Diagnosis of Bacterial Pathogens in the Urine of Urinary-Tract-Infection Patients Using Surface-Enhanced Raman Spectroscopy
Abstract
1. Introduction
2. Results
2.1. Establishment of Reference Raman Spectrum
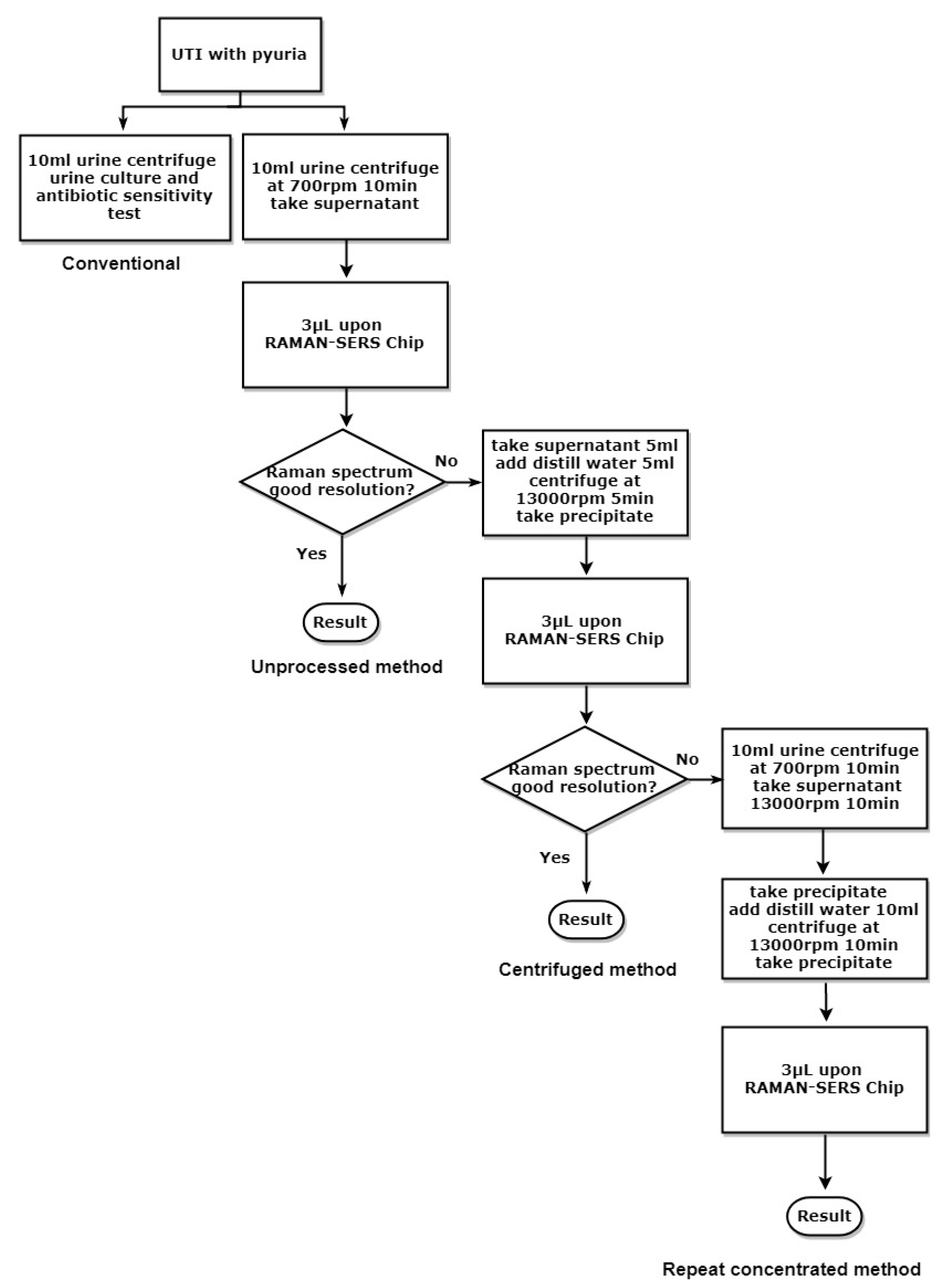
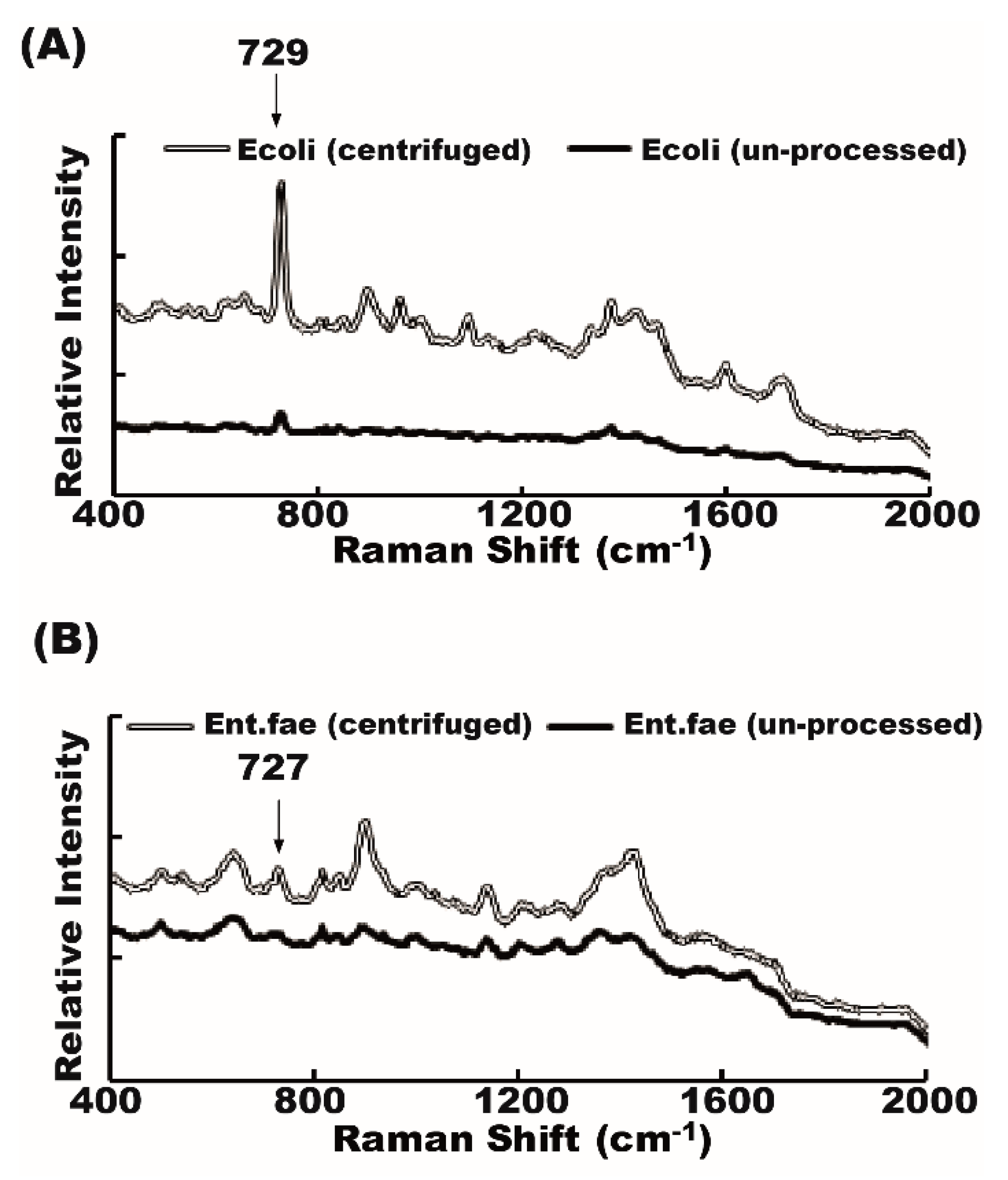
2.2. Raman Spectrum of Bacteria from Unprocessed and Centrifuged Urine Samples
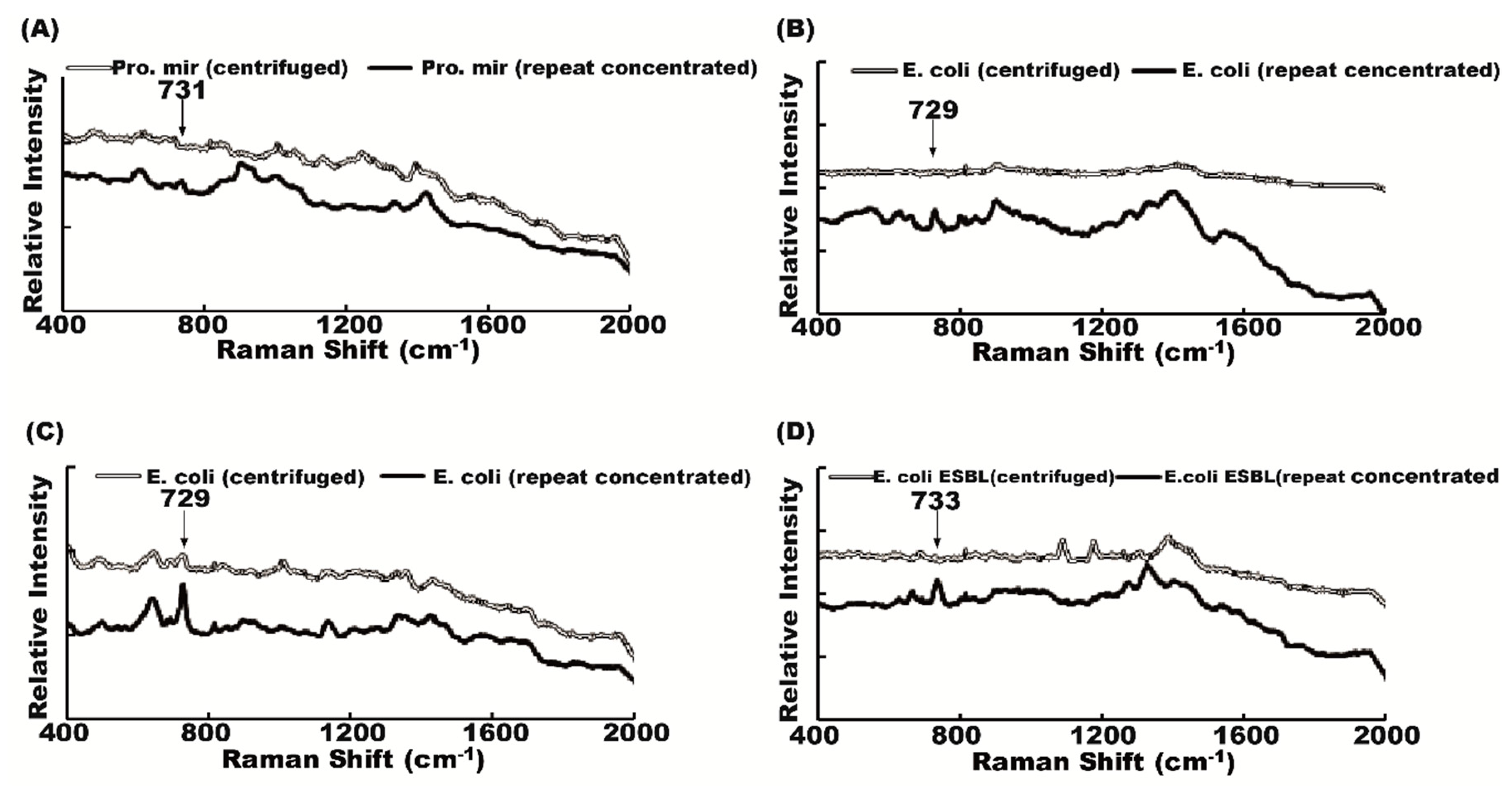
2.3. Effect of Repeated Concentration on Raman Spectrum
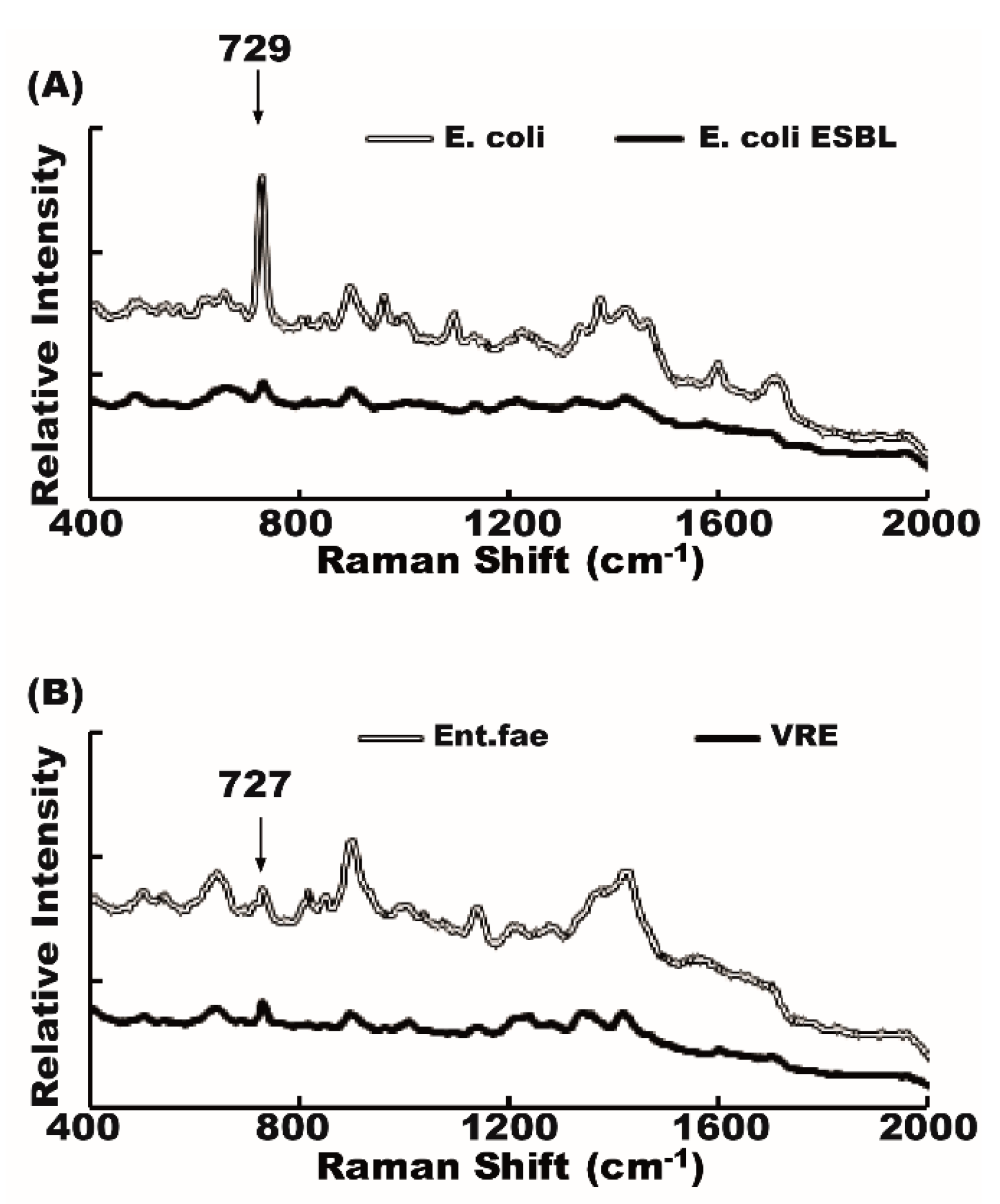
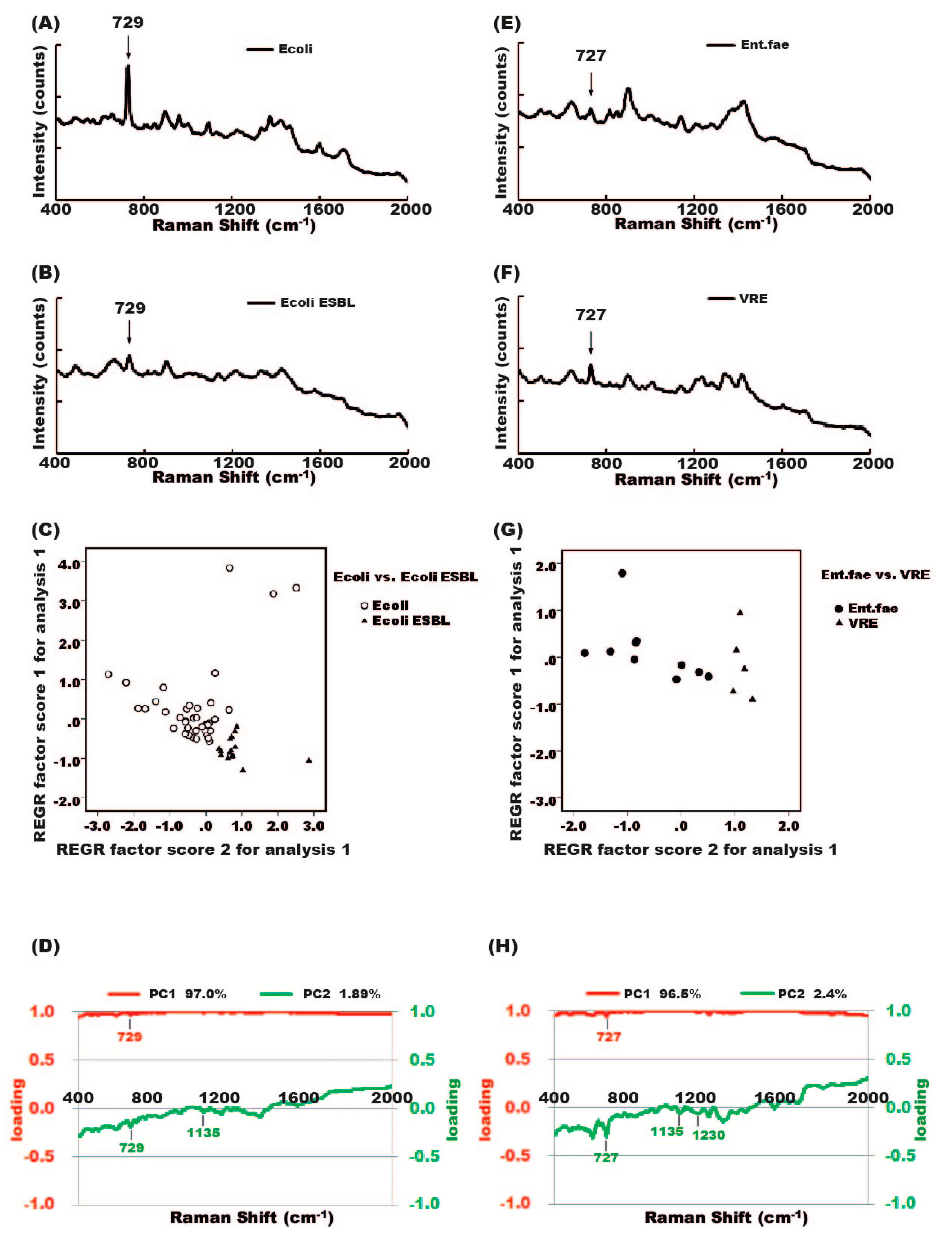
2.4. Raman Spectra of Antibiotic-Susceptible and Antibiotic-Resistant Bacteria
2.5. Differentiation between Antibiotic-Susceptible and Antibiotic-Resistant Bacteria Strains Using Principal Component Analysis (PCA)
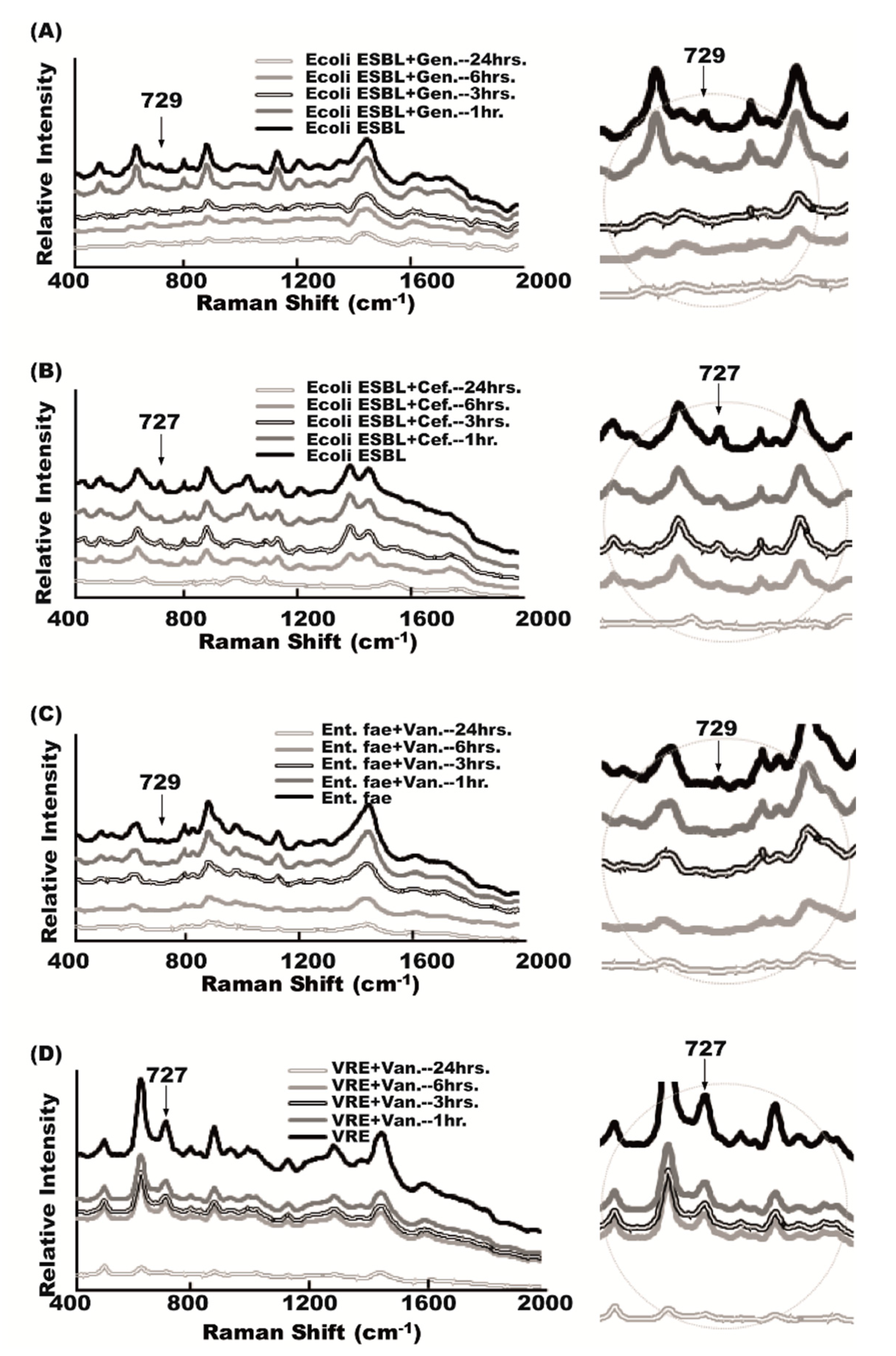
2.6. Antibiotic Effect on Raman Spectra
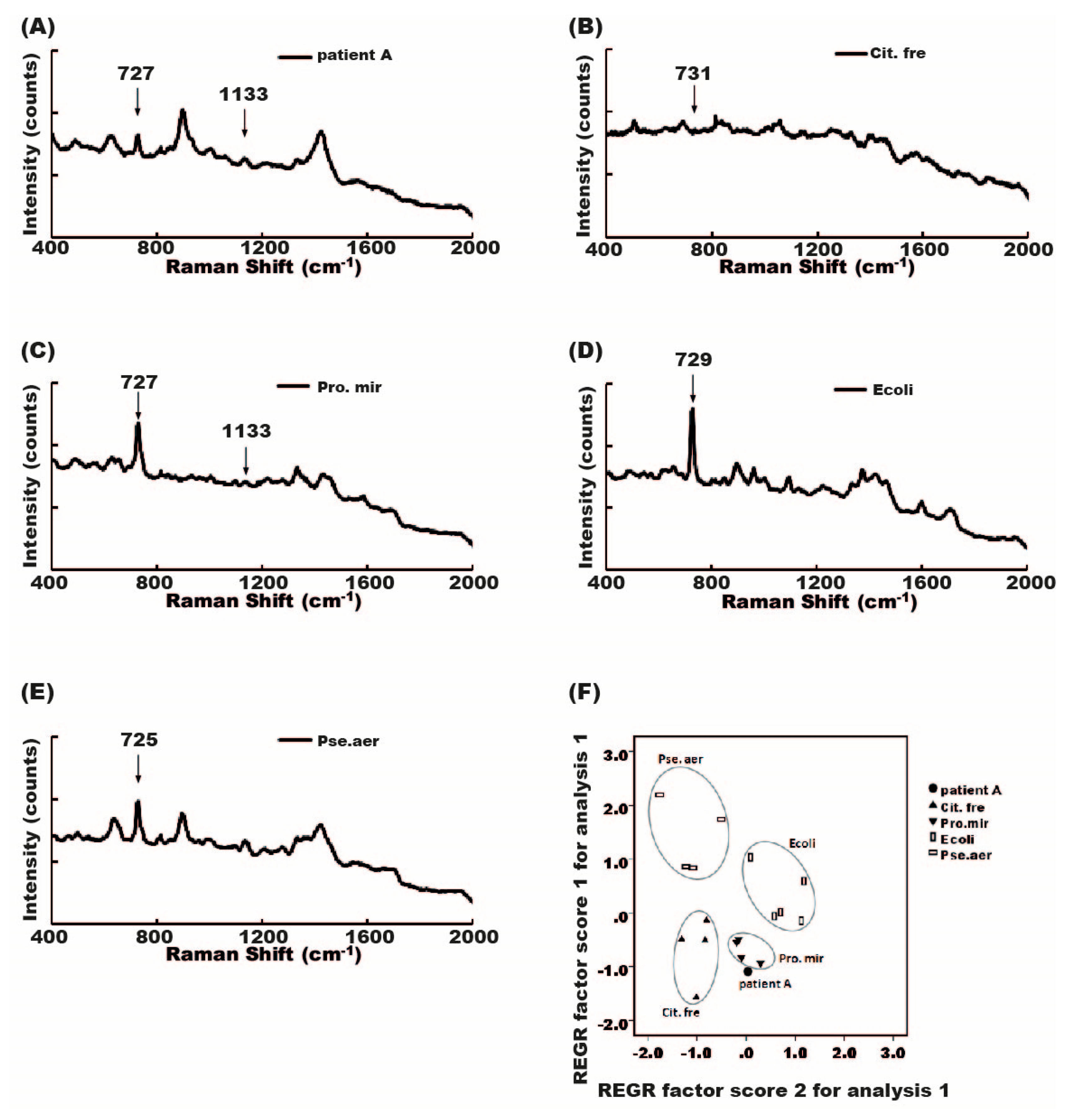
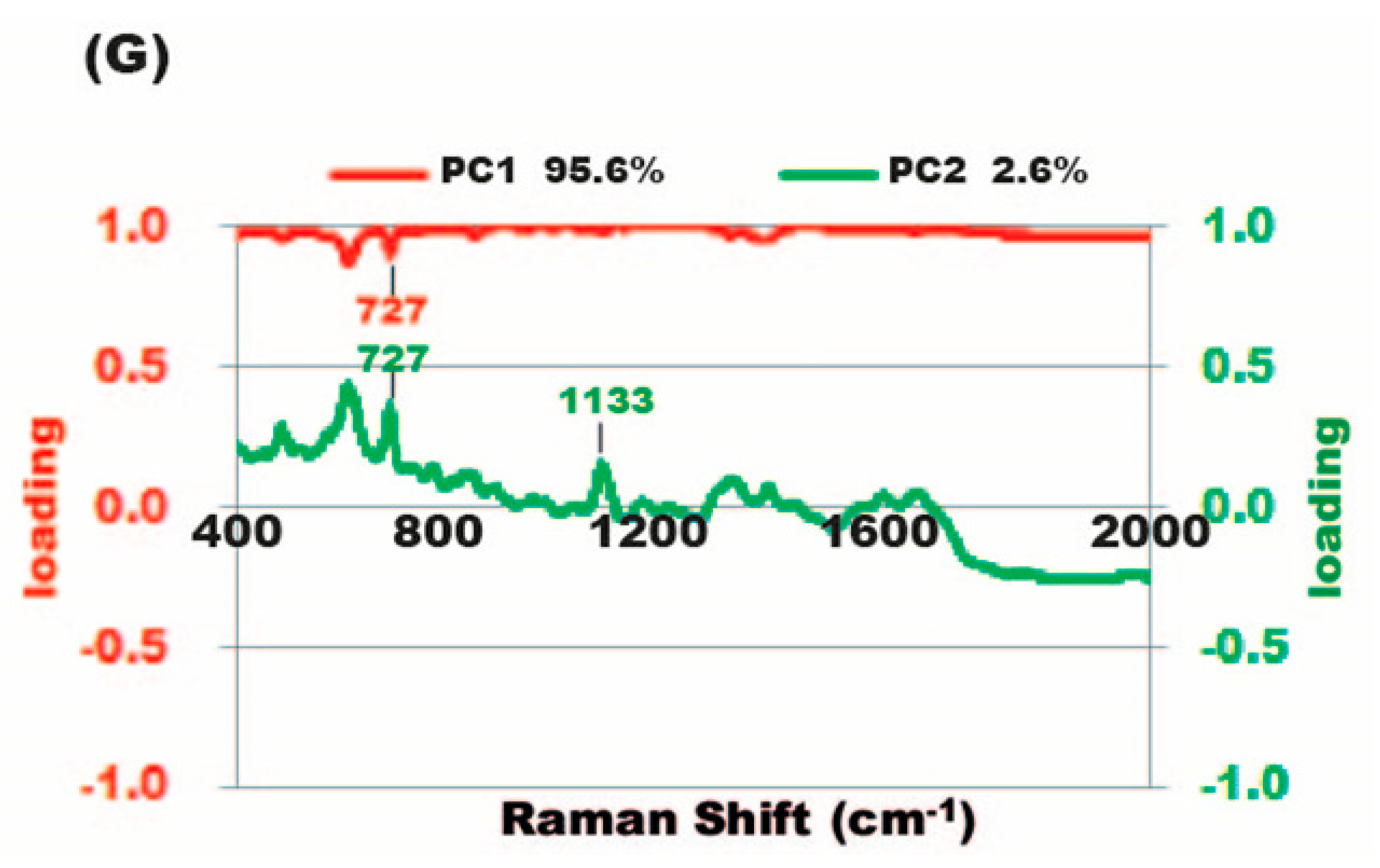
2.7. Diagnosis of Mixed-Flora Infections
3. Discussion
4. Materials and Methods
4.1. Samples
4.2. Sample Processing for SERS Studies
4.3. Conventional Bacterial Culture
4.4. Antimicrobial Susceptibility Test after Urine Culture
4.5. Raman Spectrum of Different Bacteria
4.6. PCA
4.7. Antibiotic-Susceptibility Test
