SumSec: Accurate Prediction of Sumoylation Sites Using Predicted Secondary Structure
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset Description
2.2. Predicted Secondary Structure
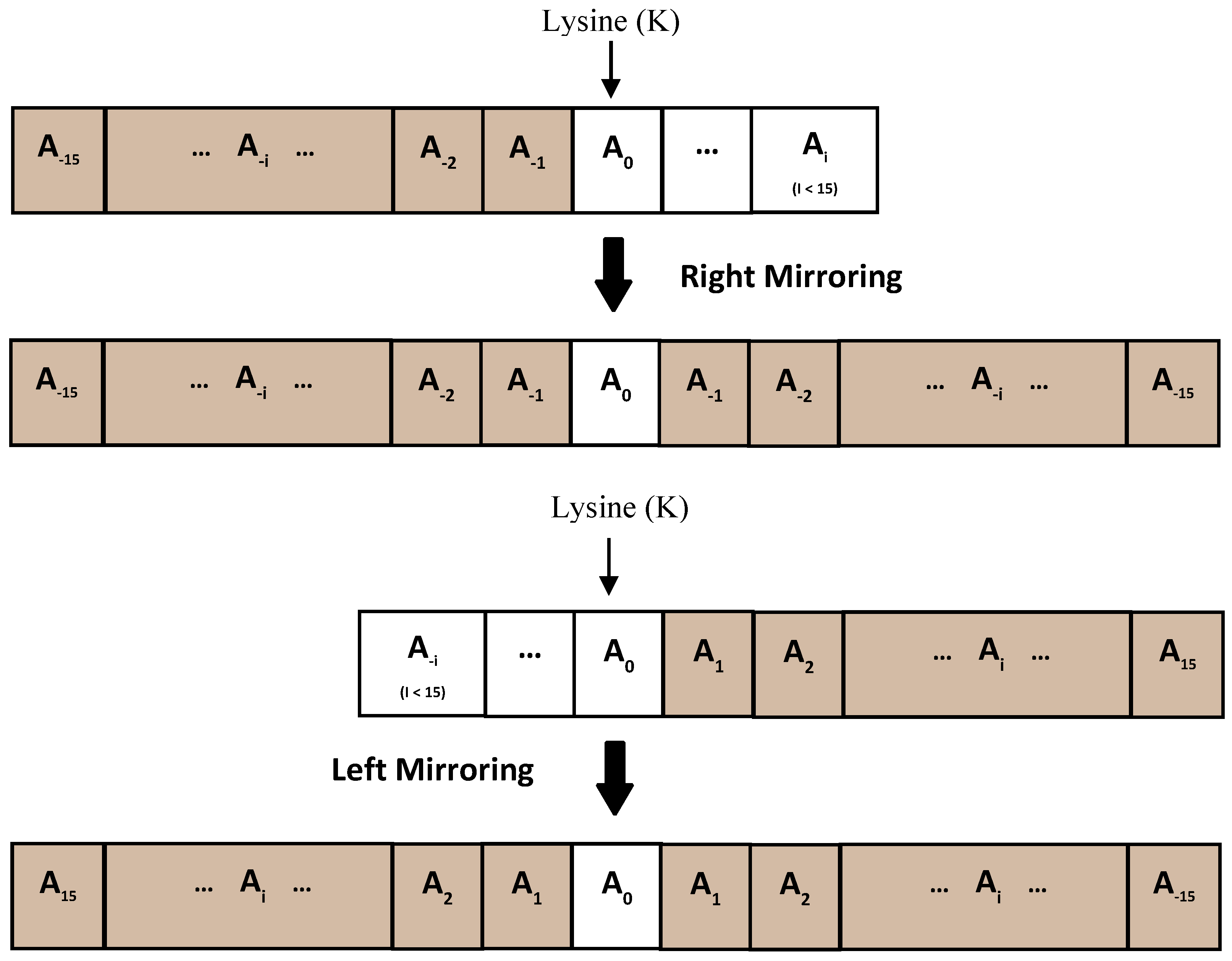
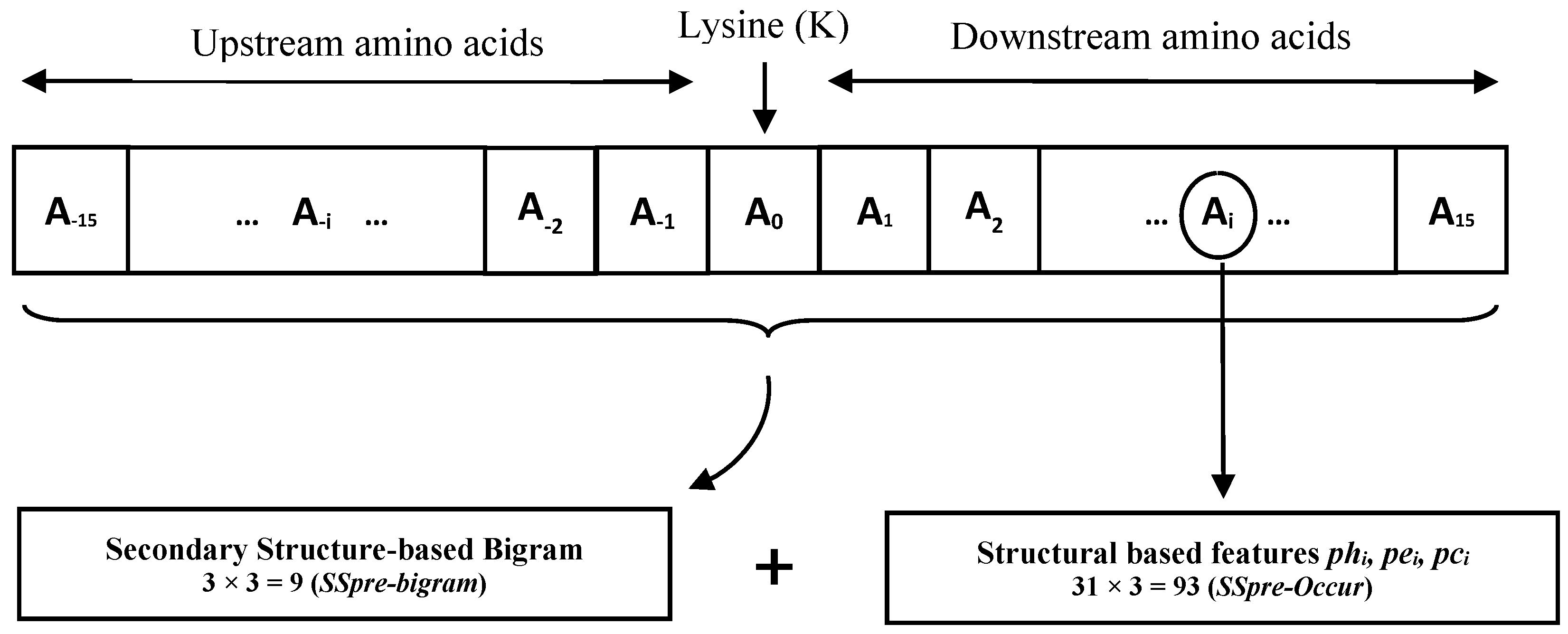
2.3. Feature Extraction
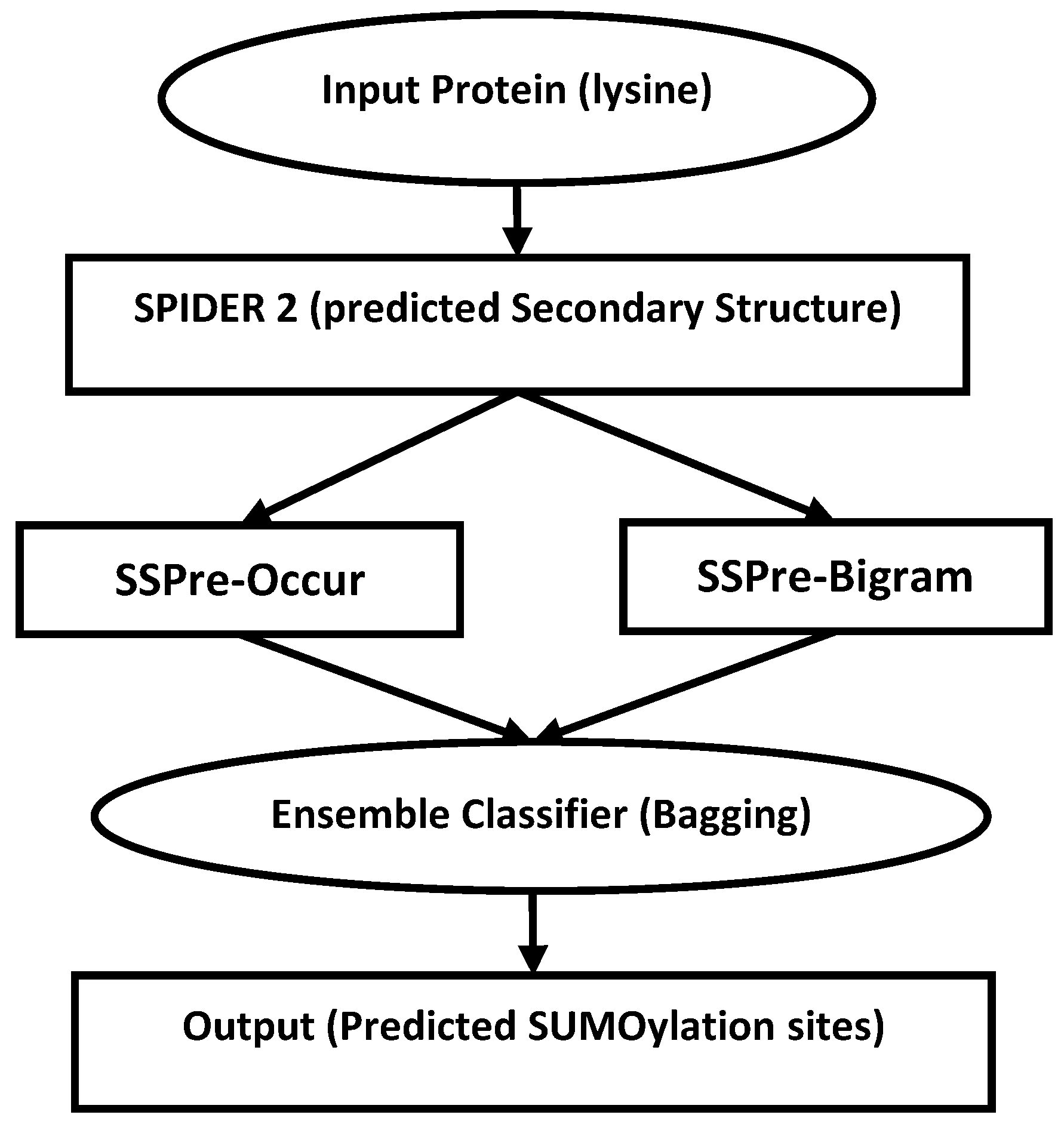
2.4. Classification
2.5. Evaluation Method and Performance Measurements
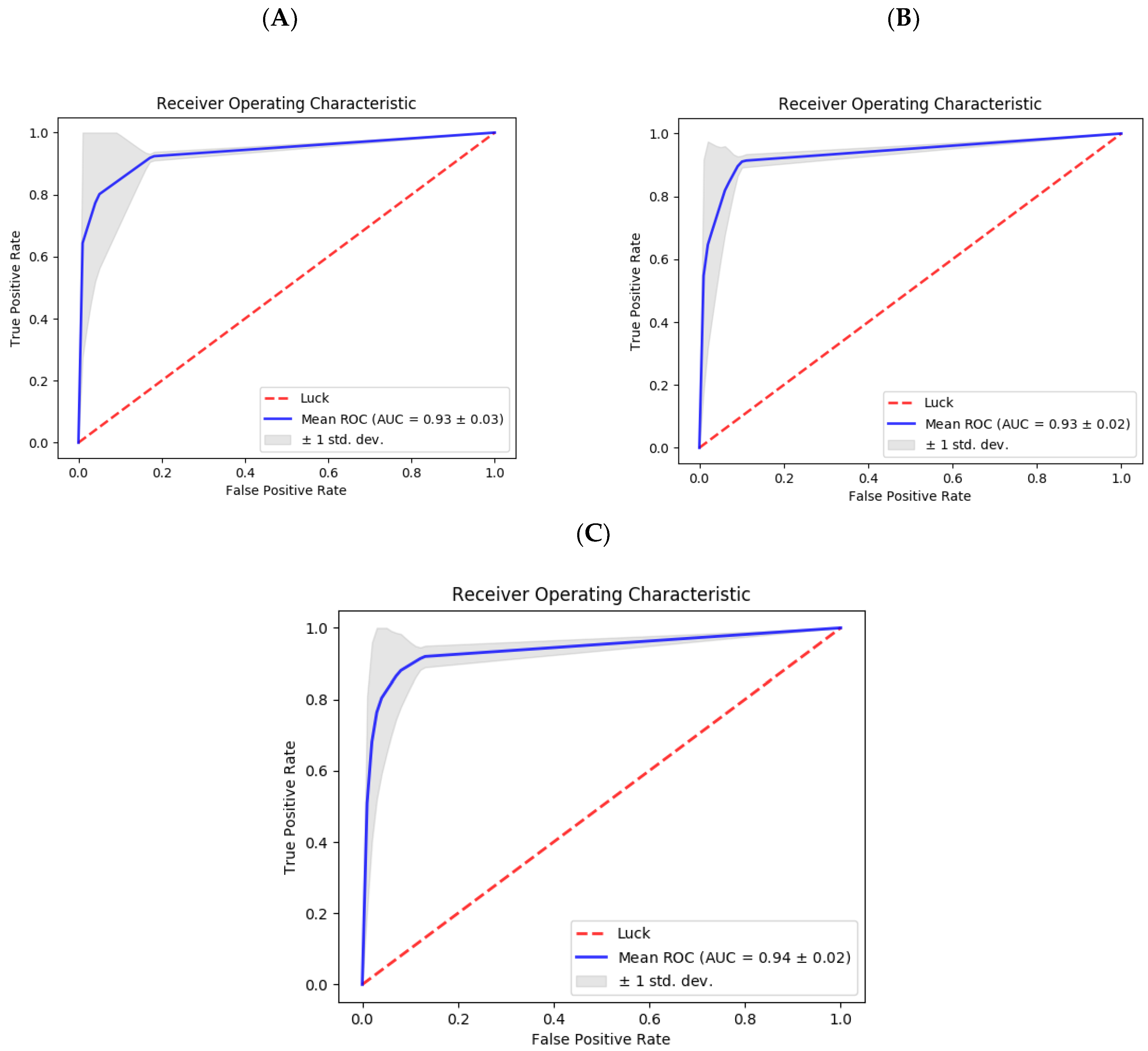
3. Results and Discussion
4. Conclusions and Future Work
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| PTM | post-translation modification |
| SumSec | Sumoylation predictor using Structural-based features |
| ASA | accessible surface area |
| FN | false negative |
| TN | true negative |
| TP | true positive |
| FP | false positive |
| MCC | Matthew’s correlation coefficient |
| AUC | area under curve |
| ROC | receiver operating characteristic |
References
- Mann, M.; Jensen, O.N. Proteomic analysis of post-translational modifications. Nat. Biotechnol. 2003, 21, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Jensen, O.N. Interpreting the protein language using proteomics. Nat. Rev. Mol. Cell Biol. 2006, 7, 391. [Google Scholar] [CrossRef] [PubMed]
- Hendriks, I.A.; D’Souza, R.C.J.; Yang, B.; Verlaan-de Vries, M.; Mann, M.; Vertegaal, A.C.O. Uncovering global sumoylation signaling networks in a site-specific manner. Nat. Struct. Mol. Biol. 2014, 21, 927–936. [Google Scholar] [CrossRef] [PubMed]
- Pawson, T.; Scott, J.D. Protein phosphorylation in signaling—50 years and counting. Trends Biochem. Sci. 2005, 30, 286–290. [Google Scholar] [CrossRef]
- Kessler, B.M.; Edelmann, M.J. Ptms in conversation: Activity and function of deubiquitinating enzymes regulated via post-translational modifications. Cell Biochem. Biophys. 2011, 60, 21–38. [Google Scholar] [CrossRef]
