Structural Dynamics of DPP-4 and Its Influence on the Projection of Bioactive Ligands
Abstract
:1. Introduction
2. Results and Discussion
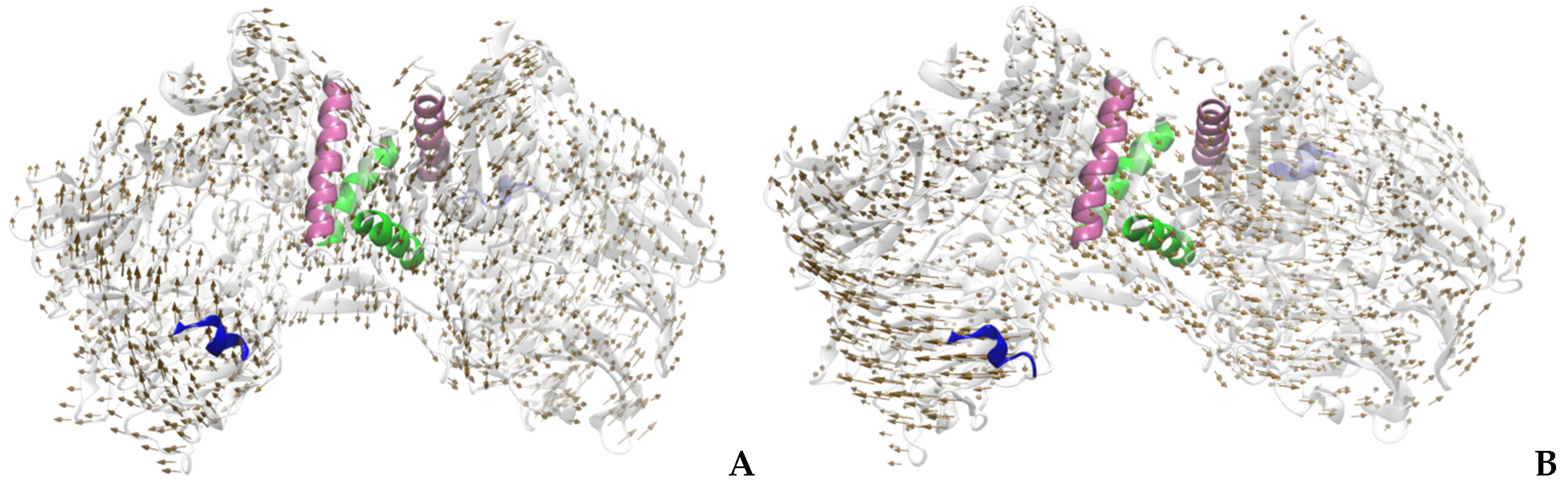
2.1. Calculation of Normal Modes
2.2. Interactions between DPP-4 and the Inhibitor N7F
2.3. Search for a Binding Site and Druggable Regions with FTSite and FTMap
3. Materials and Methods
3.1. Protocol Overview-Calculation of Normal Modes (NM)
3.2. Protocol Overview-Interactions Between DPP-4 and the N7F Inhibitor
3.3. Protocol Overview-Search for Binding Sites FTSite and FTMap
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- International Diabetes Federation (IDF). Estimated number of people with diabetes worldwide and per region in 2015 and 2040 (20–79 years). In IDF Diabetes Atlas, 7th ed.; International Diabetes Federation: Brussels, Belgium, 2015; p. 13. [Google Scholar]
- Chaudhury, A.; Duvoor, C.; Reddy Dendi, V.S.; Kraleti, S.; Chada, A.; Ravilla, R.; Marco, A.; Shekhawat, N.S.; Montales, M.T.; Kuriakose, K.; et al. Clinical Review of Antidiabetic Drugs: Implications for Type 2 Diabetes Mellitus Management. Front. Endocrinol. (Lausanne) 2017, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Maiseyeu, A.; Davis, S.N.; Rajagopalan, S. DPP4 in Cardiometabolic Disease Recent Insights From the Laboratory and Clinical Trials of DPP4 Inhibition. Circ. Res. 2015, 116, 1491–1504. [Google Scholar] [CrossRef] [PubMed]
- Lupher, M.L.; Rao, N.; Eck, M., Jr.; Band, H. The Cbl protooncoprotein: A negative regulator of immune receptor signal transduction. Immunol. Today. 1999, 20, 375–382. [Google Scholar] [CrossRef]
- Bjelke, J.R.; Christensen, J.; Branner, S.; Wagtmann, N.; Olsen, C.; Kanstrup, A.B.; Rasmussen, H.B. Tyrosine 547 Constitutes an Essential Part of the Catalytic Mechanism of Dipeptidyl Peptidase IV. J. Biol. Chem. 2004, 279, 34691–34697. [Google Scholar] [CrossRef] [PubMed]
- Pantaleao, S.Q.; Maltarollo, V.G.; Araujo, S.C.; Gertrudes, J.C.; Honorio, K.M. Molecular docking studies and 2D analyses of DPP-4 inhibitors as candidates in the treatment of diabetes. Mol. Biosyst. 2015, 11, 3188–3193. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Hu, Y.; Liu, T. Recent advances in non-peptidomimetic dipeptidyl peptidase 4 inhibitors: Medicinal chemistry and preclinical aspects. Curr. Med. Chem. 2012, 19, 3982–3999. [Google Scholar] [CrossRef] [PubMed]