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sahm, D.F.; Thornsberry, C.; Mayfield, D.C.; Jones, M.E.; Karlowsky, J.A. Multidrug-resistant urinary tract isolates of Escherichia coli: Prevalence and patient demographics in the United States in 2000. Antimicrob. Agents Chemother. 2001, 45, 1402–1406. [Google Scholar] [CrossRef] [PubMed]
- Nordenstam, G.; Sundh, V.; Lincoln, K.; Svanborg, A.; Eden, C.S. Bacteriuria in representative population samples of persons aged 72-79 years. Am. J. Epidemiol. 1989, 130, 1176–1186. [Google Scholar] [CrossRef]
- Manges, A.R.; Johnson, J.R.; Foxman, B.; O’Bryan, T.T.; Fullerton, K.E.; Riley, L.W. Widespread distribution of urinary tract infections caused by a multidrug-resistant Escherichia coli clonal group. N. Engl. J. Med. 2001, 345, 1007–1013. [Google Scholar] [CrossRef] [PubMed]
- Graham, J.C.; Galloway, A. ACP Best Practice No 167: The laboratory diagnosis of urinary tract infection. J. Clin. Pathol. 2001, 54, 911–919. [Google Scholar] [CrossRef] [PubMed]
- Seng, P.; Abat, C.; Rolain, J.M.; Colson, P.; Lagier, J.C.; Gouriet, F.; Fournier, P.E.; Drancourt, M.; La Scola, B.; Raoult, D. Identification of rare pathogenic bacteria in a clinical microbiology laboratory: Impact of matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2013, 51, 2182–2194. [Google Scholar] [CrossRef] [PubMed]
