopolysaccharide (LPS)-Induced Nitric Oxide Production
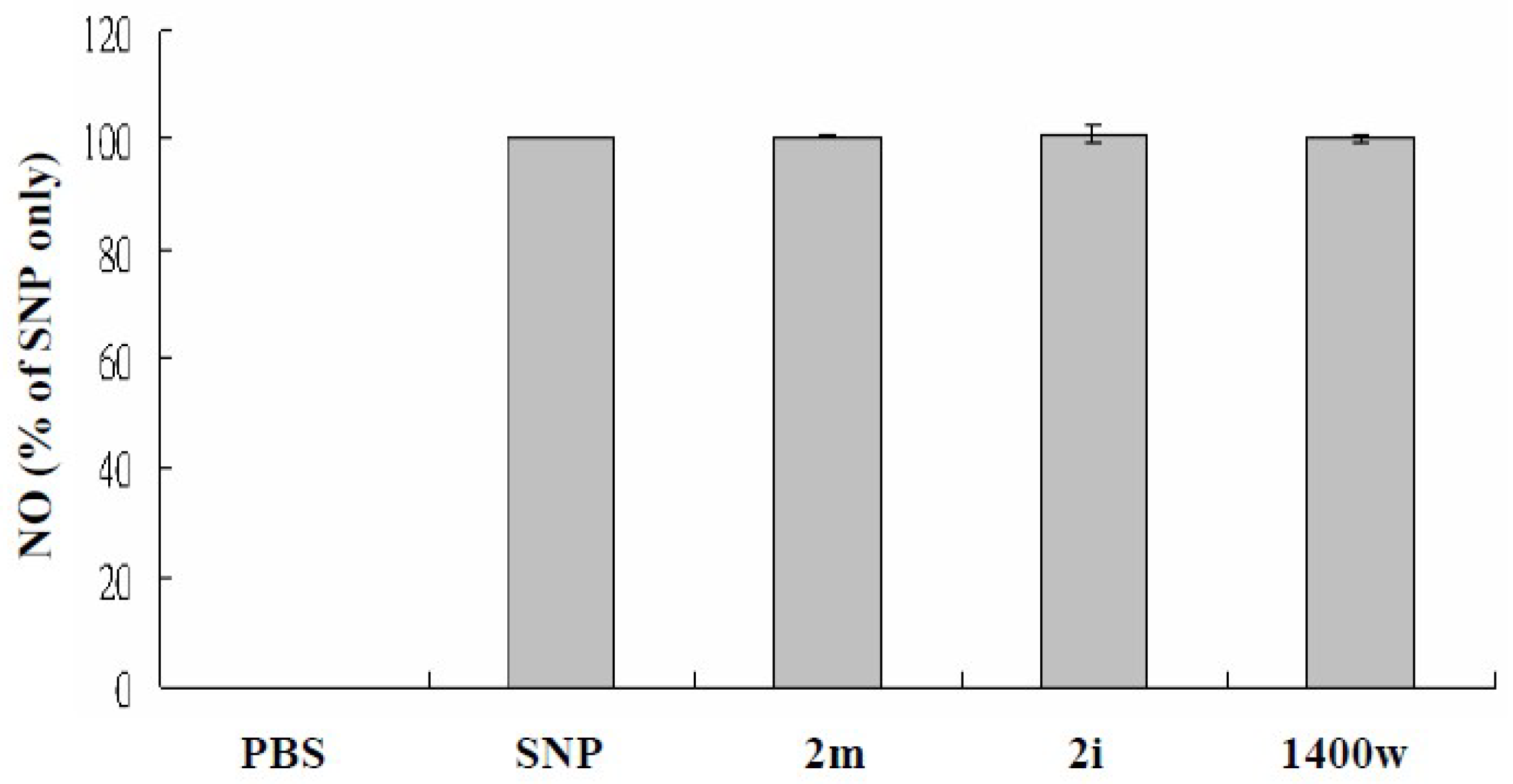
3.2. Nitric Oxide (NO)-Scavenging Effects
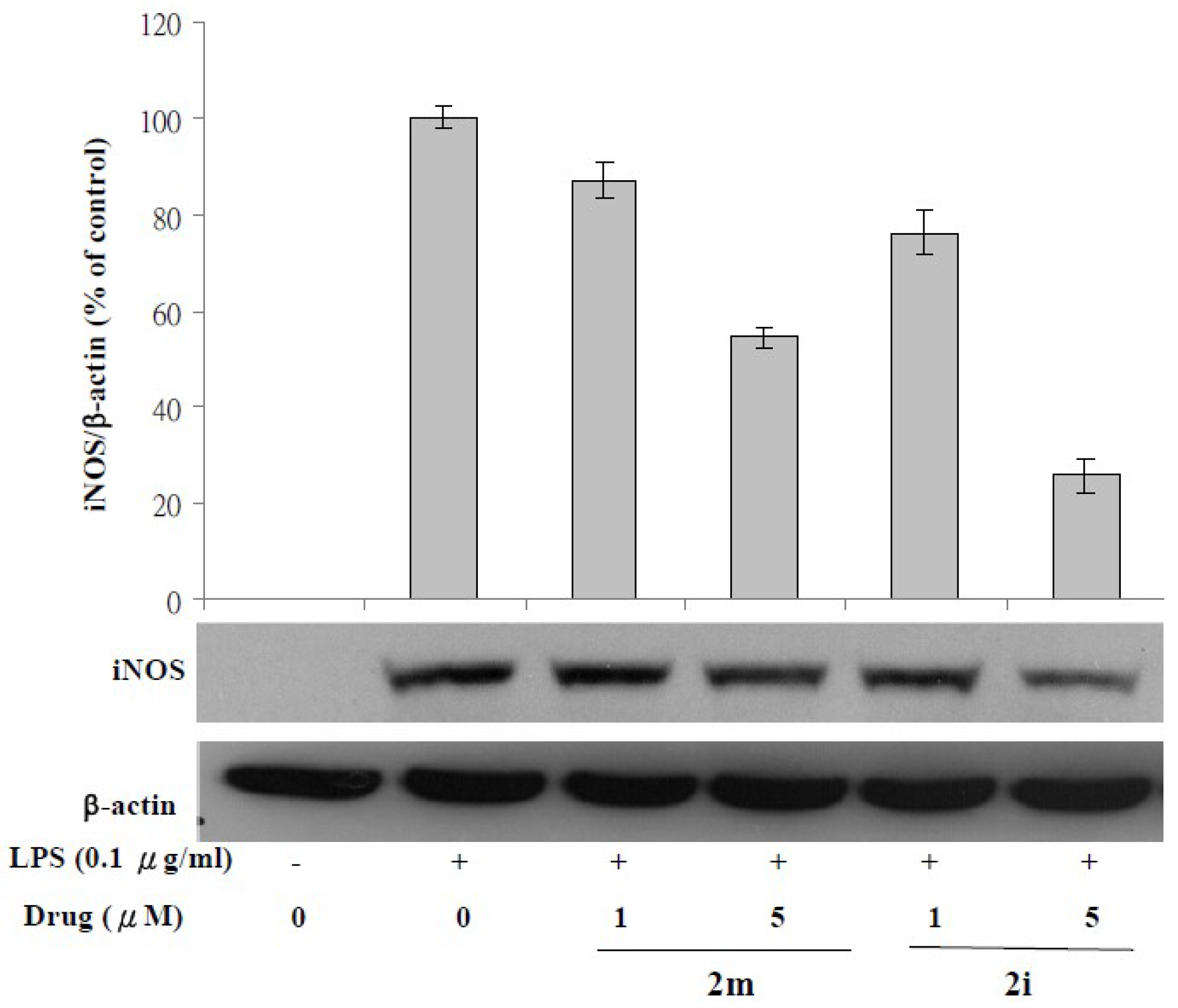
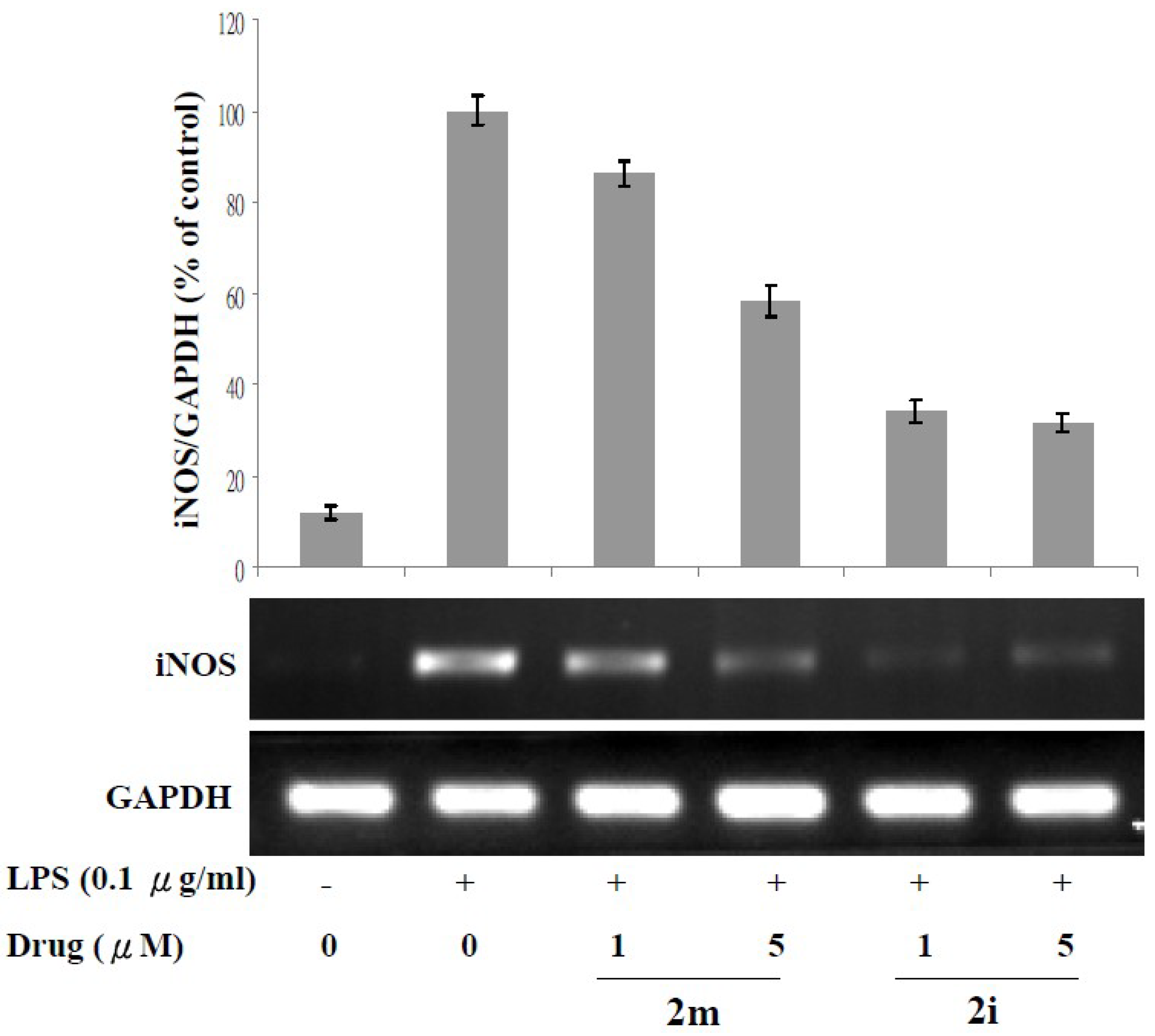
3.3. Lipopolysaccharide (LPS)-Induced iNOS Protein and mRNA Expression
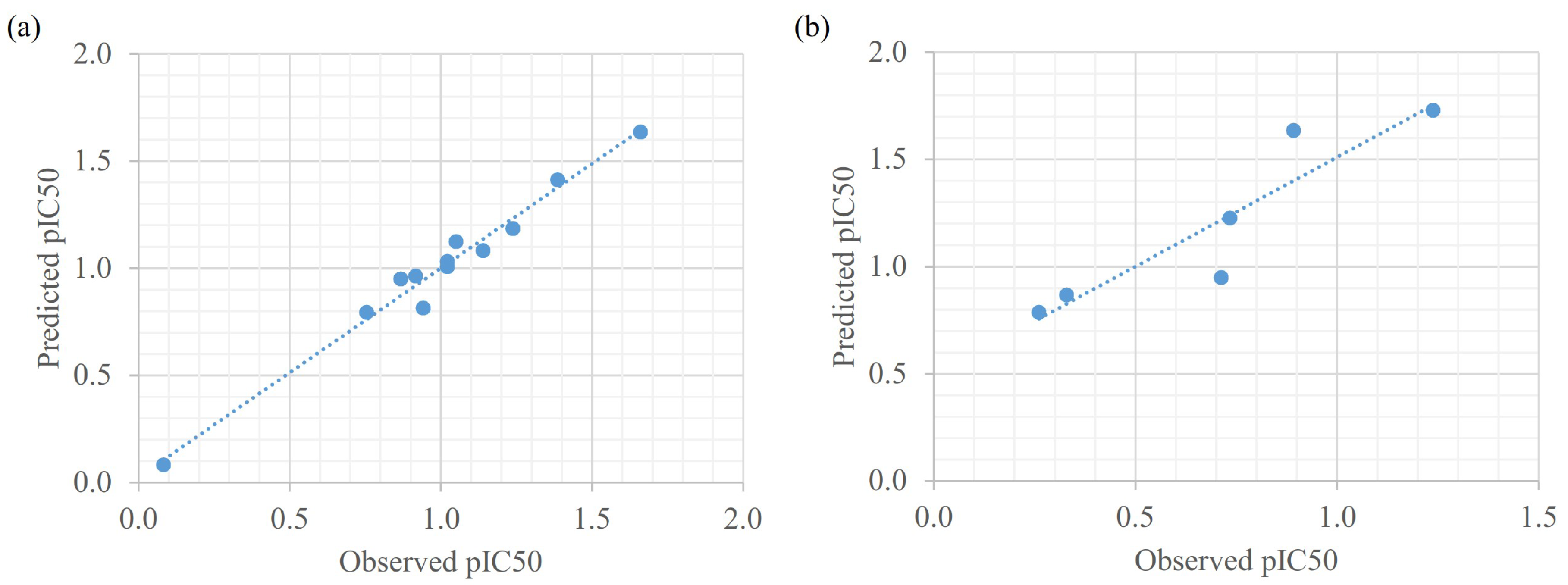
3.4. Quantitative Structure-Activity Relationship (QSAR)
4. Experimental
4.1. General Information
4.1.1. 3-Amino-4-(2-methoxyphenylamino)-1H-pyrazolo[4,3-c]quinoline (2c)
4.1.2. 3-Amino-4-(3-methoxyphenylamino)-1H-pyrazolo[4,3-c]quinoline (2e)
4.1.3. 3-Amino-4-(3-acetylphenylamino)-1H-pyrazolo[4,3-c]quinoline (2f)
4.1.4. 3-Amino-4-(4-methylthiophenylamino)-1H-pyrazolo[4,3-c]quinoline (2q)
4.1.5. 3-Amino-4-(3,4,5-trimethoxyphenylamino)-1H-pyrazolo[4,3-c]quinoline (2r)
4.2. Cell Culture
4.2.1. Nitrite Measurement
4.2.2. Cell Viability
4.2.3. NO-Scavenging Activity
4.2.4. Western Blotting
4.2.5. RNA Extraction and Determination of iNOS mRNA Expression
4.2.6. Quantitative Structure-Activity Relationship (QSAR)
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Singer, I.I.; Kawka, D.W.; Scott, S.; Weidner, J.R.; Mumford, R.A.; Riehl, T.E.; Stenson, W.F. Expression of inducible nitric oxide synthase and nitrotyrosine in colonic epithelium in inflammatory bowel disease. Gastroenterology 1996, 111, 871–885. [Google Scholar] [CrossRef]
- Nguyen, T.; Brunson, D.; Crespi, C.L.; Penman, B.W.; Wishnok, J.S.; Tannenbaum, S.R. DNA damage and mutation in human cells exposed to nitric oxide in vitro. Proc. Natl. Acad. Sci. USA 1992, 1, 3030–3034. [Google Scholar] [CrossRef]