- Tanwar, O.; Deora, G.S.; Tanwar, L.; Kumar, G.; Janardhan, S.; Alam, M.M.; Shaquiquzzaman, M.; Akhter, M. Novel hydrazine derivatives as selective DPP-IV inhibitors: Findings from virtual screening and validation through molecular dynamics simulations. J. Mol. Model. 2014, 20, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Barrett, A.J.; Rawlings, N.D.; Woessner, J.F. Cysteine, serine and threonine peptidases. In Handbook of Proteolytic Enzymes, 2nd ed.; Academic Press: Cambridge, MA, USA, 2004; Volume l2, p. 1045. [Google Scholar]
- Thoma, R.; Loffler, B.; Stihle, M.; Huber, W.; Ruf, A.; Hennig, M. Structural basis of proline-specific exopeptidase activity as observed in human dipeptidyl peptidase-IV. Structure 2003, 11, 947–959. [Google Scholar] [CrossRef]
- Rasmussen, H.B.; Branner, S.; Wiberg, F.C.; Wagtmann, N. Crystal structure of human dipeptidyl peptidase IV/CD26 in complex with a substrate analog. Nat. Struct. Mol. Biol. 2002, 10, 19–25. [Google Scholar] [CrossRef] [PubMed]
- De Meester, I.; Scharpé, S.; Vanham, G.; Bosmans, E.; Heyligen, H.; Vanhoof, G.; Corte, G. Antibody binding profile of purified and cell-bound CD26.Designation of BT5/9 and TA5.9 to the CD26 cluster. Immunology 1999, 188, 145–158. [Google Scholar]
- Weihofen, W.A.; Liu, J.; Reutter, W.; Saenger, W.; Fan, H. Crystal structure of CD26/dipeptidyl-peptidase IV in complex with adenosine deaminase reveals a highly amphiphilic interface. J. Biol. Chem. 2004, 279, 43330–43335. [Google Scholar] [CrossRef] [PubMed]
- Sutton, J.M.; Clark, D.E.; Dunsdon, S.J.; Fenton, G.; Fillmore, A.; Harris, N.V.; Higgs, C.; Hurley, C.A.; Krintel, S.L.; MacKenzie, R.E.; et al. Novel heterocyclic DPP-4 inhibitors for the treatment of type 2 diabetes. Bioorg. Med. Chem. Lett. 2012, 22, 1464–1468. [Google Scholar] [CrossRef] [PubMed]
- Sutton, J.M.; Clark, D.E.; Dunsdon, S.J.; Fenton, G.; Fillmore, A.; Harris, N.V.; Higgs, C.; Hurley, C.A.; Krintel, S.L.; MacKenzie, R.E.; et al. Erratum to Novel heterocyclic DPP-4 inhibitors for the treatment of type 2 diabetes. Bioorg. Med. Chem. Lett. 2012, 22, 2359. [Google Scholar] [CrossRef]
- Berman, H.; Henrick, K.; Nakamura, H. Announcing the worldwide Protein Data Bank. Nat. Struct. Mol. Biol. 2003, 10, 980. [Google Scholar] [CrossRef] [PubMed]