- Huber, S.C.; Hardin, S.C. Numerous posttranslational modifications provide opportunities for the intricate regulation of metabolic enzymes at multiple levels. Curr. Opin. Plant Biol. 2004, 7, 318–322. [Google Scholar] [CrossRef] [PubMed]
- Swaney, D.L.; Beltrao, P.; Starita, L.; Guo, A.; Rush, J.; Fields, S.; Krogan, N.J.; Villén, J. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat. Methods 2013, 10, 676. [Google Scholar] [CrossRef]
- Duan, G.; Walther, D. The roles of post-translational modifications in the context of protein interaction networks. PLOS Comput. Biol. 2015, 11, e1004049. [Google Scholar] [CrossRef]
- Flotho, A.; Melchior, F. Sumoylation: A regulatory protein modification in health and disease. Annu. Rev. Biochem. 2013, 82, 357–385. [Google Scholar] [CrossRef]
- Steffan, J.S.; Agrawal, N.; Pallos, J.; Rockabrand, E.; Trotman, L.C.; Slepko, N.; Illes, K.; Lukacsovich, T.; Zhu, Y.-Z.; Cattaneo, E. SUMO modification of Huntingtin and Huntington’s disease pathology. Science 2004, 304, 100–104. [Google Scholar] [CrossRef]
- Krumova, P.; Weishaupt, J.H. Sumoylation in neurodegenerative diseases. Cell. Mol. Life Sci. 2012, 70, 2123–2138. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.; Sakurai, M.; Matsuzaki, S.; Arancio, O.; Fraser, P. Sumo and alzheimer’s disease. NeuroMol. Med. 2013, 15, 720–736. [Google Scholar] [CrossRef] [PubMed]
- Shuai, K.; Liu, B. Regulation of jak–stat signalling in the immune system. Nat. Rev. Immunol. 2003, 3, 900–911. [Google Scholar] [CrossRef] [PubMed]