- Rees, D.D.; Palmer, R.M.J.; Moncada, S. Role of Endothelium-derived nitric oxide in the regulation of blood pressure. Proc. Natl. Acad. Sci. USA 1989, 86, 3375–3378. [Google Scholar] [CrossRef] [PubMed]
- Marletta, M.A. Nitric oxide synthase structure and mechanism. J. Biol. Chem. 1993, 268, 12231–12234. [Google Scholar] [PubMed]
- Grisham, M.B.; Jourd, H.D.; Wink, D.A.I. Physiological chemistry of nitric oxide and its metabolites: Implications in inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 1999, 276, 315–321. [Google Scholar] [CrossRef]
- Morikawa, A.; Koide, N.; Kato, Y.; Sugiyama, T.; Chakravortty, D.; Yoshida, T.; Yokochi, T. Augmentation of nitric oxide production by gamma interferon in a mouse vascular endothelial cell line and its modulation by tumor necrosis factor alpha and lipopolysaccharide. Infect. Immun. 2000, 68, 6209–6214. [Google Scholar] [CrossRef] [PubMed]
- Bach, D.H.; Liu, J.Y.; Kim, W.K.; Hong, J.Y.; Park, S.H.; Kim, D.; Qin, S.N.; Luu, T.T.; Park, H.J.; Xu, Y.N.; et al. Synthesis and biological activity of new phthalimides as potential anti-inflammatory agents. Bioorg. Med. Chem. 2017, 25, 3396–3405. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Xie, C.; Ma, Y.; Liu, J.; Xiang, M.; Ye, X.; Zheng, H.; Chen, Z.; Xu, Q.; Chen, T.; et al. Synthesis and biological evaluation of novel 5-benzylidenethiazolidine-2,4-dione derivatives for the treatment of inflammatory diseases. J. Med. Chem. 2011, 54, 2060–2068. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; He, L.; Lei, L.; Liang, X.; Lei, K.; Zhang, R.; Yang, Z.; Chen, L. Synthesis and biological evaluation of 5-nitropyrimidine-2,4-dione analogues as inhibitors of nitric oxide and iNOS activity. Chem. Biol. Drug Des. 2015, 85, 296–299. