Mycobacterium tuberculosis H37Rv LpqG Protein Peptides Can Inhibit Mycobacterial Entry through Specific Interactions
Abstract
1. Introduction
2. Results
2.1. Bioinformatics Analysis
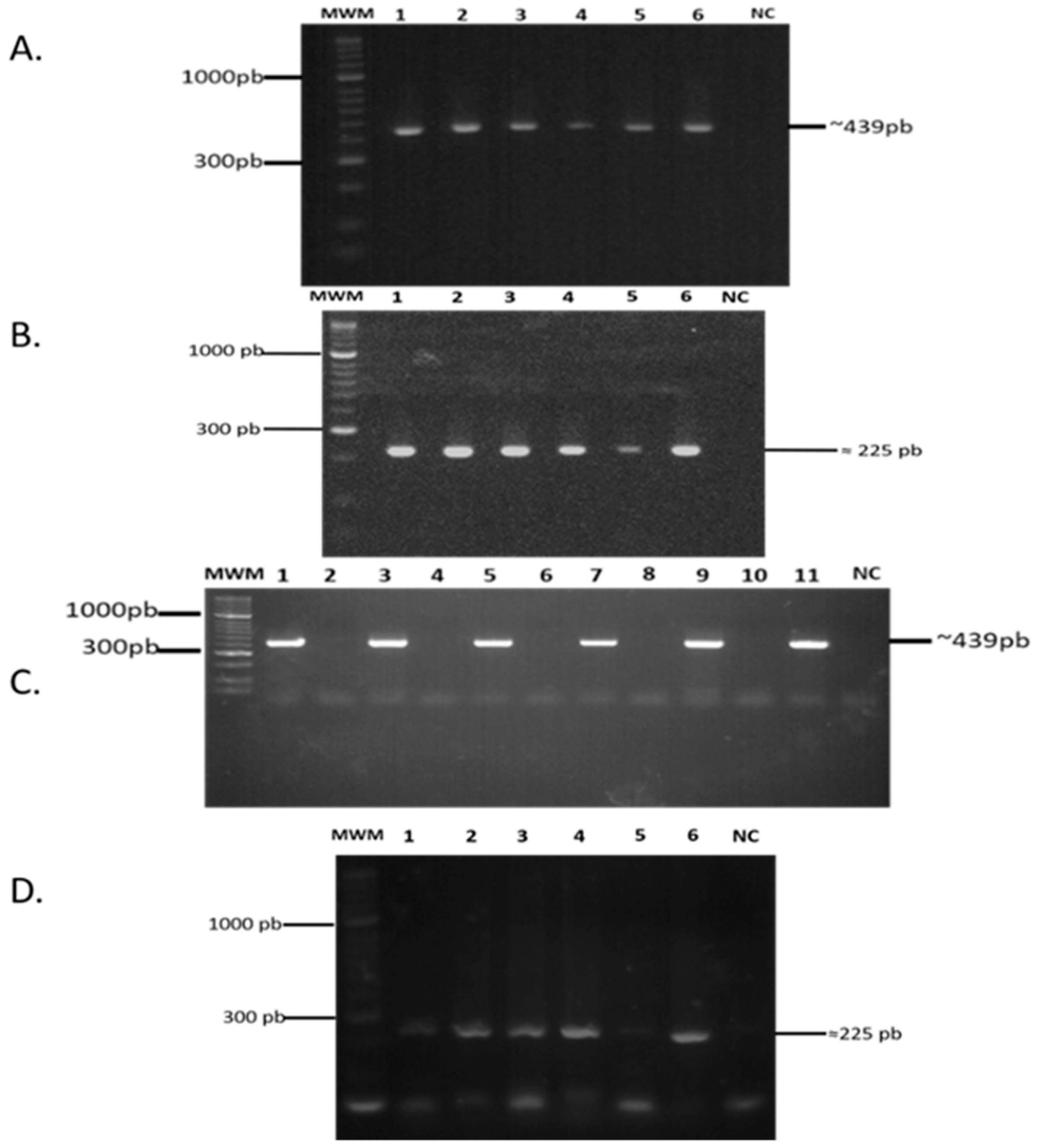
2.2. lpqG Gene Presence and Transcription
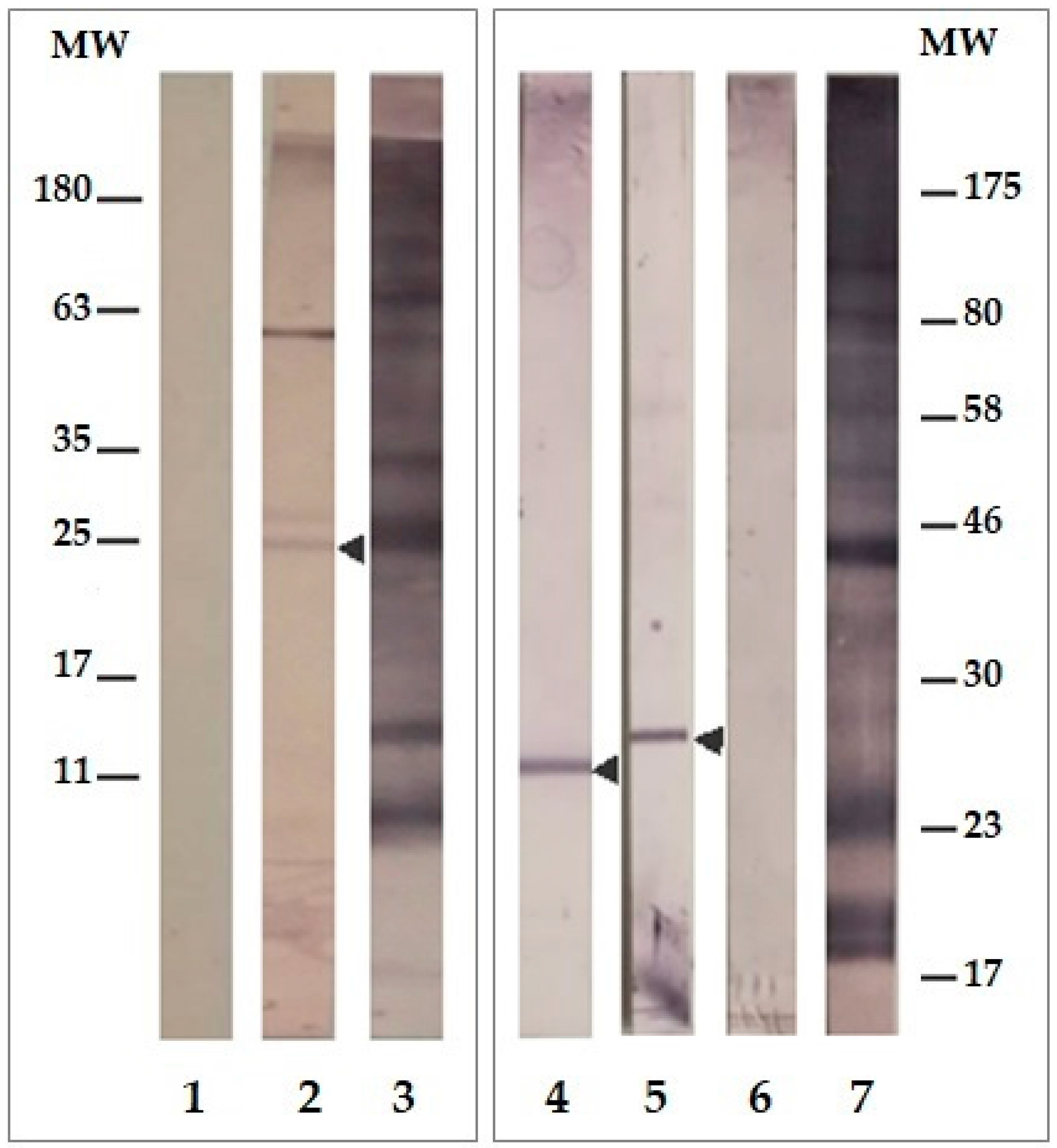
2.3. LpqG Protein Was Present on Mtb Surface
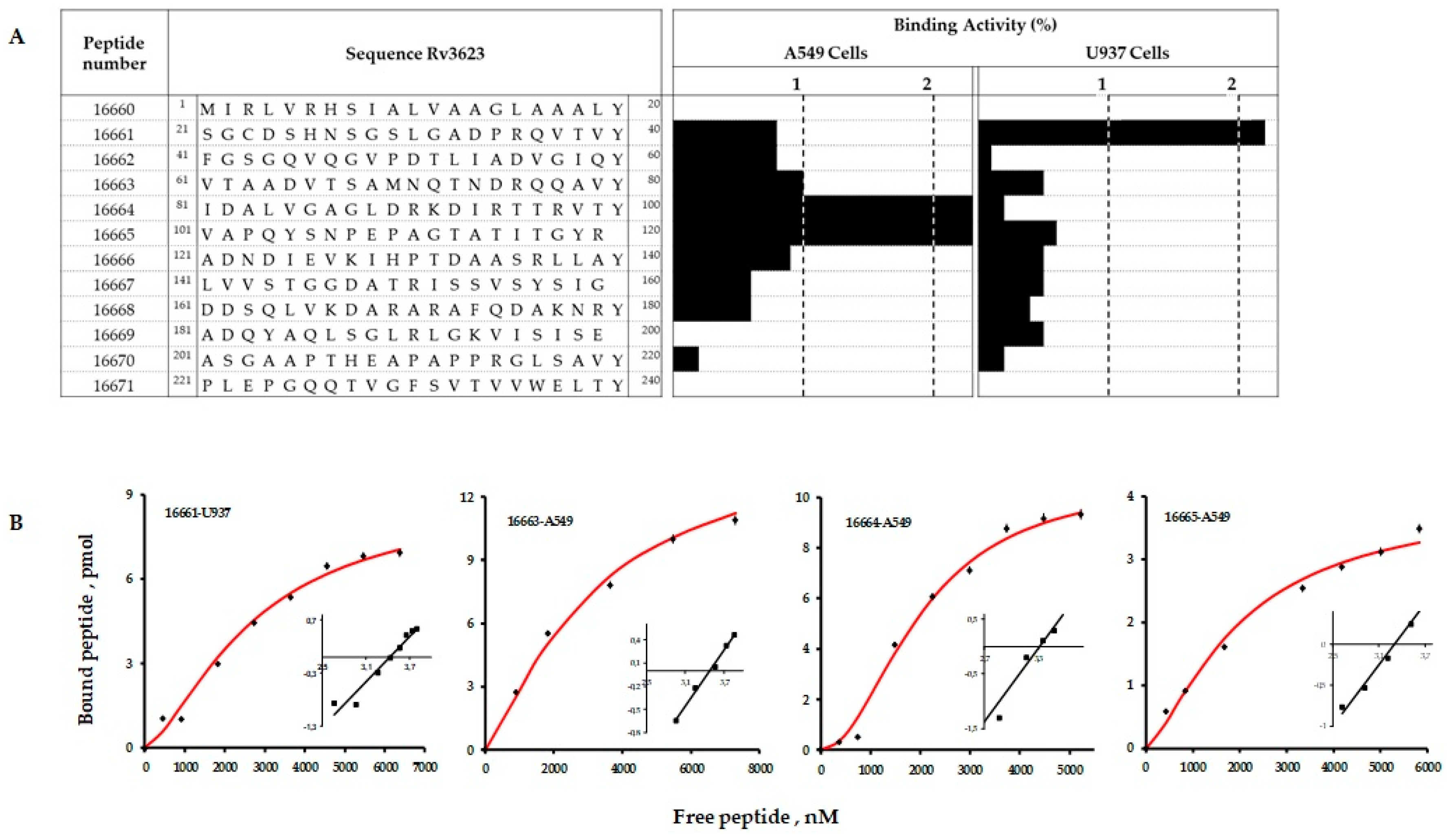
2.4. Synthetic Peptides Bound Specifically to A549 and U937 Cells
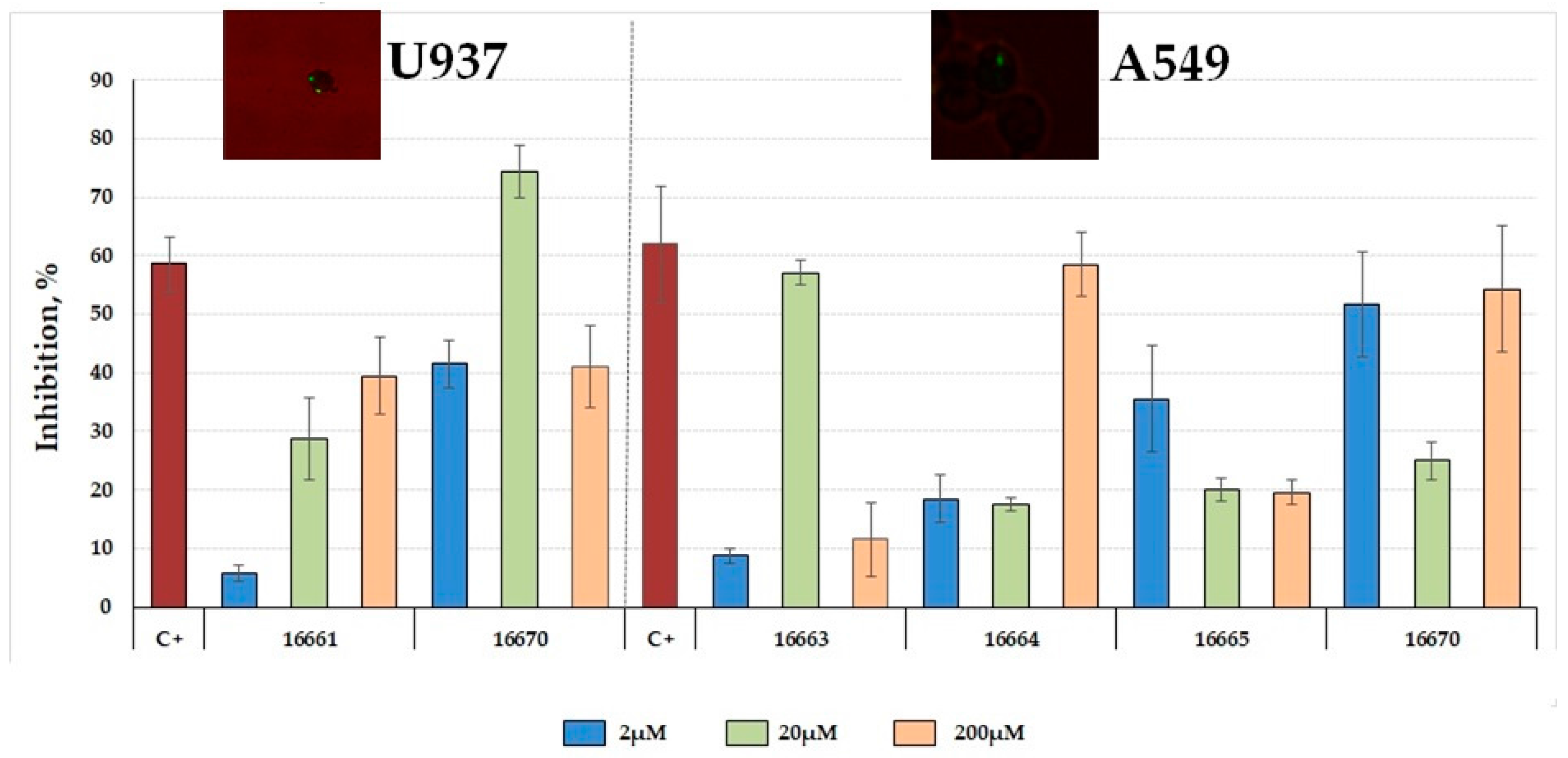
2.5. Inhibiting Mycobacterium tuberculosis Entry to Target Cells
2.6. LpqG Peptide Antigenicity
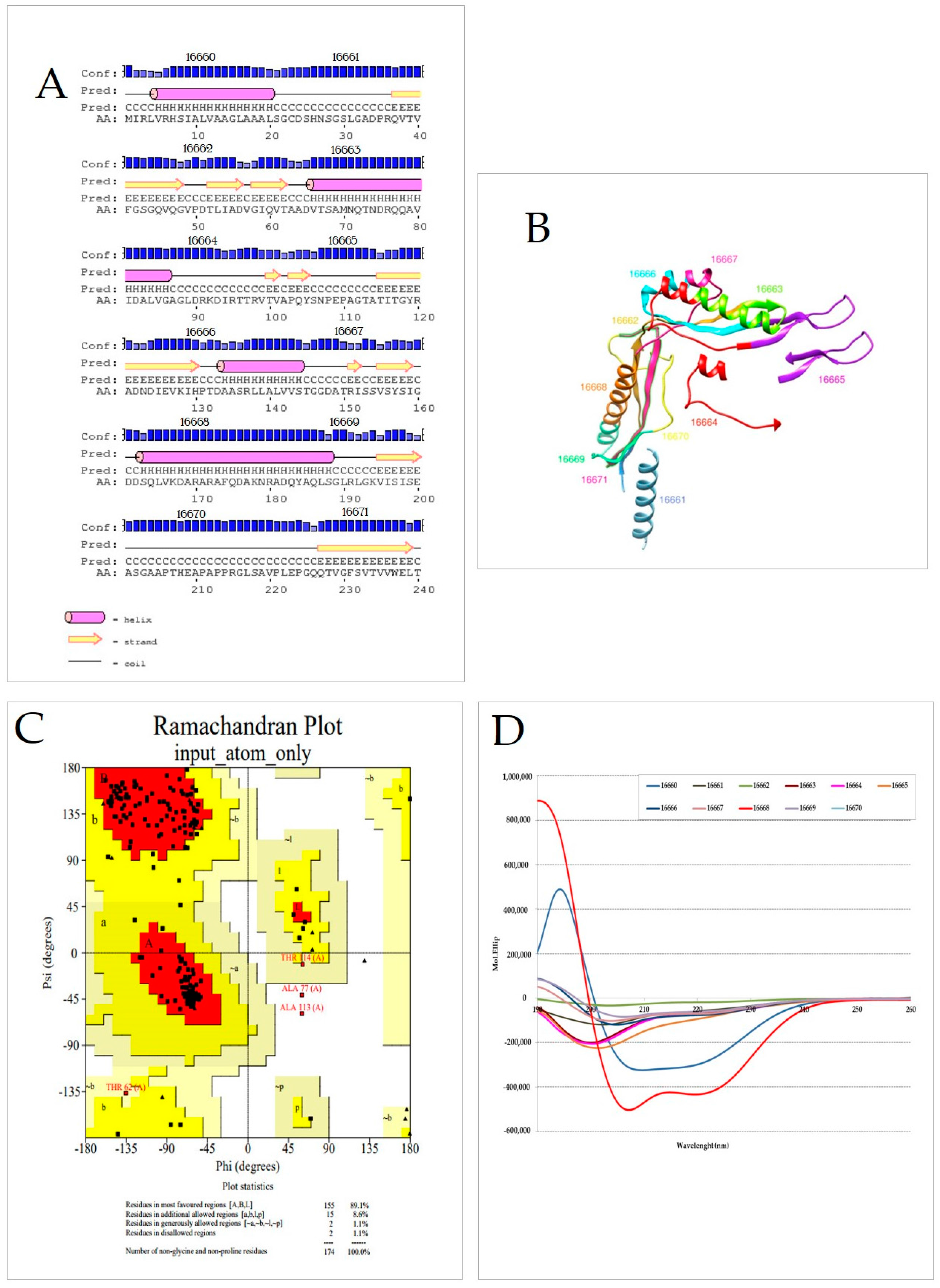
2.7. LpqG Protein Peptides’ Secondary Structure
3. Discussion
4. Materials and Methods
4.1. Bioinformatics Analysis of the LpqG Protein
4.2. lpqG gene Presence and Transcription
4.3. LpqG Translation in Mycobacterium tuberculosis H37Rv
4.4. Identifying Synthetic Peptides Having High Specific LpqG Binding to Target Cells
4.5. Inhibiting Mycobacterial Entry to Infection Target Cells
4.6. Evaluating LpqG Peptide Antigenicity
4.7. Determining LpqG Peptide Secondary Structure
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ernst, J.D. The immunological life cycle of tuberculosis. Nat. Rev. Immunol. 2012, 12, 581–591. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Global Tuberculosis Report 2017; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Mangtani, P.; Abubakar, I.; Ariti, C.; Beynon, R.; Pimpin, L.; Fine, P.E.; Rodrigues, LC; Smith, P.G.; Lipman, M.; Whiting, P.F.; et al. Protection by BCG vaccine against tuberculosis: A systematic review of randomized controlled trials. Clin. Infect. Dis. 2014, 58, 470–480. [Google Scholar] [CrossRef] [PubMed]
