iTRAQ-Based Identification of Proteins Related to Lignin Synthesis in the Pear Pollinated with Pollen from Different Varieties
Abstract
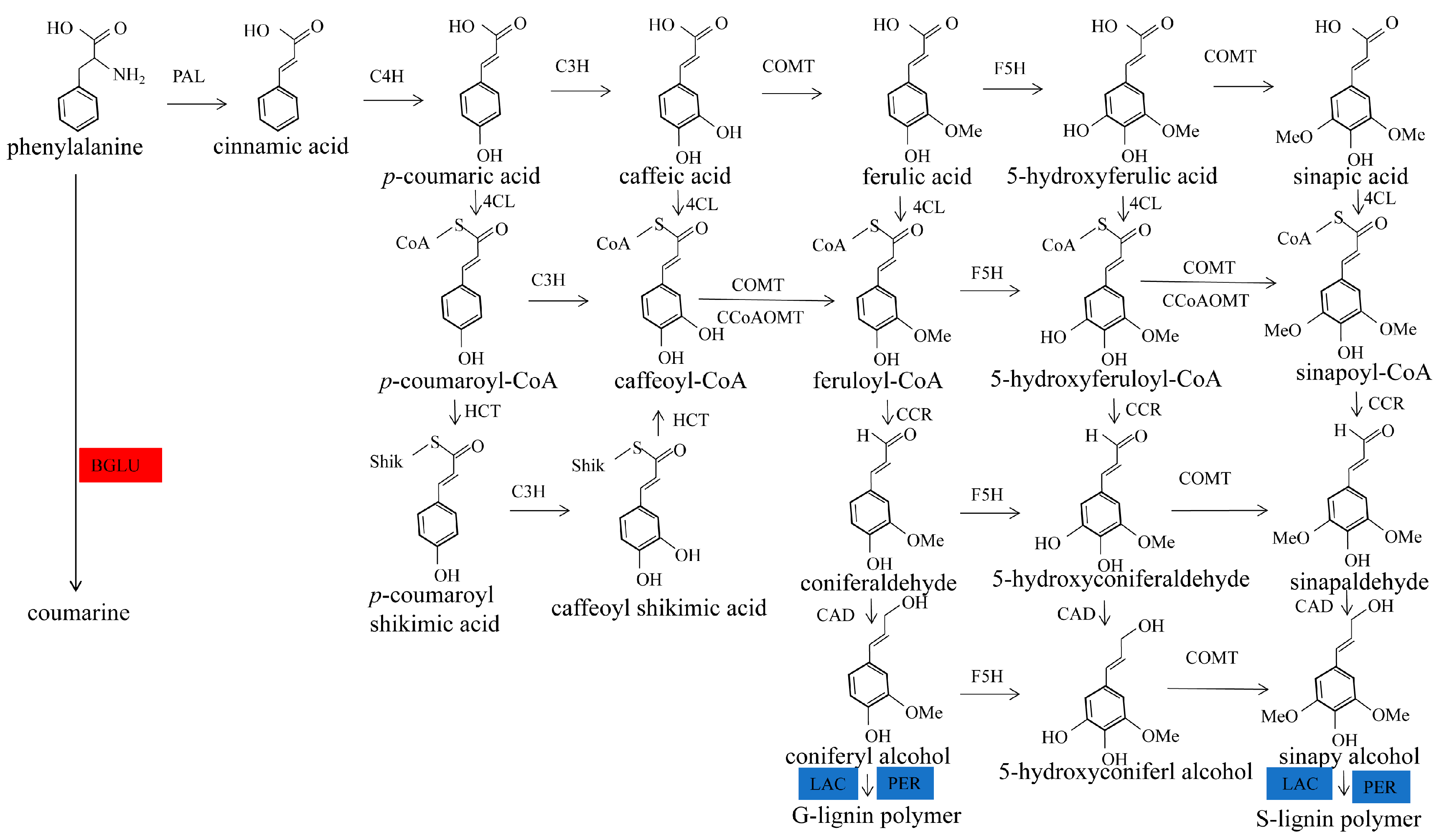
1. Introduction
2. Results
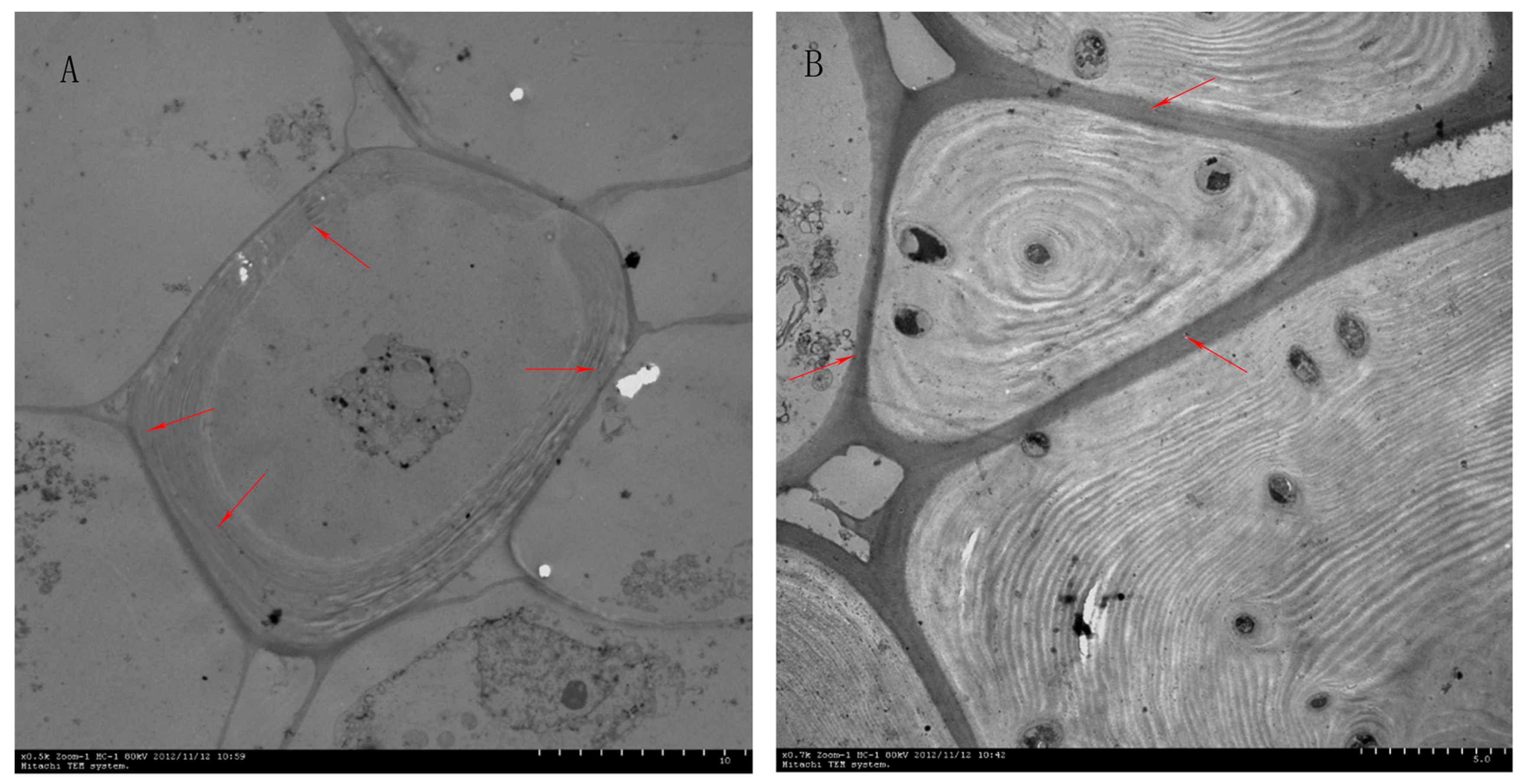
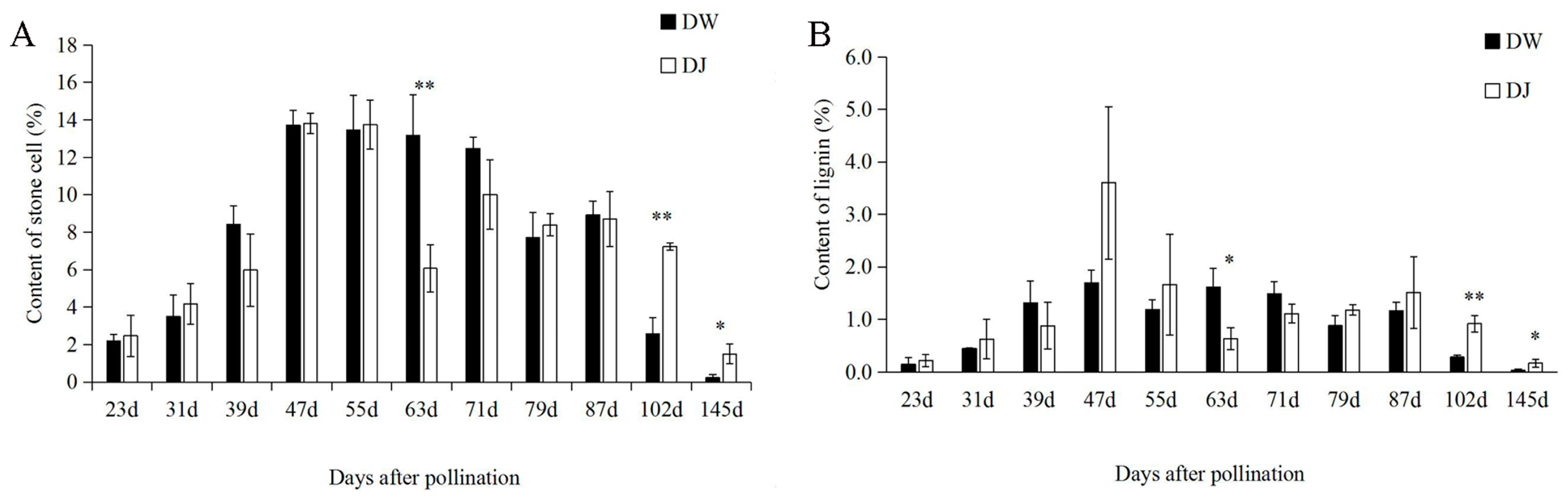
2.1. Effect of Different Pollinations on Dangshan Su Stone Cell Content and Lignin Content
2.2. Protein Identification and Quantification
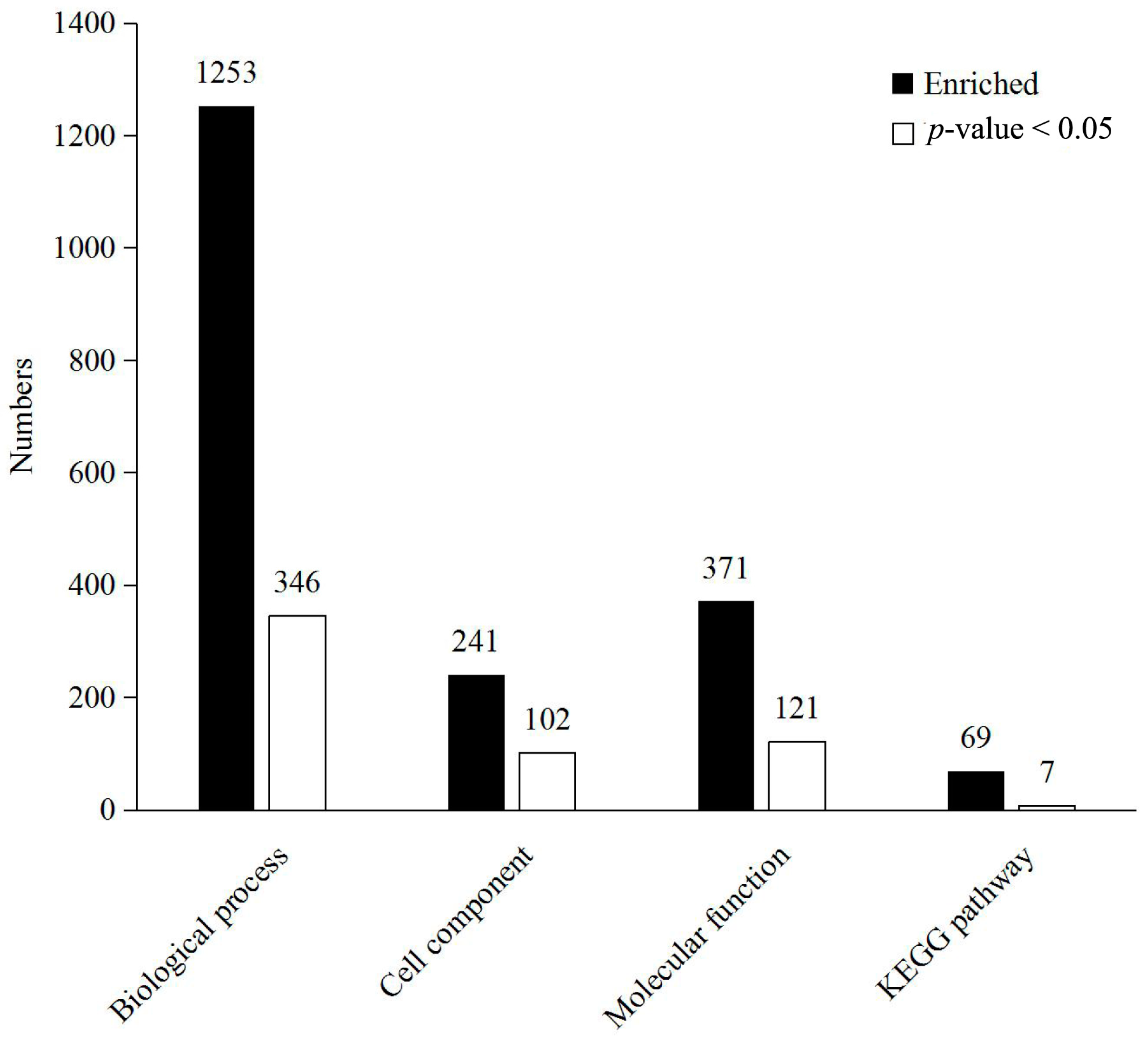
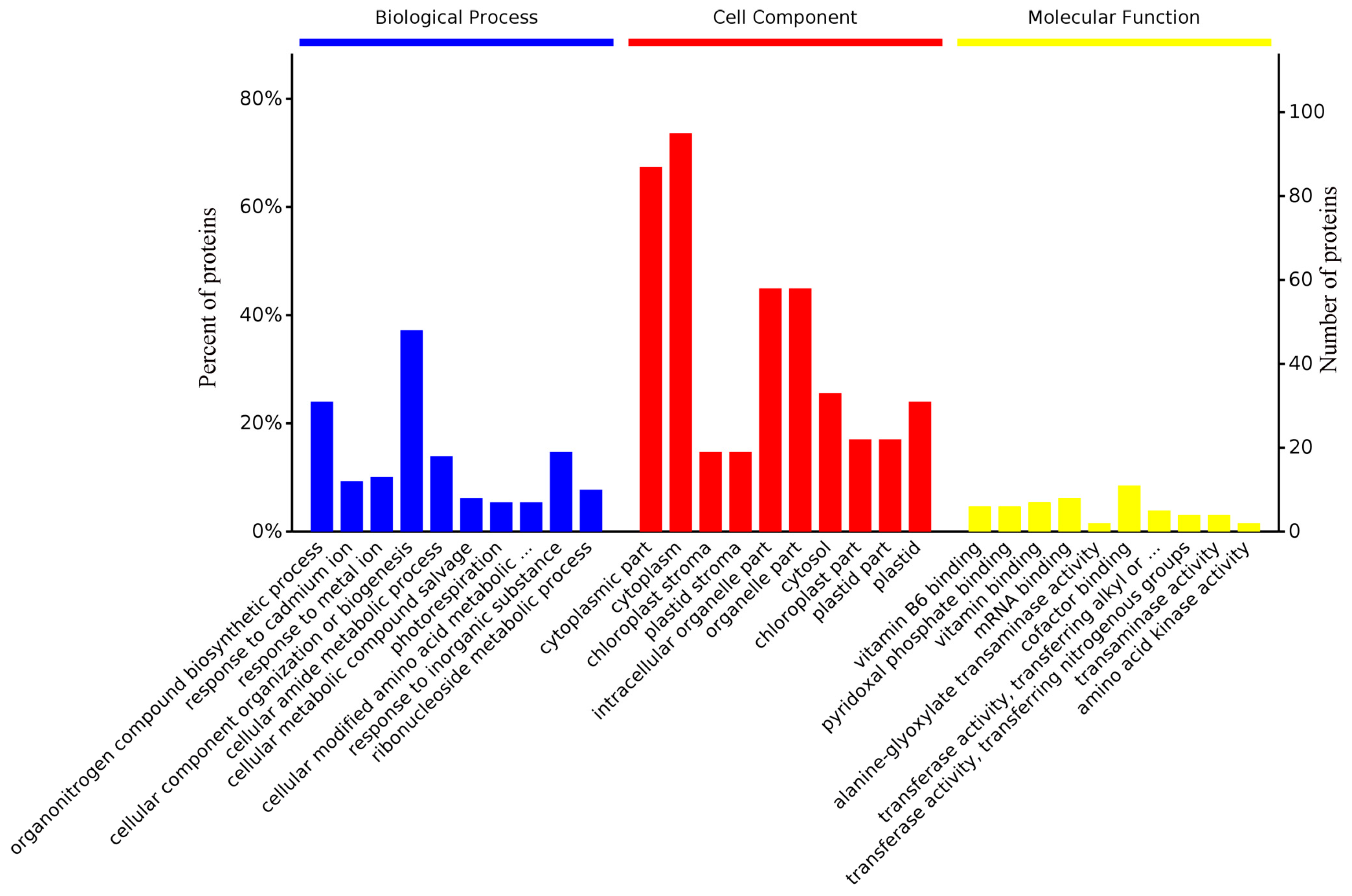
2.3. Functional Analysis and Annotation of Differentially Expressed Proteins
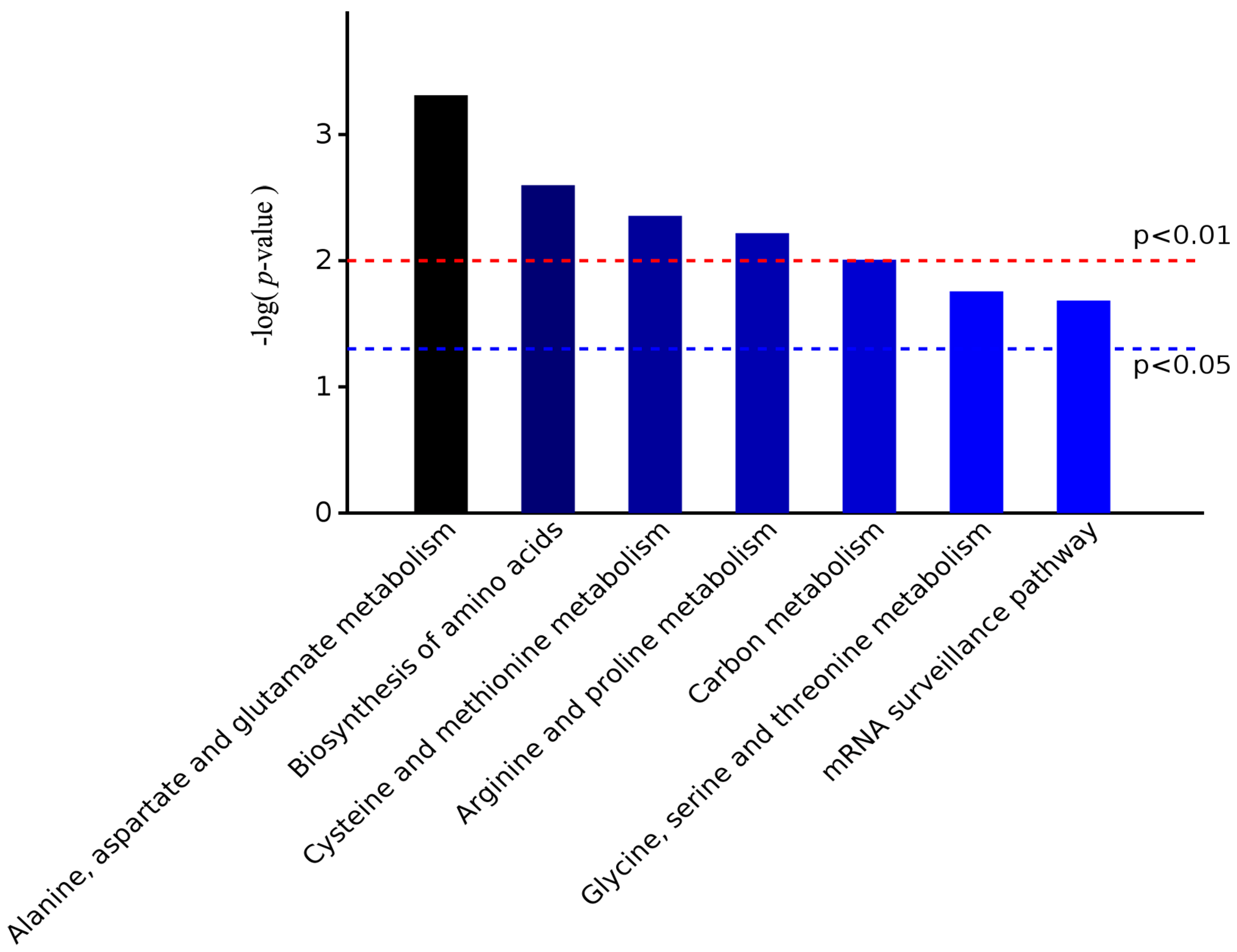
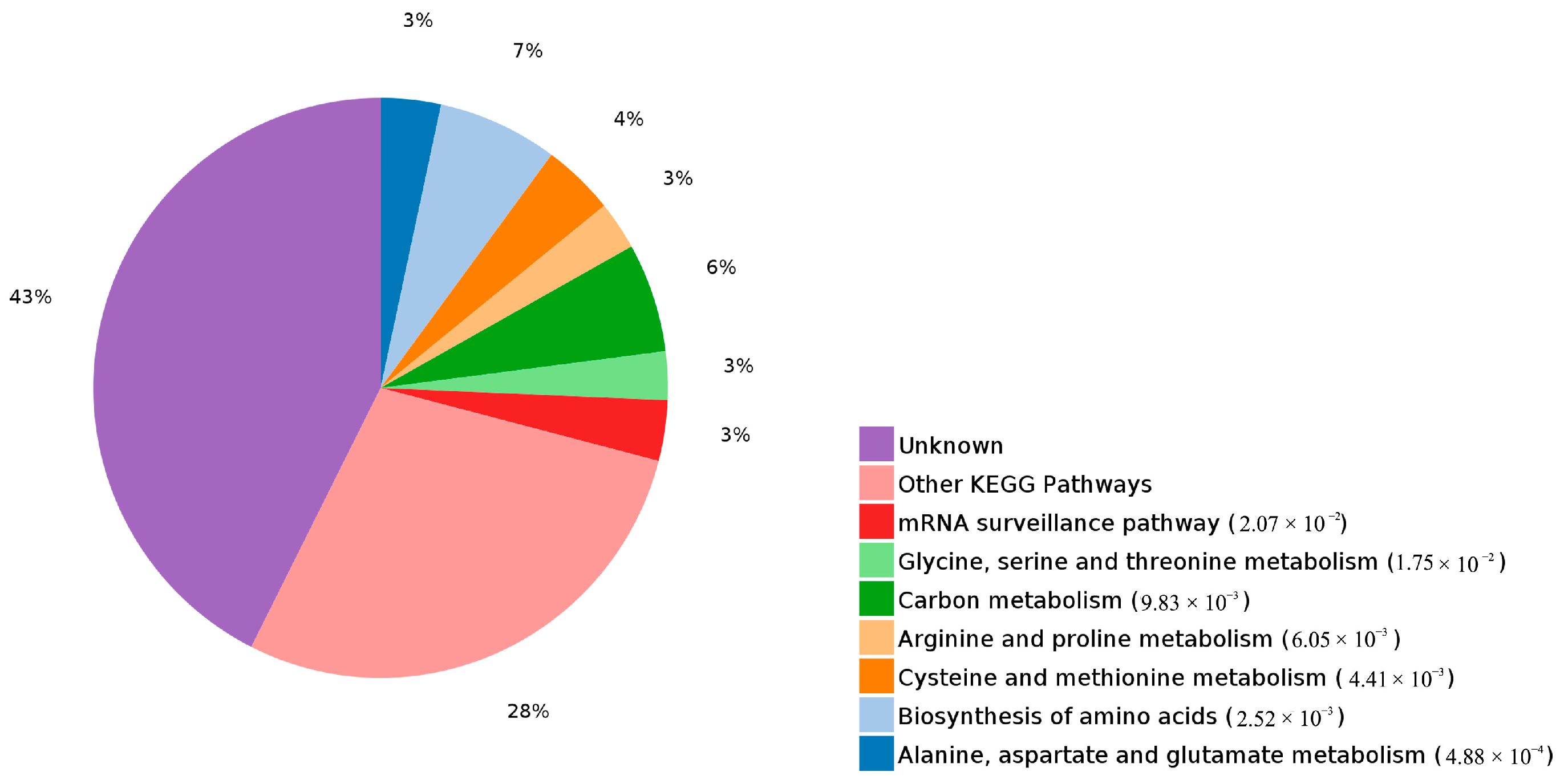
2.4. Functional Annotation of Differentially Expressed Proteins Involves in Lignin Biosynthesis
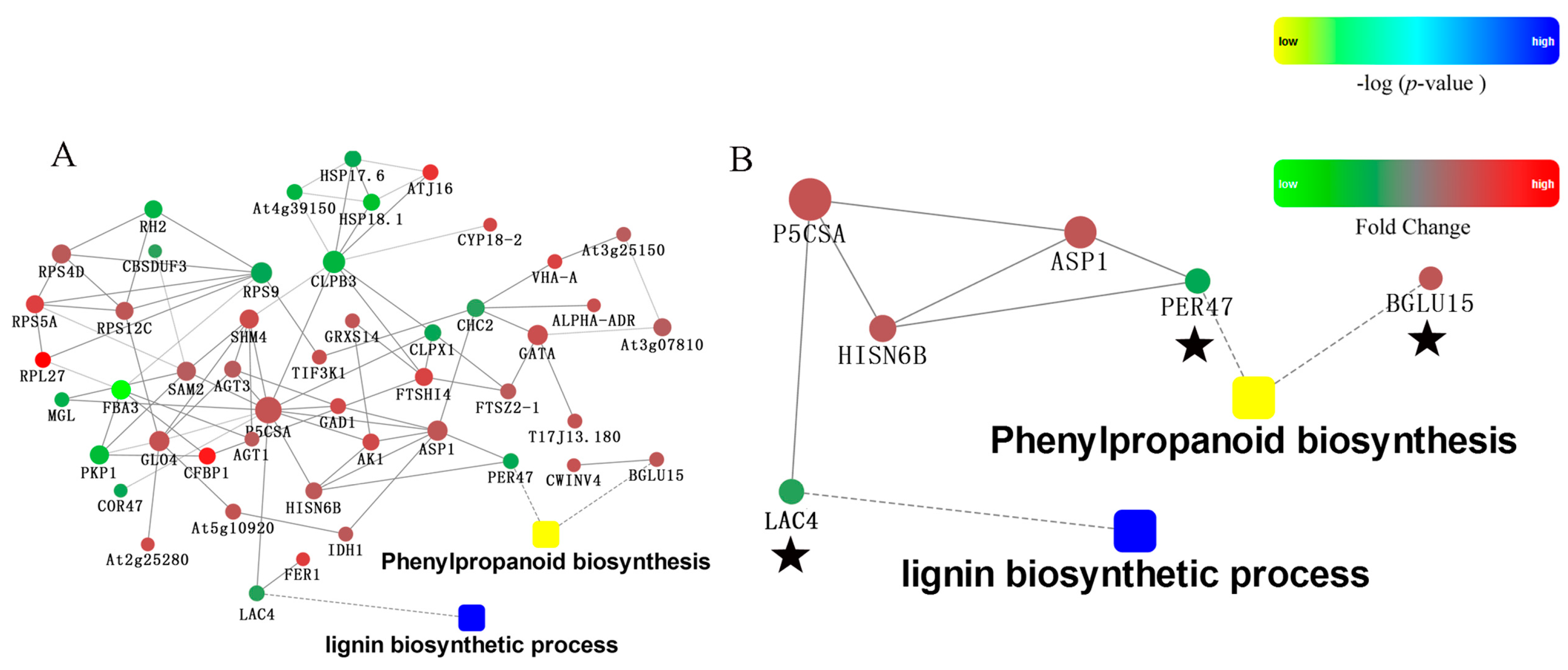
2.5. Protein-Protein Interaction Analysis of Differently Pollinated Dangshan Su
2.6. Verification of Differentially Expressed Genes by qRT-PCR
3. Discussion
4. Materials and Methods
4.1. Processing and Collection of Pear Fruits
4.2. Determination of Stone Cell Content
4.3. Determination of Lignin Content
4.4. Protein Extraction
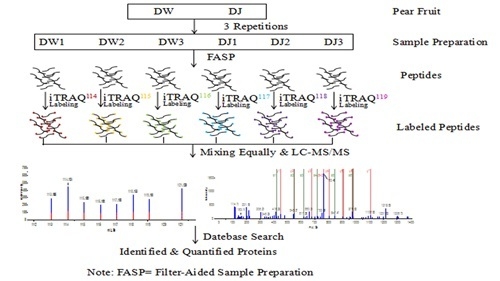
4.5. Protein Samples Preparation and Labelling
4.6. Peptide Fractionation and Quantitative Proteomic Analysis by LC-MS/MS
4.7. Protein Identification and Quantification
4.8. Bioinformatics Analysis
4.9. qRT-PCR Analysis of the Differentially Expressed Proteins