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Pei, H.; Lei, L.; He, L.; Chen, J.; Liang, X.; Peng, A.; Ye, H.; Xiang, M.; Chen, L. Structural exploration, synthesis and pharmacological evaluation of novel 5-benzylidenethiazolidine-2,4-dione derivatives as iNOS inhibitors against inflammatory diseases. Eur. J. Med. Chem. 2015, 92, 178–190. [Google Scholar] [CrossRef] [PubMed]
- Mahapatra, D.K.; Bharti, S.K.; Asata, V. Chalcone derivatives: Anti-inflammatory potential and molecular targets perspectives. Curr. Top. Med. Chem. 2017, 17, 3146–3169. [Google Scholar] [CrossRef] [PubMed]
- Bluhm, U.; Boucher, J.L.; Buss, U.; Clement, B.; Friedrich, F.; Girreser, U.; Heber, D.; Lam, T.; Lepoivre, M.; Rostaie-Gerylow, M.; et al. Synthesis and evaluation of pyrido[1,2-a]pyrimidines as inhibitors of nitric oxide synthases. Eur. J. Med. Chem. 2009, 44, 2877–2887. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.L.; Lin, J.; Lv, X.Y.; Feng, J.H.; Zhang, X.Q.; Wang, H.; Ye, W.C. Synthesis and biological evaluation of clovamide analogues as potent anti-neuroinflammatory agents in vitro and in vivo. Eur. J. Med. Chem. 2018, 151, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Tseng, C.H.; Lin, C.S.; Shih, P.K.; Tsao, L.T.; Wang, J.P.; Cheng, C.M.; Tzeng, C.C.; Chen, Y.L. Furo[3,2:3,4]naphtho[1,2-d]imidazole derivatives as potential inhibitors of inflammatory factors in sepsis. Bioorg. Med. Chem. 2009, 17, 6773–6779. [Google Scholar] [CrossRef] [PubMed]
- Mekheimer, R. A convenient synthesis of new substituted pyrazolo[4,3-c]quinolines with potentialantiinflammatory activity. Pharmazie 1994, 49, 486–489. [Google Scholar] [PubMed]