- Rajapandiyan, P.; Yang, J. Sensitive cylindrical SERS substrate array for rapid microanalysis of nucleobases. Anal. Chem. 2012, 84, 10277–10282. [Google Scholar] [CrossRef] [PubMed]
- Zavaleta, C.L.; Kircher, M.F.; Gambhir, S.S. Raman’s “effect” on molecular imaging. J. Nucl. Med. 2011, 52, 1839–1844. [Google Scholar] [CrossRef] [PubMed]
- Camden, J.P.; Dieringer, J.A.; Zhao, J.; Van Duyne, R.P. Controlled plasmonic nanostructures for surface-enhanced spectroscopy and sensing. Acc. Chem. Res. 2008, 41, 1653–1661. [Google Scholar] [CrossRef]
- Lombardi, J.R.; Birke, R.L. A unified view of surface-enhanced Raman scattering. Acc. Chem. Res. 2009, 42, 734–742. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.Y.; Han, Y.Y.; Shih, P.H.; Lian, W.N.; Wang, H.H.; Lin, C.H.; Hsueh, P.R.; Wang, J.K.; Wang, Y.L. Rapid bacterial antibiotic susceptibility test based on simple surface-enhanced Raman spectroscopic biomarkers. Sci. Rep. 2016, 6, 23375. [Google Scholar] [CrossRef] [PubMed]
- Tien, N.; Chen, H.C.; Gau, S.L.; Lin, T.H.; Lin, H.S.; You, B.J.; Tsai, P.C.; Chen, I.R.; Tsai, M.F.; Wang, I.K.; et al. Diagnosis of bacterial pathogens in the dialysate of peritoneal dialysis patients with peritonitis using surface-enhanced Raman spectroscopy. Clin. Chim. Acta 2016, 461, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Boyaci, I.H.; Temiz, H.T.; Uysal, R.S.; Velioglu, H.M.; Yadegari, R.J.; Rishkan, M.M. A novel method for discrimination of beef and horsemeat using Raman spectroscopy. Food Chem. 2014, 148, 37–41. [Google Scholar] [CrossRef]
- Blackie, E.J.; Le Ru, E.C.; Etchegoin, P.G. Single-molecule surface-enhanced Raman spectroscopy of nonresonant molecules. J. Am. Chem. Soc. 2009, 131, 14466–14472. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.Y.; Tsai, K.T.; Wang, H.H.; Chen, Y.; Chen, Y.H.; Chao, Y.C.; Chang, H.H.; Lin, C.H.; Wang, J.K.; Wang, Y.L. Functionalized arrays of Raman-enhancing nanoparticles for capture and culture-free analysis of bacteria in human blood. Nat. Commun. 2011, 2, 538. [Google Scholar] [CrossRef] [PubMed]
- Schroder, U.C.; Ramoji, A.; Glaser, U.; Sachse, S.; Leiterer, C.; Csaki, A.; Hubner, U.; Fritzsche, W.; Pfister, W.; Bauer, M.; et al. Combined dielectrophoresis-Raman setup for the classification of pathogens recovered from the urinary tract. Anal. Chem. 2013, 85, 10717–10724. [Google Scholar] [CrossRef]
- Draz, M.S.; Lu, X. Development of a loop mediated isothermal amplification (LAMP)—surface enhanced Raman spectroscopy (SERS) assay for the detection of salmonella enterica serotype enteritidis. Theranostics 2016, 6, 522–532. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Premasiri, W.R.; Ziegler, L.D. Surface enhanced Raman spectroscopy of Chlamydia trachomatis and Neisseria gonorrhoeae for diagnostics, and extra-cellular metabolomics and biochemical monitoring. Sci. Rep. 2018, 8, 5163. [Google Scholar] [CrossRef]
- Dinarelli, S.; Girasole, M.; Kasas, S.; Longo, G. Nanotools and molecular techniques to rapidly identify and fight bacterial infections. J. Microbiol. Methods 2017, 138, 72–81. [Google Scholar] [CrossRef]
- Han, X.X.; Huang, G.G.; Zhao, B.; Ozaki, Y. Label-free highly sensitive detection of proteins in aqueous solutions using surface-enhanced Raman scattering. Anal. Chem. 2009, 81, 3329–3333. [Google Scholar] [CrossRef]
- Sengupta, A.; Mujacic, M.; Davis, E.J. Detection of bacteria by surface-enhanced Raman spectroscopy. Anal. Bioanal. Chem. 2006, 386, 1379–1386. [Google Scholar] [CrossRef]
- Wragg, R.; Harris, A.; Patel, M.; Robb, A.; Chandran, H.; McCarthy, L. Extended spectrum beta lactamase (ESBL) producing bacteria urinary tract infections and complex pediatric urology. J. Pediatr. Surg. 2017, 52, 286–288. [Google Scholar] [CrossRef]
- Lee, Y.C.; Hsiao, C.Y.; Hung, M.C.; Hung, S.C.; Wang, H.P.; Huang, Y.J.; Wang, J.T. Bacteremic urinary tract infection caused by multidrug-resistant enterobacteriaceae are associated with severe sepsis at admission: Implication for empirical therapy. Medicine (Baltimore) 2016, 95, e3694. [Google Scholar] [CrossRef]