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Franklin-Tong, N.V.E.; Franklin, F.C.H. Gametophytic self-incompatibility inhibits pollen tube growth using different mechanisms. Trends Plant Sci. 2003, 8, 598–605. [Google Scholar] [CrossRef]
- Lāce, B.; Lācis, G. Evaluation of pear (Pyrus communis L.) cultivars in Latvia. Hortic. Sci. 2015, 42, 107–113. [Google Scholar] [CrossRef]
- Wang, S.; Song, C.; Li, L.; Zhang, Y. Physiological function of polyamines during blossoming and fruit setting in apple. Acta Hortic. Sinica 1996, 23, 319–325. [Google Scholar]
- Rosianski, Y.; Freiman, Z.E.; Cochavi, S.M.; Yablovitza, Z.; Kerem, Z.; Flaishman, M.A. Advanced analysis of developmental and ripening characteristics of pollinated common-type fig (Ficus carica L.). Sci. Hortic. 2016, 198, 98–106. [Google Scholar] [CrossRef]
- Cheng, X.; Yan, C.; Zhang, J.; Ma, C.; Li, S.; Jin, Q.; Zhang, N.; Cao, Y.; Lin, Y.; Cai, Y. The Effect of Different Pollination on the Expression of Dangshan Su Pear MicroRNA. Biomed. Res. Int. 2017, 2, 1–18. [Google Scholar] [CrossRef]
- Jin, Q.; Yan, C.; Qiu, J.; Zhang, N.; Lin, Y.; Cai, Y. Structural characterization and deposition of stone cell lignin in Dangshan Su pear. Sci. Hortic. 2013, 155, 123–130. [Google Scholar] [CrossRef]
- Yan, C.; Yin, M.; Zhang, N.; Jin, Q.; Fang, Z.; Lin, Y.; Cai, Y. Stone cell distribution and lignin structure in various pear varieties. Sci. Hortic. 2014, 174, 142–150. [Google Scholar] [CrossRef]