- Cheng, K.W.; Tseng, C.H.; Yang, C.N.; Tzeng, C.C.; Cheng, T.C.; Leu, Y.L.; Chuang, Y.C.; Wang, J.Y.; Lu, Y.C.; Chen, Y.L.; et al. Specific inhibition of bacterial β-glucuronidase by pyrazolo[4,3-c]quinoline derivatives via a pH-dependent manner to suppress chemotherapy- induced intestinal toxicity. J. Med. Chem. 2017, 60, 9222–9238. [Google Scholar] [CrossRef] [PubMed]
- Mekheimer, R.A. Fused quinoline heterocycles I. First example of the 2,4-diazidoquinoline-3-carbonitrile and 1-aryl-1,5-dihydro-1,2,3,4,5,6-hexaazaacephenanthrylenes ring systems. J. Chem. Soc. Perkin Trans. 1 1999, 2183–2188. [Google Scholar] [CrossRef]
- Green, L.C.; Wagner, D.A.; Glogowski, J.; Skipper, P.L.; Wishnok, J.S.; Tannenbaum, S.R. Analysis of nitrate, nitrite, and [15N]nitrate in biological fluids. Anal. Biochem. 1982, 126, 131–138. [Google Scholar] [CrossRef]
- Alderton, W.K.; Cooper, C.E.; Knowles, R.G. Nitric oxide synthases: Structure, function and inhibition. Biochem. J. 2001, 357, 593–615. [Google Scholar] [CrossRef] [PubMed]
- Marcocci, L.; Maguire, J.J.; Droy-Lefaix, M.T.; Packer, L. The nitric oxide-scavenging properties of Ginkgo biloba extract EGb 761. Biochem. Biophys. Res. Commun. 1994, 201, 748–755. [Google Scholar] [CrossRef] [PubMed]
- Ronchetti, D.; Impagnatiello, F.; Guzzetta, M.; Gasparini, L.; Borgatti, M.; Gambari, R.; Ongini, E. Modulation of iNOS expression by a nitric oxide-realeasing derivatives of the natural antioxidant ferulic acid in activatived RAW 264.7 macrophages. Eur. J. Pharmacol. 2006, 532, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.J.; Chen, S.M.; Chen, W.C.; Chang, Y.M.; Lee, T.H. Selective inducible nitric oxide synthase suppression by new bracteanolides from Murdannia bracteata. J. Ethnopharmacol. 2007, 112, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Mardini, I.A.; FitzGerald, G.A. Selective inhibitors of cyclooxygenase-2: A growing class of antiinflammatory drugs. Mol. Interv. 2001, 1, 30–38. [Google Scholar] [PubMed]