- Tseng, W.P.; Chen, Y.C.; Chen, S.Y.; Chen, S.Y.; Chang, S.C. Risk for subsequent infection and mortality after hospitalization among patients with multidrug-resistant gram-negative bacteria colonization or infection. Antimicrob. Resist. Infect. Control. 2018, 7, 93. [Google Scholar] [CrossRef] [PubMed]
- Cheng, I.F.; Chang, H.C.; Chen, T.Y.; Hu, C.; Yang, F.L. Rapid (<5 min) identification of pathogen in human blood by electrokinetic concentration and surface-enhanced Raman spectroscopy. Sci. Rep. 2013, 3, 2365. [Google Scholar]
- Pahlow, S.; Meisel, S.; Cialla-May, D.; Weber, K.; Rosch, P.; Popp, J. Isolation and identification of bacteria by means of Raman spectroscopy. Adv. Drug Deliv. Rev. 2015, 89, 105–120. [Google Scholar] [CrossRef] [PubMed]
- Cheng, I.F.; Chen, T.Y.; Chao, W.C. Increasing local density and purity of molecules/bacteria on a sensing surface from diluted blood using 3D hybrid electrokinetics. Biomicrofluidics 2016, 10, 034116. [Google Scholar] [CrossRef] [PubMed]
- Puchberger-Enengl, D.; Podszun, S.; Heinz, H.; Hermann, C.; Vulto, P.; Urban, G.A. Microfluidic concentration of bacteria by on-chip electrophoresis. Biomicrofluidics 2011, 5, 044111. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.C.; Tien, N.; Yang, J.S.; Lu, C.C.; Tsai, F.J.; Huang, T.J.; Wang, I.K. Class 1 integrons and plasmid-mediated multiple resistance genes of the Campylobacter species from pediatric patient of a university hospital in Taiwan. Gut Pathogens 2017, 9, 50. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.T.; Chi, C.Y.; Ho, C.M.; Lin, P.C.; Chou, C.H.; Wang, J.H.; Wang, J.H.; Lin, H.C.; Tien, N.; Lin, K.H.; et al. Correlation of virulence genes to clinical manifestations and outcome in patients with Streptococcus dysgalactiae subspecies equisimilis bacteremia. J. Microbiol. Immunol. Infect. 2014, 47, 462–468. [Google Scholar] [CrossRef] [PubMed]
- Chun, K.; Syndergaard, C.; Damas, C.; Trubey, R.; Mukindaraj, A.; Qian, S.; Jin, X.; Breslow, S.; Niemz, A. Sepsis pathogen identification. J. Lab. Autom. 2015, 20, 539–561. [Google Scholar] [CrossRef] [PubMed]
- Andrews, J.M. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 2001, 48 (Suppl. 1), 5–16. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are available from the authors. |
| Conventional | Raman SERS | |
|---|---|---|
| Pathogen Result | ||
| Single bacteria | 97 | 97 |
| Mixed flora (two kinds) | 7 | failure to detect |
| Mixed flora (three kinds) | 4 | failure to detect |