- Baucher, M.; Halpin, C.; Petit-Conil, M.; Boerjan, W. Lignin: Genetic engineering and impact on pulping. Crit. Rev. Biochem. Mol. Biol. 2003, 38, 305–350. [Google Scholar] [CrossRef]
- Jaini, R.; Wang, P.; Dudareva, N.; Chapple, C.; Morgan, J.A. Targeted Metabolomics of the Phenylpropanoid Pathway in Arabidopsis thaliana using Reversed Phase Liquid Chromatography Coupled with Tandem Mass Spectrometry. Phytochem. Anal. 2017, 28, 267–276. [Google Scholar] [CrossRef]
- Novo, M.; Silvar, C.; Merino, F.; Martínez-Cortés, T.; Lu, F.; Ralph, J.; Pomar, F. Deciphering the role of the phenylpropanoid metabolism in the tolerance of Capsicum annuum L. to Verticillium dahliae Kleb. Plant Sci. 2017, 258, 12–20. [Google Scholar] [CrossRef]
- Li, X.; Zang, C.; Ge, H.; Zhang, J.; Grierson, D.; Yin, X.; Chen, K. Involvement of PAL, C4H, and 4CL in Chilling Injury-induced Flesh Lignification of Loquat Fruit. HortScience 2017, 52, 127–131. [Google Scholar] [CrossRef]
- Shi, R.; Sun, Y.; Li, Q.; Heber, S.; Sederoff, R.; Chiang, V.L. Towards a systems approach for lignin biosynthesis in Populus trichocarpa: Transcript abundance and specificity of the monolignol biosynthetic genes. Plant Cell Physiol. 2009, 51, 144–163. [Google Scholar] [CrossRef]
- Guo, W.; Li, J.; Miao, Y.; He, X.; Hu, Q.; Guo, K.; Zhu, L.; Zhang, X. An ethylene response-related factor, GbERF1—Like, from Gossypium barbadense, improves resistance to Verticillium dahliae, via activating lignin synthesis. Plant Mol. Biol. 2016, 91, 305–318. [Google Scholar] [CrossRef]