- Yang, H.; Yin, P.; Shi, Z.; Ma, Y.; Zhao, C.; Zheng, J.; Chen, T. Sinomenine, a COX-2 inhibitor, induces cell cycle arrest and inhibits growth of human coloncarcinoma cells in vitro and in vivo. Oncol. Lett. 2016, 11, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Halawany, A.M.E.; Sayed, N.S.E.; Abdallah, H.M.; Dine, R.S.E. Protective effects of gingerol on streptozotocin-induced sporadic Alzheimer’s disease: Emphasis on inhibition of β-amyloid, COX-2, alpha-, beta-secretases and APH1a. Sci. Rep. 2017, 7, 2902. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tian, D.; Zhu, C.; Ren, L. Demethoxycurcumin preserves renovascular function by downregulating COX-2 expression in hypertension. Oxid. Med. Cell. Longev. 2016, 2016, 9045736. [Google Scholar] [CrossRef] [PubMed]
- Tung, C.W. Prediction of pupylation sites using the composition of k-spaced amino acid pairs. J. Theor. Biol. 2013, 336, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Tung, C.W.; Wu, M.T.; Chen, Y.K.; Wu, C.C.; Chen, W.C.; Li, H.P.; Chou, S.H.; Wu, D.C.; Wu, I.C. Identification of biomarkers for esophageal squamous cell carcinoma using feature selection and decision tree methods. Sci. World J. 2013, 2013, 782031. [Google Scholar] [CrossRef] [PubMed]
- Tseng, C.H.; Tung, C.W.; Wu, C.H.; Tzeng, C.C.; Chen, Y.H.; Hwang, T.L.; Chen, Y.L. Discovery of indeno[1,2-c]quinoline derivatives as potent dual antituberculosis and anti-inflammatory agents. Molecules 2017, 22, 1001. [Google Scholar] [CrossRef] [PubMed]