| Raman Sample Method | ||
| unprocessed method | 93 | |
| repeat concentrated method | 4 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tien, N.; Lin, T.-H.; Hung, Z.-C.; Lin, H.-S.; Wang, I.-K.; Chen, H.-C.; Chang, C.-T. Diagnosis of Bacterial Pathogens in the Urine of Urinary-Tract-Infection Patients Using Surface-Enhanced Raman Spectroscopy. Molecules 2018, 23, 3374. https://doi.org/10.3390/molecules23123374
Tien N, Lin T-H, Hung Z-C, Lin H-S, Wang I-K, Chen H-C, Chang C-T. Diagnosis of Bacterial Pathogens in the Urine of Urinary-Tract-Infection Patients Using Surface-Enhanced Raman Spectroscopy. Molecules. 2018; 23(12):3374. https://doi.org/10.3390/molecules23123374
Chicago/Turabian StyleTien, Ni, Tzu-Hsien Lin, Zen-Chao Hung, Hsiu-Shen Lin, I-Kuan Wang, Hung-Chih Chen, and Chiz-Tzung Chang. 2018. "Diagnosis of Bacterial Pathogens in the Urine of Urinary-Tract-Infection Patients Using Surface-Enhanced Raman Spectroscopy" Molecules 23, no. 12: 3374. https://doi.org/10.3390/molecules23123374
APA StyleTien, N., Lin, T.-H., Hung, Z.-C., Lin, H.-S., Wang, I.-K., Chen, H.-C., & Chang, C.-T. (2018). Diagnosis of Bacterial Pathogens in the Urine of Urinary-Tract-Infection Patients Using Surface-Enhanced Raman Spectroscopy. Molecules, 23(12), 3374. https://doi.org/10.3390/molecules23123374