- Straus, D.S.; Glass, C.K. Anti-inflammatory actions of ppar ligands: New insights on cellular and molecular mechanisms. Trends Immunol. 2007, 28, 551–558. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Guo, D.; Isales, C.M.; Eizirik, D.L.; Atkinson, M.; She, J.-X.; Wang, C.-Y. Sumo wrestling with type 1 diabetes. J. Mol. Med. 2005, 83, 504–513. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.I.; Baek, S.H. SUMOylation code in cancer development and metastasis. Mol. Cells 2006, 22, 247–253. [Google Scholar] [PubMed]
- Eifler, K.; Vertegaal, A.C.O. Sumoylation-mediated regulation of cell cycle progression and cancer. Trends Biochem. Sci. 2015, 40, 779–793. [Google Scholar] [CrossRef]
- Qing, G.; Lu, Q.; Xiong, Y.; Zhang, L.; Wang, H.; Li, X.; Liang, X.; Sun, T. New Opportunities and Challenges of Smart Polymers in Post-Translational Modification Proteomics. Adv. Mater. 2017, 29. [Google Scholar] [CrossRef]
- Dehzangi, A.; Lopez, Y.; Lal, S.; Taherzadeh, G.; Sattar, A.; Tsunoda, T.; Sharma, A. Improving succinylation prediction accuracy by incorporating the secondary structure via helix, strand and coil, and evolutionary information from profile bigrams. PLoS ONE 2018, 13, e0191900. [Google Scholar] [CrossRef]
- Dehzangi, A.; Lopez, Y.; Lal, S.P.; Taherzadeh, G.; Michaelson, J.; Sattar, A.; Tsunoda, T.; Sharma, A. PSSM-Suc: Accurately predicting succinylation using position specific scoring matrix into bigram for feature extraction. J. Theor. Biol. 2017, 425, 97–102. [Google Scholar] [CrossRef]
- Lopez, Y.; Dehzangi, A.; Lal, S.P.; Taherzadeh, G.; Michaelson, J.; Sattar, A.; Tsunoda, T.; Sharma, A. SucStruct: Prediction of succinylated lysine residues by using structural properties of amino acids. Anal. Biochem. 