- Palomino, J.C.; Martin, A. Drug Resistance Mechanisms in Mycobacterium tuberculosis. Antibiotics 2014, 3, 317–340. [Google Scholar] [CrossRef] [PubMed]
- Dockrell, H.M.; Smith, S.G. What Have We Learnt about BCG Vaccination in the Last 20 Years? Front. Immunol. 2017, 8, 1134. [Google Scholar] [CrossRef] [PubMed]
- Boggiano, C.; Eichelberg, K.; Ramachandra, L.; Shea, J.; Ramakrishnan, L.; Behar, S.; Ernst, J.D.; Porcelli, S.A.; Maeurer, M.; Kornfeld, H. “The Impact of Mycobacterium tuberculosis Immune Evasion on Protective Immunity: Implications for TB Vaccine Design”—Meeting report. Vaccine 2017, 35, 3433–3440. [Google Scholar] [CrossRef] [PubMed]
- Awuh, J.A.; Flo, T.H. Molecular basis of mycobacterial survival in macrophages. Cell. Mol. Life Sci. 2017, 74, 1625–1648. [Google Scholar] [CrossRef] [PubMed]
- Behar, S.M.; Divangahi, M.; Remold, H.G. Evasion of innate immunity by Mycobacterium tuberculosis: is death an exit strategy? Nat. Rev. Microbiol. 2010, 8, 668–674. [Google Scholar] [CrossRef] [PubMed]
- Dorhoi, A.; Reece, S.T.; Kaufmann, S.H.E. For better or for worse: The immune response against Mycobacterium tuberculosis balances pathology and protection. Immunol. Rev. 2011, 240, 235–251. [Google Scholar] [CrossRef] [PubMed]
- Sani, M.; Houben, E.N.; Geurtsen, J.; Pierson, J.; de Punder, K.; van Zon, M.; Wever, B.; Piersma, S.R.; Jimenez, C.R.; Daffe, M.; et al. Direct visualization by cryo-EM of the mycobacterial capsular layer: a labile structure containing ESX-1-secreted proteins. PLoS Pathog. 2010, 6, e1000794. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.; Chen, J.; Dobos, K.M.; Bradbury, E.M.; Belisle, J.T.; Chen, X. Comprehensive proteomic profiling of the membrane constituents of a Mycobacterium tuberculosis strain. Mol. Cell. Proteom. MCP 2003, 2, 1284–1296. [Google Scholar] [CrossRef] [PubMed]