- Gramatica, P.; Corradi, M.; Consonni, V. Modelling and prediction of soil sorption coefficients of non-ionic organic pesticides by molecular descriptors. Chemosphere 2000, 41, 763–777. [Google Scholar] [CrossRef]
- Hall, L.H.; Kier, L.B. Electrotopological state indices for atom types: A novel combination of electronic, topological, and valence state information. J. Chem. Inf. Comput. Sci. 1995, 35, 1039–1045. [Google Scholar] [CrossRef]
- Liu, R.; Sun, H.; So, S.S. Development of quantitative structure−property relationship models for early ADME evaluation in drug discovery. 2. Blood-brain barrier penetration. J. Chem. Inf. Comput. Sci. 2001, 41, 1623–1632. [Google Scholar] [CrossRef] [PubMed]
- Steinbeck, C.; Han, Y.; Kuhn, S.; Horlacher, O.; Luttmann, E.; Willighagen, E. The Chemistry Development Kit (CDK): An open-source Java library for Chemo- and Bioinformatics. J. Chem. Inf. Comput. Sci. 2003, 43, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Todeschini, R.; Consonni, V. Molecular Descriptors for Chemoinformatics; Wiley VCH: Weinheim, Germany, 2009. [Google Scholar]
- Roehm, N.W.; Rodgers, G.H.; Hatfield, S.M.; Glasebrook, A.L. An improved colorimetric assay for cell proliferation and viability utilizing the tetrazolium salt XTT. J. Immunol. Methods 1991, 142, 257–265. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Yap, C.W. PaDEL-descriptor: An open source software to calculate molecular descriptors and fingerprints. J. Comput. Chem. 2011, 32, 1466–1474. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds reported herein are available from the authors. |