2017, 527, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Qiu, W.R.; Sun, B.Q.; Xiao, X.; Xu, Z.C.; Jia, J.H.; Chou, K.C. iKcr-PseEns: Identify lysine crotonylation sites in histone proteins with pseudo components and ensemble classifier. Genomics 2017, 110, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.M.; Saha, S.; Rahman, M.M.; Shatabda, S.; Farid, D.M.; Dehzangi, A. iProtGly-SS: identifying protein glycation sites using sequence and structure based features. Proteins Struct. Funct. Bioinform. 2018, 86, 777–789. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.M.; Yang, S.; Zhou, Y.; Mollah, M.N.H. Succinsite: A computational tool for the prediction of protein succinylation sites by exploiting the amino acid patterns and properties. Mol. BioSyst. 2016, 12, 786–795. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Liu, Z.; Xiao, X.; Liu, B.; Chou, K.-C. Psuc-lys: Predict lysine succinylation sites in proteins with pseaac and ensemble random forest approach. J. Theor. Biol. 2016, 394, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Ding, Y.-X.; Deng, N.-Y.; Liu, L.-M. Prediction of sumoylation sites in proteins using linear discriminant analysis. Gene 2016, 576, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-Z.; Chen, Z.; Gong, Y.-A.; Ying, G. Sumohydro: A novel method for the prediction of sumoylation sites based on hydrophobic properties. PLoS ONE 2012, 7, e39195. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y. Gly-PseAAC: Identifying protein lysine glycation through sequences. Gene 2017, 602, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Taherzadeh, G.; Yang, Y.; Xu, H.; Xue, Y.; Liew, A.W.C.; Zhou, Y. Predicting lysine-malonylation sites of proteins using sequence and predicted structural features. J. Comput. Chem. 2018, 39, 1757–1763. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Zhou, F.; Fu, C.; Xu, Y.; Yao, X. Sumosp: A web server for sumoylation site prediction. Nucleic Acids Res. 2006, 34, W254–W257. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; He, Y.; Qiang, B.; Yuan, J.; Peng, X.; Pan, X.-M. A novel method for high accuracy sumoylation site prediction from protein sequences. BMC Bioinform. 2008, 9, 8. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Gao, X.; Jin, C.; Zhu, M.; Wang, X.; Shaw, A.; Wen, L.; Yao, X.; Xue, Y. Systematic study of protein sumoylation: Development of a site-specific predictor of SUMOsp 2.0. Proteomics 2009, 9, 3409–3412. [Google Scholar] [CrossRef]
- Zhao, Q.; Xie, Y.; Zheng, Y.; Jiang, S.; Liu, W.; Mu, W.; Liu, Z.; Zhao, Y.; Xue, Y.; Ren, J. Gps-sumo: A tool for the prediction of sumoylation sites and sumo-interaction motifs. Nucleic Acids Res. 2014, 42, W325–W330. [Google Scholar] [CrossRef] [PubMed]
- Yavuz, A.; Sezerman, O. Predicting sumoylation sites using support vector machines based on various sequence features, conformational flexibility and disorder. BMC Genom. 2014, 15, S18. [Google Scholar] [CrossRef]
- Jia, J.; Zhang, L.; Liu, Z.; Xiao, X.; Chou, K.-C. pSumo-CD: Predicting sumoylation sites in proteins with covariance discriminant algorithm by incorporating sequence-coupled effects into general PseAAC. Bioinformatics 2016, 32, 3133–3141. [Google Scholar] [CrossRef] [PubMed]
- Lyons, J.; Paliwal, K.K.; Dehzangi, A.; Heffernan, R.; Tsunoda, T.; Sharma, A. Protein fold recognition using hmm–hmm alignment and dynamic programming. J. Theor. Biol. 2016, 393, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Saini, H.; Raicar, G.; Dehzangi, A.; Lal, S.; Sharma, A. Subcellular localization for Gram positive and Gram negative bacterial proteins using linear interpolation smoothing model. J. Theor. Biol. 2015, 386, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Dehzangi, A.; Lyons, J.; Paliwal, K.; Tsunoda, T.; Sharma, A. Predict gram-positive and gram-negative subcellular localization via incorporating evolutionary information and physicochemical features into Chou’s general PseAAC. IEEE Trans. NanoBiosci. 