- Cole, S.T.; Brosch, R.; Parkhill, J.; Garnier, T.; Churcher, C.; Harris, D.; Gordon, S.V.; Eiglmeier, K.; Gas, S.; Barry, C.E., III; et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 1998, 393, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Arora, S.; Kosalai, K.; Namane, A.; Pym, A.S.; Cole, S.T. Proteome analysis of the plasma membrane of Mycobacterium tuberculosis. Comp. Funct. Genom. 2002, 3, 470–483. [Google Scholar] [CrossRef] [PubMed]
- Rezwan, M.; Grau, T.; Tschumi, A.; Sander, P. Lipoprotein synthesis in mycobacteria. Microbiology 2007, 153, 652–658. [Google Scholar] [CrossRef] [PubMed]
- Sutcliffe, I.C.; Harrington, D.J. Lipoproteins of Mycobacterium tuberculosis: An abundant and functionally diverse class of cell envelope components. FEMS Microbiol. Rev. 2004, 28, 645–659. [Google Scholar] [CrossRef] [PubMed]
- Malen, H.; Pathak, S.; Søfteland, T.; de Souza, G.A.; Wiker, H.G. Definition of novel cell envelope associated proteins in Triton X-114 extracts of Mycobacterium tuberculosis H37Rv. BMC Microbiol. 2010, 10, 132. [Google Scholar] [CrossRef] [PubMed]
- Malen, H.; Berven, F.S.; Softeland, T.; Arntzen, M.O.; D’Santos, C.S.; De Souza, G.A.; Wiker, H.G. Membrane and membrane-associated proteins in Triton X-114 extracts of Mycobacterium bovis BCG identified using a combination of gel-based and gel-free fractionation strategies. Proteomics 2008, 8, 1859–1870. [Google Scholar] [CrossRef] [PubMed]
- Ocampo, M.; Patarroyo, M.A.; Vanegas, M.; Alba, M.P.; Patarroyo, M.E. Functional, biochemical and 3D studies of Mycobacterium tuberculosis protein peptides for an effective anti-tuberculosis vaccine. Crit. Rev. Microbiol. 2013, 40, 117–145. [Google Scholar] [CrossRef] [PubMed]