| Compound | a Percent Inhibition of NO Production at 10 μM | a IC50 ( μM) | a Cell Survival Rate at 10 μM |
|---|---|---|---|
| 2a | 81.66 ± 0.99 | 0.39 ± 0.08 | 9.09 ± 0.24 |
| 2b | 103.44 ± 6.55 | 0.42 ± 0.11 | 53.48 ± 1.53 |
| 2c | 82.23 ± 6.49 | 0.49 ± 0.14 | 47.31 ± 1.67 |
| 2d | 93.52 ± 7.57 | 0.77 ± 0.23 | 81.70 ± 0.09 |
| 2e | 101.43 ± 1.31 | 0.72 ± 0.18 | 94.32 ± 0.49 |
| 2f | 99.14 ± 2.16 | 0.29 ± 0.06 | 94.37 ± 0.36 |
| 2g | 103.72 ± 6.71 | 0.32 ± 0.05 | 81.61 ± 0.14 |
| 2h | 94.84 ± 9.47 | 0.41 ± 0.07 | 89.99 ± 0.02 |
| 2i | 101.72 ± 2.63 | 0.19 ± 0.05 | 87.84 ± 0.15 |
| 2j | 95.13 ± 3.75 | 0.92 ± 0.05 | 90.26 ± 0.05 |
| 2k | 90.83 ± 11.94 | 0.48 ± 0.14 | 90.57 ± 0.03 |
| 2l | 102.01 ± 6.49 | 0.29 ± 0.08 | 87.80 ± 0.11 |
| 2m | 87.68 ± 9.93 | 0.22 ± 0.07 | 85.42 ± 0.77 |
| 2n | 106.88 ± 2.48 | 0.36 ± 0.09 | 69.14 ± 0.62 |
| 2o | 100.03 ± 3.02 | 0.47 ± 0.07 | 84.53 ± 0.04 |
| 2p | 89.11 ± 0.86 | 0.40 ± 0.09 | 90.86 ± 0.32 |
| 2q | 102.29 ± 7.01 | 0.36 ± 0.08 | 89.62 ± 0.05 |
| 2r | 107.16 ± 2.58 | 0.35 ± 0.06 | 85.24 ± 0.04 |
| 1400w b | 102.15 ± 2.16 | 0.20 ± 0.02 | 99.54 ± 2.51 |
| Coefficient | Estimate Std. Error | t-Value | Pr (>|t|) | ||
|---|---|---|---|---|---|
| (Intercept) | 1.258 | 0.196 | 6.42 | 3.60 × 10−4 | *** |
| SsNH2 | −0.262 | 0.017 | −15.16 | 1.30 × 10−6 | *** |
| SHBint9 | 0.049 | 0.005 | 9.21 | 3.70 × 10−5 | *** |
| nHBAcc | 0.226 | 0.038 | 5.96 | 5.60 × 10−4 | *** |
| AATSC4m | −0.037 | 0.011 | −3.46 | 1.06 × 10−2 | * |
| Compound | Observed pIC50 | Predicted pIC50 | Error |
|---|---|---|---|
| 2j | 0.083 | 0.083 | 0.000 |
| 2o | 0.755 | 0.794 | −0.039 |
| 2b | 0.868 | 0.950 | −0.083 |
| 2p | 0.916 | 0.963 | −0.047 |
| 2a | 0.942 | 0.814 | 0.128 |