2015, 14, 915–926. [Google Scholar] [CrossRef] [PubMed]
- Paliwal, K.K.; Sharma, A.; Lyons, J.; Dehzangi, A. Improving protein fold recognition using the amalgamation of evolutionary-based and structural based information. BMC Bioinform. 2014, 15, S12. [Google Scholar] [CrossRef] [PubMed]
- Lyons, J.; Dehzangi, A.; Heffernan, R.; Sharma, A.; Paliwal, K.; Sattar, A.; Zhou, Y.; Yang, Y. Predicting backbone cα angles and dihedrals from protein sequences by stacked sparse auto-encoder deep neural network. J. Comput. Chem. 2014, 35, 2040–2046. [Google Scholar] [CrossRef]
- Sharma, A.; Lyons, J.; Dehzangi, A.; Paliwal, K.K. A feature extraction technique using bi-gram probabilities of position specific scoring matrix for protein fold recognition. J. Theor. Biol. 2013, 320, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Taherzadeh, G.; Yang, Y.D.; Zhang, T.; Liew, A.W.C.; Zhou, Y.Q. Sequence-based prediction of protein–peptide binding sites using support vector machine. J. Comput. Chem. 2016, 37, 1223–1229. [Google Scholar] [CrossRef] [PubMed]
- Taherzadeh, G.; Zhou, Y.; Liew, A.W.-C.; Yang, Y. Sequence-based prediction of protein–carbohydrate binding sites using support vector machines. J. Chem. Inf. Model. 2016, 56, 2115–2122. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.Y.; Shatabda, S.; Dehzangi, A. Idnaprot-es: Identification of DNA-Binding Proteins Using Evolutionary and Structural Features. Available online: https://www.nature.com/articles/s41598-017-14945-1 (accessed on 18 October 2018).