- Díaz, D.P.; Ocampo, M.; Pabón, L.; Herrera, C.; Patarroyo, M.A.; Muñoz, M.; Patarroyo, M.E. Mycobacterium tuberculosis PE9 protein has high activity binding peptides which inhibit target cell invasion. Int. J. Biol. Macromol. 2016, 86, 646–655. [Google Scholar]
- Díaz, D.P.; Ocampo, M.; Varela, Y.; Curtidor, H.; Patarroyo, M.A.; Patarroyo, M.E. Identifying and characterising PPE7 (Rv0354c) high activity binding peptides and their role in inhibiting cell invasion. Mol. Cell. Biochem. 2017, 430, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, D.C.; Ocampo, M.; Reyes, C.; Arevalo-Pinzon, G.; Munoz, M.; Patarroyo, M.A.; Patarroyo, M.E. Cell-Peptide Specific Interaction Can Inhibit Mycobacterium tuberculosis H37Rv Infection. J. Cell. Biochem. 2016, 117, 946–958. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Cowley, A.; Uludag, M.; Gur, T.; McWilliam, H.; Squizzato, S.; Park, Y.M.; Buso, N.; Lopez, R. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 2015, 43, 580–584. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Human Press: New York, NY, USA, 2005; pp. 571–607. [Google Scholar]
- Kyte, J.; Doolittle, F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Shen, H.B.; Chou, K.C. Gpos-mPLoc: A top-down approach to improve the quality of predicting subcellular localization of Gram-positive bacterial proteins. Protein Pept. Lett. 2009, 16, 1478–1484. [Google Scholar] [CrossRef] [PubMed]