| 2n | 1.022 | 1.032 | −0.010 |
| 2q | 1.022 | 1.007 | 0.015 |
| 2r | 1.050 | 1.125 | −0.075 |
| 2g | 1.139 | 1.082 | 0.058 |
| 2l | 1.238 | 1.185 | 0.053 |
| 2m | 1.386 | 1.413 | −0.027 |
| 2i | 1.661 | 1.636 | 0.025 |
| Compound | Observed pIC50 | Predicted pIC50 | Error |
|---|---|---|---|
| 2d | 0.261 | 0.786 | −0.524 |
| 2e | 0.329 | 0.868 | −0.540 |
| 2c | 0.713 | 0.949 | −0.236 |
| 2k | 0.734 | 1.227 | −0.493 |
| 2h | 0.892 | 1.635 | −0.743 |
| 2f | 1.238 | 1.729 | −0.491 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tseng, C.-H.; Tung, C.-W.; Peng, S.-I.; Chen, Y.-L.; Tzeng, C.-C.; Cheng, C.-M. Discovery of Pyrazolo[4,3-c]quinolines Derivatives as Potential Anti-Inflammatory Agents through Inhibiting of NO Production. Molecules 2018, 23, 1036. https://doi.org/10.3390/molecules23051036
Tseng C-H, Tung C-W, Peng S-I, Chen Y-L, Tzeng C-C, Cheng C-M. Discovery of Pyrazolo[4,3-c]quinolines Derivatives as Potential Anti-Inflammatory Agents through Inhibiting of NO Production. Molecules. 2018; 23(5):1036. https://doi.org/10.3390/molecules23051036
Chicago/Turabian StyleTseng, Chih-Hua, Chun-Wei Tung, Shin-I Peng, Yeh-Long Chen, Cherng-Chyi Tzeng, and Chih-Mei Cheng. 2018. "Discovery of Pyrazolo[4,3-c]quinolines Derivatives as Potential Anti-Inflammatory Agents through Inhibiting of NO Production" Molecules 23, no. 5: 1036. https://doi.org/10.3390/molecules23051036
APA StyleTseng, C.-H., Tung, C.-W., Peng, S.-I., Chen, Y.-L., Tzeng, C.-C., & Cheng, C.-M. (2018). Discovery of Pyrazolo[4,3-c]quinolines Derivatives as Potential Anti-Inflammatory Agents through Inhibiting of NO Production. Molecules, 23(5), 1036. https://doi.org/10.3390/molecules23051036