- Saini, H.; Raicar, G.; Sharma, A.; Lal, S.; Dehzangi, A.; Lyons, J.; Paliwal, K.K.; Imoto, S.; Miyano, S. Probabilistic expression of spatially varied amino acid dimers into general form of Chou’s pseudo amino acid composition for protein fold recognition. J. Theor. Biol. 2015, 380, 291–298. [Google Scholar] [CrossRef]
- Shen, H.-B.; Chou, K.-C. Virus-mploc: A fusion classifier for viral protein subcellular location prediction by incorporating multiple sites. J. Biomol. Struct. Dyn. 2010, 28, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Dehzangi, A.; Paliwal, K.; Lyons, J.; Sharma, A.; Sattar, A. Proposing a highly accurate protein structural class predictor using segmentation-based features. BMC Genom. 2014, 15, S2. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, Y.; Gao, T.; Pan, Z.; Cheng, H.; Yang, Q.; Cheng, Z.; Guo, A.; Ren, J.; Xue, Y. CPLM: A database of protein lysine modifications. Nucleic Acids Res. 2014, 42, D531–D536. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed]
- Yen, S.J.; Lee, Y.S. Under-sampling approaches for improving prediction of the minority class in an imbalanced dataset. Lect. Notes Control Inf. 2006, 344, 731–740. [Google Scholar]
- Chothia, C.; Levitt, M.; Richardson, D. Structure of proteins: Packing of alpha-helices and pleated sheets. Proc. Natl. Acad. Sci. USA 1977, 74, 4130–4134. [Google Scholar] [CrossRef]
- Heffernan, R.; Paliwal, K.; Lyons, J.; Dehzangi, A.; Sharma, A.; Wang, J.; Sattar, A.; Yang, Y.; Zhou, Y. Improving prediction of secondary structure, local backbone angles, and solvent accessible surface area of proteins by iterative deep learning. Sci. Rep. 2015, 5, 11476. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Shi, X.-H.; Wang, P.; He, Z.; Feng, K.-Y.; Hu, L.; Kong, X.; Li, Y.-X.; Cai, Y.-D.; Chou, K.-C. Analysis and prediction of the metabolic stability of proteins based on their sequential features, subcellular locations and interaction networks. PLoS ONE 2010, 5, e10972. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Feng, K.-Y.; Cai, Y.-D.; Chou, K.-C.; Li, H.-P. Predicting the network of substrate-enzyme-product triads by combining compound similarity and functional domain composition. BMC Bioinform. 2010, 11, 293. [Google Scholar] [CrossRef] [PubMed]
- Dehzangi, A.; Paliwal, K.; Lyons, J.; Sharma, A.; Sattar, A. A segmentation-based method to extract structural and evolutionary features for protein fold recognition. IEEE/ACM Trans. Comput. Biol. Bioinform. 2014, 11, 510–519. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Yang, J.; Song, J.-N.; Chou, K.-C.; Shen, H.-B. Improving the accuracy of predicting disulfide connectivity by feature selection. J. Comput. Chem. 2010, 31, 1478–1485. [Google Scholar] [CrossRef] [PubMed]
- Heffernan, R.; Dehzangi, A.; Lyons, J.; Paliwal, K.; Sharma, A.; Wang, J.; Sattar, A.; Zhou, Y.; Yang, Y. Highly accurate sequence-based prediction of half-sphere exposures of amino acid residues in proteins. Bioinformatics 2015, 32, 843–849. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Heffernan, R.; Paliwal, K.; Lyons, J.; Dehzangi, A.; Sharma, A.; Wang, J.; Sattar, A.; Zhou, Y. Spider2: A package to predict secondary structure, accessible surface area, and main-chain torsional angles by deep neural networks. In Prediction of Protein Secondary Structure; Springer: New York, NY, USA, 2017; pp. 55–63. [Google Scholar]
- Chakravarty, S.; Varadarajan, R. Residue depth: A novel parameter for the analysis of protein structure and stability. Structure 1999, 7, 723–732. [Google Scholar] [CrossRef]
- Pollastri, G.; Baldi, P.; Fariselli, P.; Casadio, R. Prediction of coordination number and relative solvent accessibility in proteins. Proteins: Struct. Funct. Genet. 2002, 47, 142–153. [Google Scholar] [CrossRef]
- Craveur, P.; Rebehmed, J.; de Brevern, A.G. Ptm-sd: A database of structurally resolved and annotated posttranslational modifications in proteins. Database 2014, 2014, bau041. [Google Scholar] [CrossRef]
- Liu, Z.; Xiao, X.; Qiu, W.-R.; Chou, K.-C. Idna-methyl: Identifying dna methylation sites via pseudo trinucleotide composition. Anal. Biochem. 2015, 474, 69–77. [Google Scholar] [CrossRef]