- Käll, L.; Krogh, A.; Sonnhammer, E.L. Advantages of combined transmembrane topology and signal peptide prediction—The Phobius web server. Nucleic Acids Res. 2007, 35, 429–432. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Benkert, P.; Biasini, M.; Schwede, T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 2011, 27, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL Workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2006, 22, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Kelly, S.M.; Jess, T.J.; Price, N.C. How to study proteins by circular dichroism. BBA Proteins Proteom. 2005, 10, 119–139. [Google Scholar] [CrossRef] [PubMed]
- Sreerama, N.; Woody, R.W. Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Analy. Biochem. 2000, 15, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Babu, M.M.; Priya, M.L.; Selvan, A.T.; Madera, M.; Gough, J.; Aravind, L.; Sankaran, K. A database of bacterial lipoproteins (DOLOP) with functional assignments to predicted lipoproteins. J. Bacteriol. 2006, 188, 2761–2773. [Google Scholar] [CrossRef] [PubMed]
- Steentoft, C.; Vakhrushev, S.Y.; Joshi, H.J.; Kong, Y.; Vester-Christensen, M.B.; Schjoldager, K.T.; Lavrsen, K.; Dabelsteen, S.; Pedersen, N.B.; Marcos-Silva, L.; et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013, 32, 1478–1488. [Google Scholar] [CrossRef] [PubMed]
- Patarroyo, M.E.; Bermudez, A.; Patarroyo, M.A. Structural and immunological principles leading to chemically synthesized, multiantigenic, multistage, minimal subunit-based vaccine development. Chem. Rev. 2011, 111, 3459–3507. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed]
- Bairoch, A.; Apweiler, R.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M.; et al. The Universal Protein Resource (UniProt). Nucleic Acids Res. 2005, 33, D154–D159. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.C.; Shen, H.B. Cell-PLoc: A package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Szafron, D.; Greiner, R.; Lu, P.; Wishart, D.S.; Poulin, B.; Anvik, J.; Macdonell, C.; Eisner, R. Predicting subcellular localization of proteins using machine-learned classifiers. Bioinformatics 2004, 20, 547–556. [Google Scholar] [CrossRef] [PubMed]
- Gardy, J.L.; Laird, M.R.; Chen, F.; Rey, S.; Walsh, C.J.; Ester, M.; Brinkman, F.S. PSORTb v.2.0: Expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics 2005, 21, 617–623. [Google Scholar] [CrossRef] [PubMed]
- Restrepo-Montoya, D.; Vizcaíno, C.; Niño, L.F.; Ocampo, M.; Patarroyo, M.E.; Patarroyo, M.A. Validating subcellular localization prediction tools with mycobacterial proteins. BMC Bioinform. 2009, 10, 134. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Bendtsen, J.D.; Nielsen, H.; Widdick, D.; Palmer, T.; Brunak, S. Prediction of twin-arginine signal peptides. BMC Bioinform. 2005, 6, 167. [Google Scholar] [CrossRef] [PubMed]
- Juncker, A.S.; Willenbrock, H.; Von Heijne, G.; Brunak, S.; Nielsen, H.; Krogh, A. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 2003, 12, 1652–1662. [Google Scholar] [CrossRef] [PubMed]
- Bendtsen, J.D.; Kiemer, L.; Fausboll, A.; Brunak, S. Non-classical protein secretion in bacteria. BMC Microbiol. 2005, 5, 58. [Google Scholar] [CrossRef] [PubMed]
- Sonnhammer, E.L.; von Heijne, G.; Krogh, A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. ISMB-98 Proc. 1998, 6, 175–182. [Google Scholar]
- Larsen, J.E.; Lund, O.; Nielsen, M. Improved method for predicting linear B-cell epitopes. Immunome Res. 2006, 2, 2. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Andreatta, M.; Karosiene, E.; Rasmussen, M.; Stryhn, A.; Buus, S.; Nielsen, M. Accurate pan-specific prediction of peptide-MHC class II binding affinity with improved binding core identification. Immunogenetics 2015, 67, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Rezwan, M.; Laneelle, M.A.; Sander, P.; Daffe, M. Breaking down the wall: Fractionation of mycobacteria. J. Microbiol. Methods 2007, 68, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Bermudez, L.E.; Goodman, J. Mycobacterium tuberculosis invades and replicates within type II alveolar cells. Infect. Immun. 1996, 64, 1400–1406. [Google Scholar]
- Cascante, J.; Pascal, I.; Eguía, V.; Hueto, J. Diagnosis of tuberculosis infection. SciELO 2007, 30, 49–65. [Google Scholar]
- Jones, D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999, 292, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Gallo Cassarino, T.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef] [PubMed]
- Salomon-Ferrer, R.; Gotz, A.W.; Poole, D.; Le Grand, S.; Walker, R.C. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald. J. Chem. Theory Comput. 2013, 9, 3878–3888. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |

| Protein | Rv3623 | |
| TubercuList description | probable conserved lipoprotein LPQG | |
| Molecular weight | 24,836.80 Da | |
| Theoretical pI | 5.52 | |
| Instability index | 21.64 stable | |
| Aliphatic index | 93.54 | |
| Grand average of hydropathicity (GRAVY) | −0.055 | |
| Subcellular localisation | PA-SUB v.2.5. | Not cytoplasm |
| Not extracellular | ||
| Not plasma membrane | ||
| Gpos-Ploc | Plasma membrane | |
| PSORTb v.2.0.4 | Unknown | |
| LipoP 1.0 | Score | 25.1723 |
| Cleavage site | 22–23 Pos + 2 = D | |
| Phobius | TM/SP/prediction | 0/Y/n9-20c25/26o |
| TMHMM 2.0 | ExpAA | 0.24 |
| First60 | 0.24 | |
| PredHel | 0 | |
| Topology | outer | |