- Durrant, J.D.; McCammon, J.A. BINANA: A Novel Algorithm for Ligand-Binding Characterization. J. Mol. Graph. Model. 2011, 29, 888–893. [Google Scholar] [CrossRef] [PubMed]
- Kozakov, D.; Grove, L.E.; Hall, D.R.; Bohnuud, T.; Mottarella, S.E.; Luo, L.; Xia, B.; Beglov, D.; Vajda, S. The FTMap family of web servers for determining and characterizing ligand-binding hot spots of proteins. Nat. Protoc. 2015, 10, 733–755. [Google Scholar] [CrossRef] [PubMed]
- Ngan, C.-H.; Hall, D.R.; Zerbe, B.; Grove, L.E.; Kozakov, D.; Vajda, S. FTSite: High accuracy detection of ligand binding sites on unbound protein structures. Bioinformatics 2012, 28, 286–287. [Google Scholar] [CrossRef] [PubMed]
- Brenke, R.; Kozakov, D.; Chuang, G.-Y.; Beglov, D.; Hall, D.; Landon, M.R.; Mattos, C.; Vajda, S. Fragment-based identification of druggable ‘hot spots’ of proteins using Fourier domain correlation techniques. Bioinformatics 2009, 25, 621–627. [Google Scholar] [CrossRef] [PubMed]
- Kozakov, D.; Hall, D.R.; Chuang, G.-Y.; Cencic, R.; Brenke, R.; Grove, L.E.; Beglov, D.; Pelletier, J.; Whitty, A.; Vajda, S. Structural conservation of druggable hot spots in protein–protein interfaces. Proc. Natl. Acad. Sci. 2011, 108, 13528–13533. [Google Scholar] [CrossRef] [PubMed]
- Bohnuud, T.; Beglov, D.; Ngan, C.H.; Zerbe, B.; Hall, D.R.; Brenke, R.; Vajda, S.; Frank-Kamenetskii, M.D.; Kozakov, D. Computational mapping reveals dramatic effect of Hoogsteen breathing on duplex DNA reactivity with formaldehyde. Nucleic Acids Res. 2012, 40, 7644–7652. [Google Scholar] [CrossRef] [PubMed]
- Batista, P.R.; Robert, C.H.; Maréchal, J.D.; Hamida-Rebaï, M.B.; Pascutti, P.G.; Bisch, P.M.; Perahia, D. Consensus modes, a robust description of protein collective motions from multiple-minima normal mode analysis—Application to the HIV-1 protease. Phys. Chem. Chem. Phys. 2010, 12, 2850–2859. [Google Scholar] [CrossRef] [PubMed]
- Skjaerven, L.; Hollup, S.M.; Reuter, N. Normal mode analysis for proteins. J. Mol. Struct. 2009, 898, 42–48. [Google Scholar] [CrossRef]
- Dobbins, S.E.; Lesk, V.I.; Sternberg, M.J.E. Insights into protein flexibility: The relationship between normal modes and conformational change upon protein–protein docking. Proc. Natl. Acad. Sci. 2008, 105, 10390–10395. [Google Scholar] [CrossRef] [PubMed]