- Chen, W.; Feng, P.; Ding, H.; Lin, H.; Chou, K.-C. Irna-methyl: Identifying n6-methyladenosine sites using pseudo nucleotide composition. Anal. Biochem. 2015, 490, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Lopez, Y.; Sharma, A.; Dehzangi, A.; Lal, S.P.; Taherzadeh, G.; Sattar, A.; Tsunoda, T. Success: Evolutionary and structural properties of amino acids prove effective for succinylation site prediction. BMC Genom. 2018, 19, 923. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Liu, Z.; Xiao, X.; Liu, B.; Chou, K.-C. Isuc-pseopt: Identifying lysine succinylation sites in proteins by incorporating sequence-coupling effects into pseudo components and optimizing imbalanced training dataset. Anal. Biochem. 2016, 497, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Dehzangi, A.; Heffernan, R.; Sharma, A.; Lyons, J.; Paliwal, K.; Sattar, A. Gram-positive and gram-negative protein subcellular localization by incorporating evolutionary-based descriptors into chou׳s general pseaac. J. Theor. Biol. 2015, 364, 284–294. [Google Scholar] [CrossRef] [PubMed]
- Paliwal, K.K.; Sharma, A.; Lyons, J.; Dehzangi, A. A tri-gram based feature extraction technique using linear probabilities of position specific scoring matrix for protein fold recognition. IEEE Trans. NanoBiosci. 2014, 13, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Dehzangi, A.; Sohrabi, S.; Heffernan, R.; Sharma, A.; Lyons, J.; Paliwal, K.; Sattar, A. Gram-positive and gram-negative subcellular localization using rotation forest and physicochemical-based features. BMC Bioinform. 2015, 16, S1. [Google Scholar] [CrossRef]
- Dehzangi, A.; Paliwal, K.; Sharma, A.; Lyons, J.; Sattar, A. Protein fold recognition using an overlapping segmentation approach and a mixture of feature extraction models. Adv. Artif. Intell. 2013, 32–43. [Google Scholar] [CrossRef]
- Nanni, L.; Brahnam, S.; Lumini, A. Prediction of protein structure classes by incorporating different protein descriptors into general chou’s pseudo amino acid composition. J. Theor. Biol. 2014, 360, 109–116. [Google Scholar] [CrossRef]
- Wei, L.; Liao, M.; Gao, X.; Zou, Q. Enhanced Protein Fold Prediction Method Through a Novel Feature Extraction Technique. IEEE Trans. NanoBiosci. 2015, 14, 649–659. [Google Scholar] [CrossRef]
- Hayat, M.; Tahir, M.; Khan, S.A. Prediction of protein structure classes using hybrid space of multi-profile bayes and bi-gram probability feature spaces. J. Theor. Biol. 2014, 346, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Zakeri, P.; Jeuris, B.; Vandebril, R.; Moreau, Y. Protein fold recognition using geometric kernel data fusion. Bioinformatics 2014, 30, 1850–1857. [Google Scholar] [CrossRef] [PubMed]
- Bbeiman, L. Bagging Predictors. Mach. Learn. 1996, 24, 123–140. [Google Scholar] [CrossRef]
- Dietterich, T.G. Ensemble methods in machine learning. In International Workshop on Multiple Classifier Systems: 2000; Springer: Berlin, Germany, 2000; pp. 1–15. [Google Scholar]
- Quinlan, J.R. C4.5: Programs for Machine Learning; Elsevier: San Mateo, CA, USA, 2014. [Google Scholar]
- Dehzangi, A.; Paliwal, K.; Sharma, A.; Dehzangi, O.; Sattar, A. A combination of feature extraction methods with an ensemble of different classifiers for protein structural class prediction problem. IEEE/ACM Trans. Comput. Biol. Bioinform. 2013, 10, 564–575. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.C. An Unprecedented Revolution in Medicinal Chemistry Driven by the Progress of Biological Science. Curr. Top. Med. Chem. 2017, 17, 2337–2358. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.-C.; Shen, H.-B. Cell-ploc: A package of web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |
| Methods | Sensitivity | Specificity | Accuracy | MCC |
|---|---|---|---|---|
| pSumo-CD | 0.536 | 0.921 | 72.8% | 0.494 |
| C-Validation 6 | 0.910 | 0.959 | 93.5% | 0.873 |
| C-Validation 8 | 0.907 | 0.963 | 93.4% | 0.872 |
| C-Validation 10 | 0.910 | 0.967 | 93.8% | 0.880 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dehzangi, A.; López, Y.; Taherzadeh, G.; Sharma, A.; Tsunoda, T. SumSec: Accurate Prediction of Sumoylation Sites Using Predicted Secondary Structure. Molecules 2018, 23, 3260. https://doi.org/10.3390/molecules23123260
Dehzangi A, López Y, Taherzadeh G, Sharma A, Tsunoda T. SumSec: Accurate Prediction of Sumoylation Sites Using Predicted Secondary Structure. Molecules. 2018; 23(12):3260. https://doi.org/10.3390/molecules23123260
Chicago/Turabian StyleDehzangi, Abdollah, Yosvany López, Ghazaleh Taherzadeh, Alok Sharma, and Tatsuhiko Tsunoda. 2018. "SumSec: Accurate Prediction of Sumoylation Sites Using Predicted Secondary Structure" Molecules 23, no. 12: 3260. https://doi.org/10.3390/molecules23123260
APA StyleDehzangi, A., López, Y., Taherzadeh, G., Sharma, A., & Tsunoda, T. (2018). SumSec: Accurate Prediction of Sumoylation Sites Using Predicted Secondary Structure. Molecules, 23(12), 3260. https://doi.org/10.3390/molecules23123260