- Li, J.M.; Huang, X.S.; Li, L.T.; Zheng, D.M.; Xue, C.; Zhang, S.L.; Wu, J. Proteome analysis of pear reveals key genes associated with fruit development and quality. Planta 2015, 241, 1363–1379. [Google Scholar] [CrossRef]
- Huang, J.; You, W.; Luo, X.; Caihuan, K. iTRAQ-Based Identification of Proteins Related to Muscle Growth in the Pacific Abalone, Haliotis discus hannai. Int. J. Mol. Sci. 2017, 18, 2237. [Google Scholar] [CrossRef]
- Ross, P.L.; Huang, Y.N.; Marchese, J.N.; Williamson, B.; Parker, K.; Hattan, S.; Khainovski, N.; Pillai, S.; Dey, S.; Daniels, S.; et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteom. 2004, 3, 1154–1169. [Google Scholar] [CrossRef]
- Wei, Y.; Zeng, B.; Zhang, H.; Chen, C.; Wu, Y.; Wang, N.; Wu, Y.; Shen, L. iTRAQ-based proteomics analysis of serum proteins in Wistar rats treated with sodium fluoride: Insight into the potential mechanism and candidate biomarkers of fluorosis. Int. J. Mol. Sci. 2016, 17, 1644. [Google Scholar] [CrossRef]
- Zhou, Y.S.; Lamrani, M.; Chan-Park, M.B.; Leong, S.S.J.; Wook, M.C.; Chen, W.N. iTRAQ-coupled two-dimensional liquid chromatography/tandem mass spectrometric analysis of protein profile in Escherichia coli incubated with human neutrophil peptide 1-potential in antimicrobial strategy. Rapid Commun. Mass Spectrom. 2010, 24, 2787–2790. [Google Scholar] [CrossRef]