| SignalP 4.0 | Signal peptide probability | 0.939 |
| Max. cleavage site probability | 0.697 | |
| Cleavage site | 25–26 | |
| NetOGlyc 4.0 | Thr: 96, 97, 100, 207 Ser: 218 | |
| U937 | A549 | |||
|---|---|---|---|---|
| HABP | 16661 | 16663 | 16664 | 16665 |
| Dissociation constant (KD), nM | 2700 | 2800 | 2000 | 2000 |
| Hill coefficient (nH) | 1.5 | 1.4 | 3.0 | 1.4 |
| Binding sites per cell | 5 × 106 | 9 × 106 | 7 × 106 | 3 × 106 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-Barinas, C.D.; Ocampo, M.; Vanegas, M.; Castañeda-Ramirez, J.J.; Patarroyo, M.A.; Patarroyo, M.E. Mycobacterium tuberculosis H37Rv LpqG Protein Peptides Can Inhibit Mycobacterial Entry through Specific Interactions. Molecules 2018, 23, 526. https://doi.org/10.3390/molecules23030526
Sánchez-Barinas CD, Ocampo M, Vanegas M, Castañeda-Ramirez JJ, Patarroyo MA, Patarroyo ME. Mycobacterium tuberculosis H37Rv LpqG Protein Peptides Can Inhibit Mycobacterial Entry through Specific Interactions. Molecules. 2018; 23(3):526. https://doi.org/10.3390/molecules23030526
Chicago/Turabian StyleSánchez-Barinas, Christian David, Marisol Ocampo, Magnolia Vanegas, Jeimmy Johana Castañeda-Ramirez, Manuel Alfonso Patarroyo, and Manuel Elkin Patarroyo. 2018. "Mycobacterium tuberculosis H37Rv LpqG Protein Peptides Can Inhibit Mycobacterial Entry through Specific Interactions" Molecules 23, no. 3: 526. https://doi.org/10.3390/molecules23030526
APA StyleSánchez-Barinas, C. D., Ocampo, M., Vanegas, M., Castañeda-Ramirez, J. J., Patarroyo, M. A., & Patarroyo, M. E. (2018). Mycobacterium tuberculosis H37Rv LpqG Protein Peptides Can Inhibit Mycobacterial Entry through Specific Interactions. Molecules, 23(3), 526. https://doi.org/10.3390/molecules23030526