- Philot, E.A.; Perahia, D.; Braz, A.S.K.; Costa, M.G.; de Scott, L.P.B. Binding sites and hydrophobic pockets in Human Thioredoxin 1 determined by normal mode analysis. J. Struct. Biol. 2013, 184, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Cheng, X.; Swails, J.M.; Yeom, M.S.; Eastman, P.K.; Lemkul, J.A.; Wei, S.; Buckner, J.; Jeong, J.C.; Qi, Y. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 2015, 12, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, H.J.C.; van der Spoel, D.; van Drunen, R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995, 91, 43–56. [Google Scholar] [CrossRef]
- Lindahl, E.; Hess, B.; van der Spoel, D.J. GROMACS 3.0: A package for molecular simulation and trajectory analysis. J. Mol. Model. 2001, 306–317. [Google Scholar] [CrossRef]
- van der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 1701–1718. [Google Scholar] [CrossRef] [PubMed]
- Hess, B.; Kutzner, C.; van der Spoel, D.; Lindahl, E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 2008, 435–447. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Cheng, X.; Islam, S.M.; Huang, L.; Rui, H.; Zhu, A.; Lee, H.S.; Qi, Y.; Han, W.; Vanommeslaeghe, K. CHARMM-GUI PDB Manipulator for Advanced Modeling and Simulations of Proteins Containing Nonstandard Residues. Adv. Protein Chem. Struct. Biol. 2014, 96, 235–265. [Google Scholar] [PubMed]
- MacKerell, A.D., Jr.; Bashford, D.; Bellot, M.; Dunbrack, R.L., Jr.; Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S.; et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 1998, 102, 3586–3616. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmuller, H.; MacKerell, A.D., Jr. CHARMM36m: An Improved Force Field for Folded and Intrinsically Disordered Proteins. Nat. Methods 2016, 14, 71–73. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
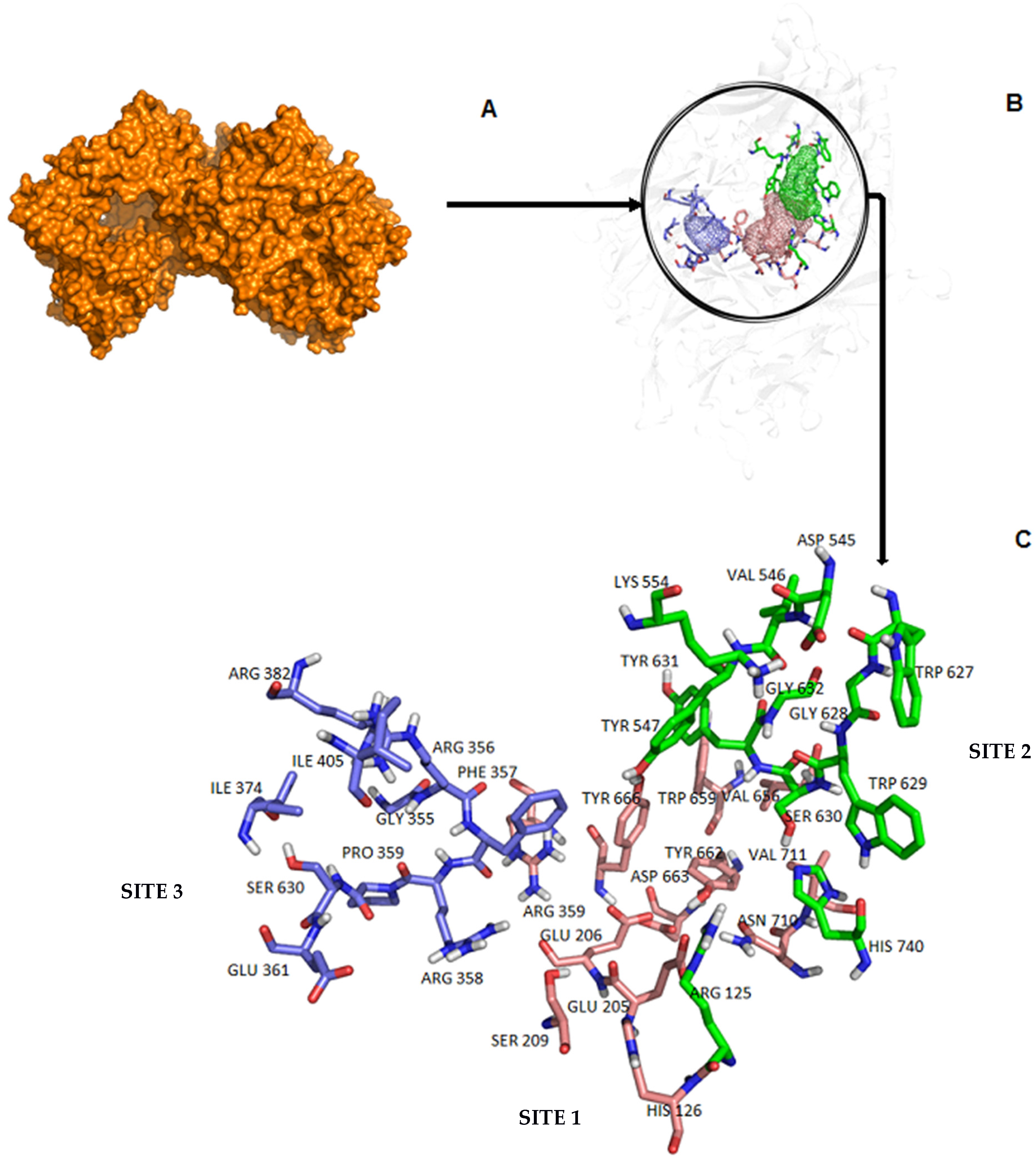



| Residue | Interaction in Chain A | Interaction in Chain B |
|---|---|---|
| Glu205 | HB/SB | SB |
| Phe357 | - | HC |
| Val546 | HC | - |
| Tyr547 | HC | HC/PIS |
| Lys554 | - | HC |
| Trp627 | HC | HC |
| Gly628 | HC | HC |
| Trp629 | HC/PIS | HC |
| Ser630 | HC | HC |
| Tyr631 | HB | HB/HC |
| Val656 | - | HC |
| Tyr662 | HC | HC/PIS |
| Asp663 | - | SB |
| Tyr666 | HC/PIT | HC |
| Val711 | HB | - |