- Zhu, M.; Simons, B.; Zhu, N.; Oppenheimer, D.G.; Chen, S. Analysis of abscisic acid responsive proteins in Brassica napus guard cells by multiplexed isobaric tagging. J. Proteom. 2010, 73, 790–805. [Google Scholar] [CrossRef]
- Cai, Y.; Li, G.; Nie, J.; Lin, Y.; Nie, F.; Zhang, J.; Xu, Y. Study of the structure and biosynthetic pathway of lignin in stone cells of pear. Sci. Hortic. 2010, 125, 374–379. [Google Scholar] [CrossRef]
- Vanholme, R.; Demedts, B.; Morree, K.; Ralph, J.; Boerjan, W. Lignin biosynthesis and structure. Plant Physiol. 2010, 153, 895–905. [Google Scholar] [CrossRef]
- Cesarino, I.; Araújo, P.; Domingues Júnior, A.P.; Mazzafera, P. An overview of lignin metabolism and its effect on biomass recalcitrance. Braz. J. Bot. 2012, 35, 303–311. [Google Scholar] [CrossRef]
- Zakzeski, J.; Bruijnincx, P.C.; Jongerius, A.L.; Weckhuysen, B.M. The catalytic valorization of lignin for the production of renewable chemicals. Chem. Rev. 2010, 110, 3552–3599. [Google Scholar] [CrossRef]
- Lin, S.Y.; Dence, C.W. Methods in Lignin Chemistry; Springer: Berlin, Germany, 1992; pp. 127–131. ISBN 978-3642740671. [Google Scholar]
- Dardick, C.D.; Callahan, A.M.; Chiozzotto, R.; Schaffer, R.J.; Piagnani, M.C.; Scorza, R. Stone formation in peach fruit exhibits spatial coordination of the lignin and flavonoid pathways and similarity to Arabidopsis dehiscence. BMC Biol. 2010, 8, 13. [Google Scholar] [CrossRef]
- Vanholme, R.; Storme, V.; Vanholme, B.; Sundin, L.; Christensen, J.H.; Goeminne, G.; Halpin, C.; Rohde, A.; Morreel, K.; Boerjan, W. A systems biology view of responses to lignin biosynthesis perturbations in Arabidopsis. Plant Cell 2012, 24, 3506–3529. [Google Scholar] [CrossRef]
- Tian, Q.; Stepaniants, S.B.; Mao, M.; Weng, L.; Feetham, M.C.; Doyle, M.J.; Yi, E.C.; Dai, H.; Thorsson, V.; Eng, J.; et al. Integrated genomic and protemomic analyses of gene expression in mammalian cells. Mol. Cell Proteom. 2004, 3, 960–969. [Google Scholar] [CrossRef]
- Beyer, A.; Hollunder, J.; Nasheuer, H.P.; Wilhelm, T. Post-transcriptional expression regulation in the yeast Saccharomyces cerevisiae on a genomic scale. Mol. Cell. Proteom. 2004, 3, 1083–1092. [Google Scholar] [CrossRef]
- Waggoner, S.A.; Liebhaber, S.A. Regulation of alpha-globin mRNA stability. Exp. Biol. Med. 2003, 228, 387–395. [Google Scholar]
- Syros, T.; Yupsanis, T.; Zafiriadis, H.; Economou, A. Activity and isoforms of peroxidases, lignin and anatomy, during adventitious rooting in cuttings of Ebenus cretica L. J. Plant Physiol. 2004, 161, 69–77. [Google Scholar] [CrossRef]
- Isaacson, T.; Damasceno, C.M.B.; Saravanan, R.S.; He, Y.; Catalá, C.; Saladié, M.; Rose, J.K.C. Sample extraction techniques for enhanced proteomic analysis of plant tissues. Nat. Protoc. 2006, 1, 769–774. [Google Scholar] [CrossRef]
- Wisniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Zhang, P.; Li, C.; Zhang, P.; Jin, C.; Pan, D.; Bao, Y. iTRAQ-Based Proteomics Reveals Novel Members Involved in Pathogen Challenge in Sea Cucumber Apostichopus japonicus. PLoS ONE 2014, 9, e100492. [Google Scholar] [CrossRef]
- Dai, C.; Cui, W.; Pan, J.; Xie, Y.; Wang, J.; Shen, W. Proteomic analysis provides insights into the molecular bases of hydrogen gas-induced cadmium resistance in Medicago sativa. J. Proteom. 2017, 152, 109–120. [Google Scholar] [CrossRef]
- Cheng, X.; Li, M.; Li, D.; Zhang, J.; Jin, Q.; Sheng, L.; Cai, Y.; Lin, Y. Characterization and analysis of CCR and CAD gene families at the whole-genome level for lignin synthesis of stone cells in pear (Pyrus bretschneideri) fruit. Biol. Open 2017, 6, 1602–1613. [Google Scholar] [CrossRef]
- Cao, Y.; Han, Y.; Li, D.; Lin, Y.; Cai, Y. MYB Transcription Factors in Chinese Pear (Pyrus bretschneideri Rehd.): Genome-Wide Identification, Classification, and Expression Profiling during Fruit Development. Front. Plant Sci. 2016, 7, 577. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
Sample Availability: Not available. |


| Accession | Protein Name | Gene | Coverage % | Peptides (95%) | Fold Change |
|---|---|---|---|---|---|
| MSTRG11809.4 | β-Glucosidase 15 | BGLU15 | 30.7 | 8 | 1.2392 |
| Pbr022326.1 | Peroxidase 47 | PER47 | 14 | 2 | 0.7949 |
| Pbr035962.1 | Laccase-4 | IRX12 | 10.6 | 2 | 0.8231 |
| Gene ID | Forward Primer (5′–3′) | Reverse Primer (5′–3′) |