| Molecule-probe | Physicochemical Characteristics |
|---|---|
| acetamide (ACD) | polar; donor and acceptor of hydrogen bond |
| acetonitrile (ACN) | polar; hydrogen acceptor binding character |
| acetone (ACT) | polar; hydrogen acceptor binding character |
| acetaldehyde (ADY) | polar; hydrogen acceptor binding character |
| methanamine (AMN) | polar; positive; donor and acceptor of hydrogen bond |
| benzaldehyde (BDY) | polar; aromatic; hydrogen acceptor binding character |
| benzene (BEN) | hydrophobic; aromatic |
| tert-butanol (BUT) | hydrophobic; aromatic |
| cyclohexane (CHX) | polar; donor and acceptor of hydrogen bond |
| N,N-dimethylformamide (DFO) | polar; hydrogen bond acceptor |
| dimethyl ether (DME) | polar; hydrogen bond acceptor |
| ethanol (EOL) | polar; donor and acceptor of hydrogen bond |
| ethane (ETH) | hydrophobic |
| phenol (PHN) | polar; aromatic; donor and acceptor of hydrogen bond |
| isopropanol (THS) | polar; hydrogen acceptor binding character |
| urea (URE) | polar; positive; donor and acceptor of hydrogen bond |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pantaleão, S.Q.; Philot, E.A.; De Resende-Lara, P.T.; Lima, A.N.; Perahia, D.; Miteva, M.A.; Scott, A.L.; Honorio, K.M. Structural Dynamics of DPP-4 and Its Influence on the Projection of Bioactive Ligands. Molecules 2018, 23, 490. https://doi.org/10.3390/molecules23020490
Pantaleão SQ, Philot EA, De Resende-Lara PT, Lima AN, Perahia D, Miteva MA, Scott AL, Honorio KM. Structural Dynamics of DPP-4 and Its Influence on the Projection of Bioactive Ligands. Molecules. 2018; 23(2):490. https://doi.org/10.3390/molecules23020490
Chicago/Turabian StylePantaleão, Simone Queiroz, Eric Allison Philot, Pedro Túlio De Resende-Lara, Angélica Nakagawa Lima, David Perahia, Maria Atanassova Miteva, Ana Ligia Scott, and Kathia Maria Honorio. 2018. "Structural Dynamics of DPP-4 and Its Influence on the Projection of Bioactive Ligands" Molecules 23, no. 2: 490. https://doi.org/10.3390/molecules23020490
APA StylePantaleão, S. Q., Philot, E. A., De Resende-Lara, P. T., Lima, A. N., Perahia, D., Miteva, M. A., Scott, A. L., & Honorio, K. M. (2018). Structural Dynamics of DPP-4 and Its Influence on the Projection of Bioactive Ligands. Molecules, 23(2), 490. https://doi.org/10.3390/molecules23020490