|---|---|---|
| Tubulin | TGGGCTTTGCTCCTCTTAC | CCTTCGTGCTCATCTTACC |
| MSTRG11809.4 | TCAACTTCACTCCTTCTGATT | CACGCACTAATAACTTATACAACA |
| Pbr022326.1 | GTAAATGGTTATGCCTTCA | TGTTCTTATTTCTCCTTTGG |
| Pbr035962.1 | GTGCACACAACATGGGGGCTTAAGA | GAAGAGATTGATTAGGTCCTTTACC |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Su, X.; Jin, Q.; Li, G.; Sun, Y.; Abdullah, M.; Cai, Y.; Lin, Y. iTRAQ-Based Identification of Proteins Related to Lignin Synthesis in the Pear Pollinated with Pollen from Different Varieties. Molecules 2018, 23, 548. https://doi.org/10.3390/molecules23030548
Li S, Su X, Jin Q, Li G, Sun Y, Abdullah M, Cai Y, Lin Y. iTRAQ-Based Identification of Proteins Related to Lignin Synthesis in the Pear Pollinated with Pollen from Different Varieties. Molecules. 2018; 23(3):548. https://doi.org/10.3390/molecules23030548
Chicago/Turabian StyleLi, Shumei, Xueqiang Su, Qing Jin, Guohui Li, Yanming Sun, Muhammad Abdullah, Yongping Cai, and Yi Lin. 2018. "iTRAQ-Based Identification of Proteins Related to Lignin Synthesis in the Pear Pollinated with Pollen from Different Varieties" Molecules 23, no. 3: 548. https://doi.org/10.3390/molecules23030548
APA StyleLi, S., Su, X., Jin, Q., Li, G., Sun, Y., Abdullah, M., Cai, Y., & Lin, Y. (2018). iTRAQ-Based Identification of Proteins Related to Lignin Synthesis in the Pear Pollinated with Pollen from Different Varieties. Molecules, 23(3), 548. https://doi.org/10.3390/molecules23030548