Semisynthesis and Inhibitory Effects of Solidagenone Derivatives on TLR-Mediated Inflammatory Responses
Abstract
1. Introduction
2. Results and Discussion
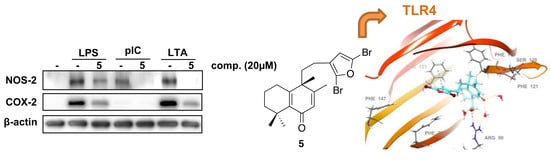
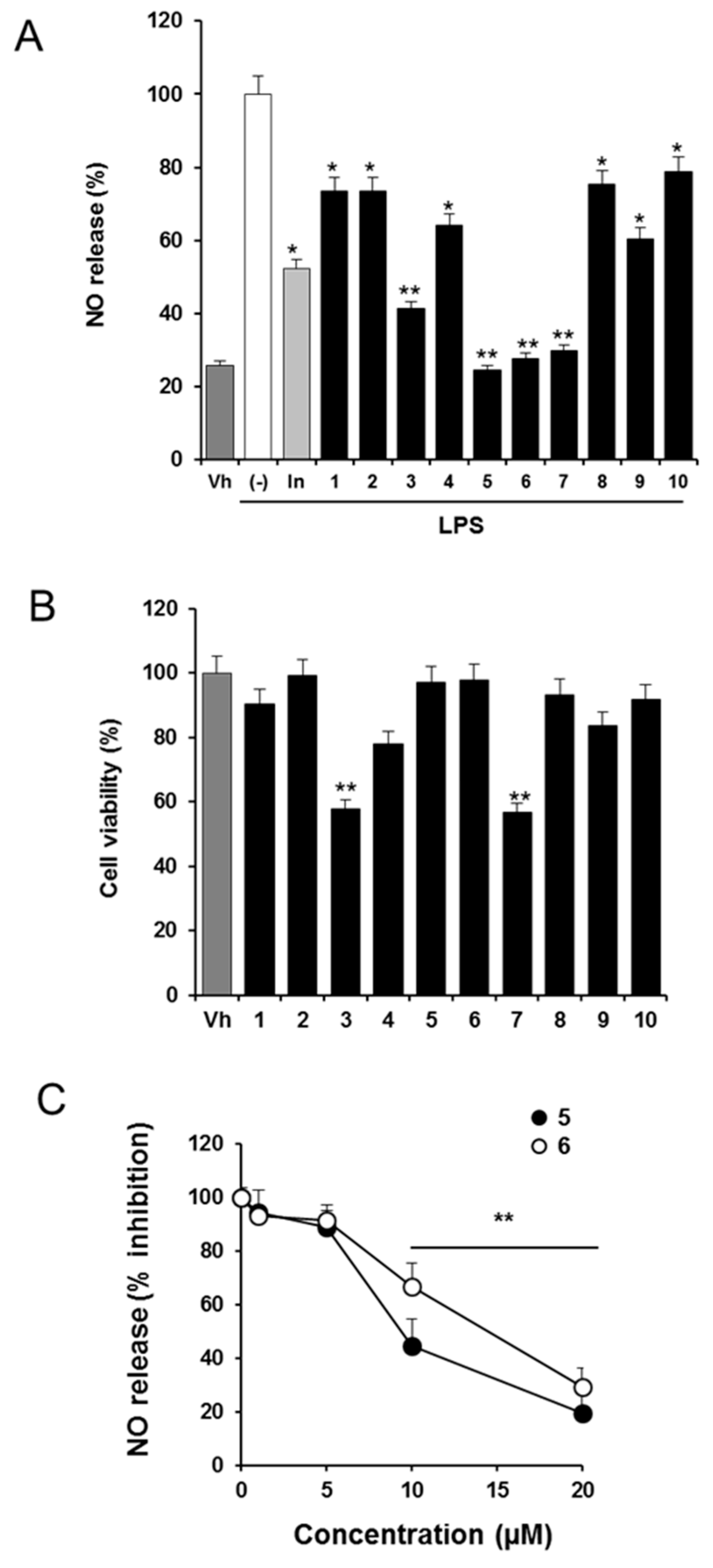
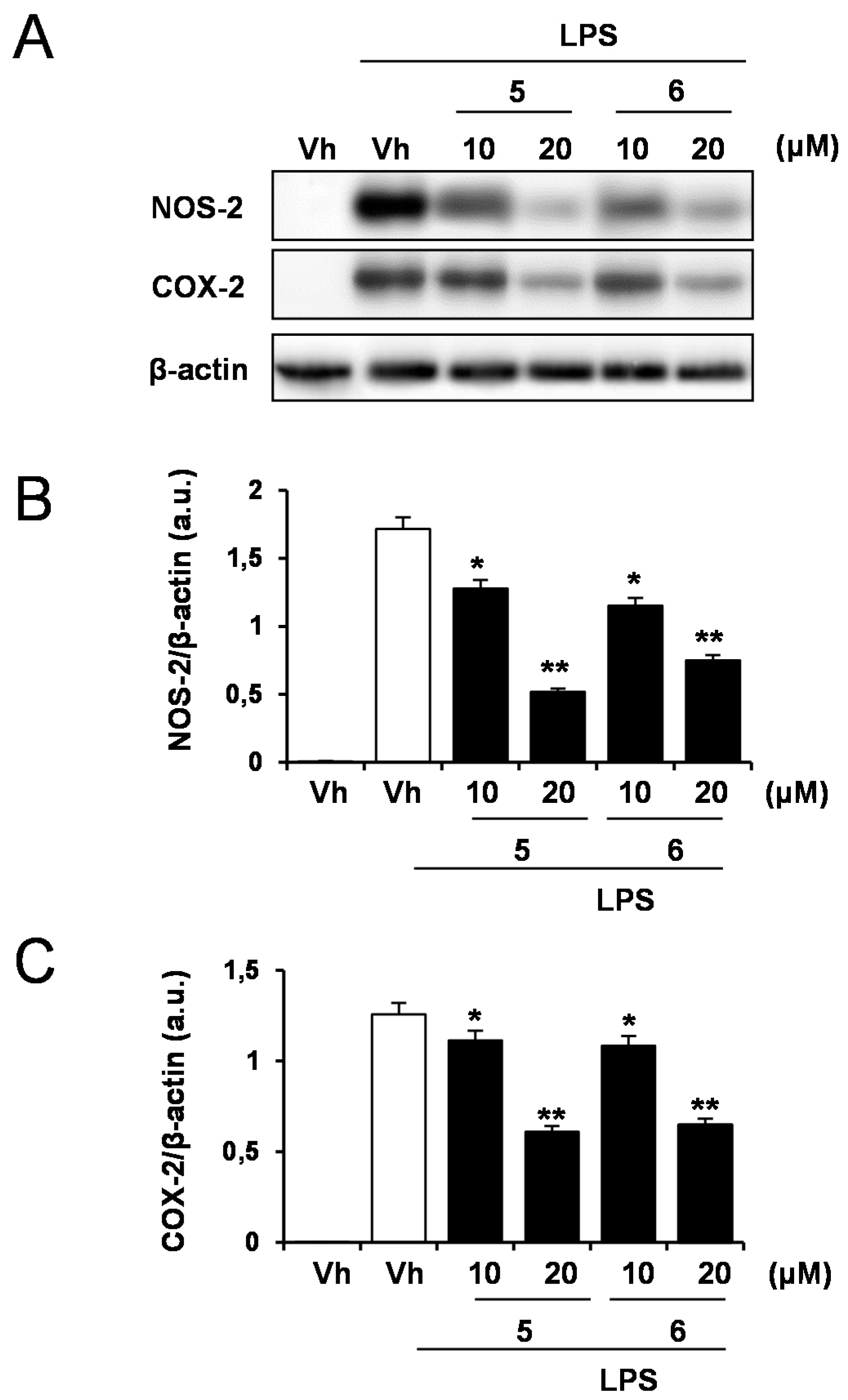
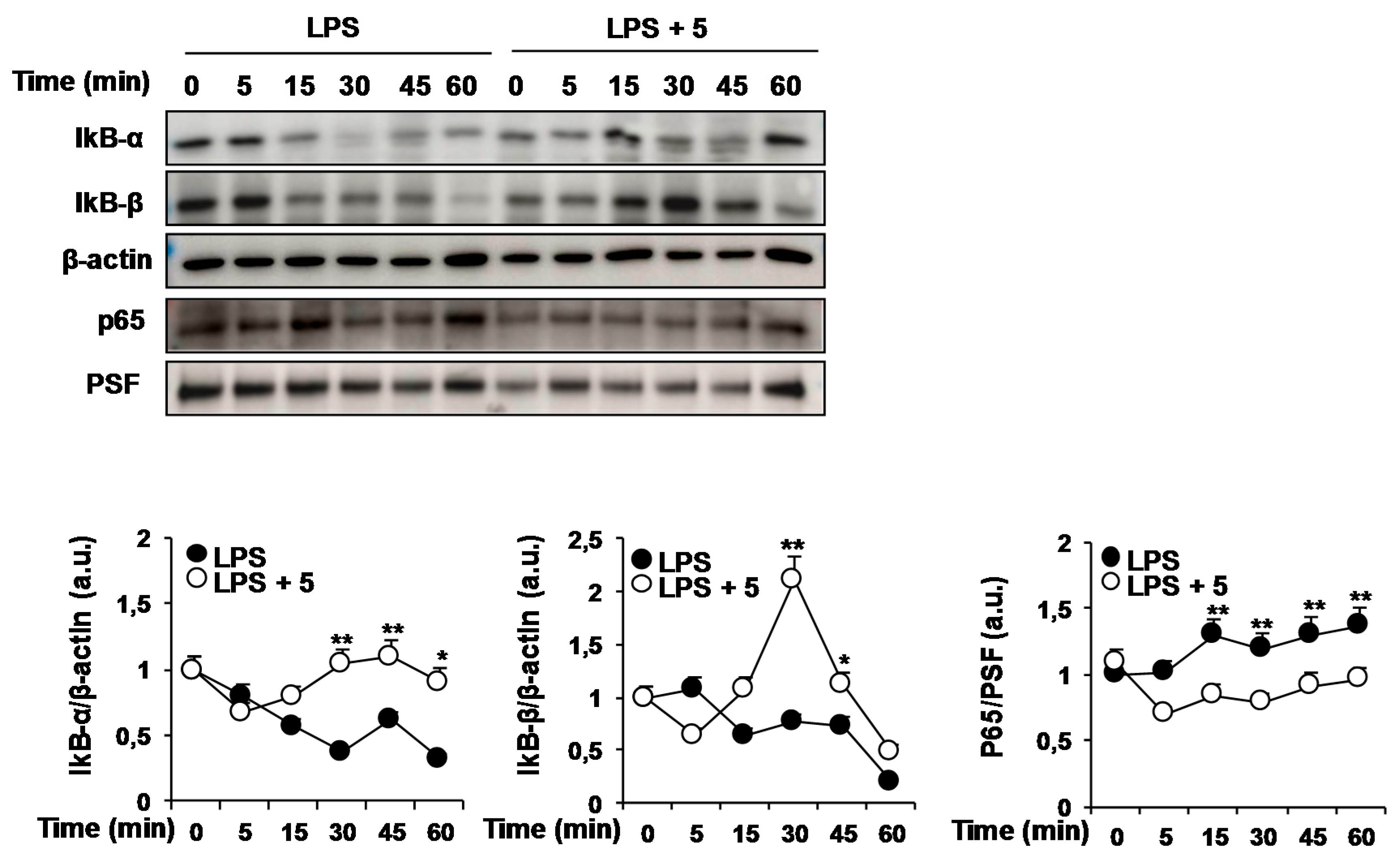
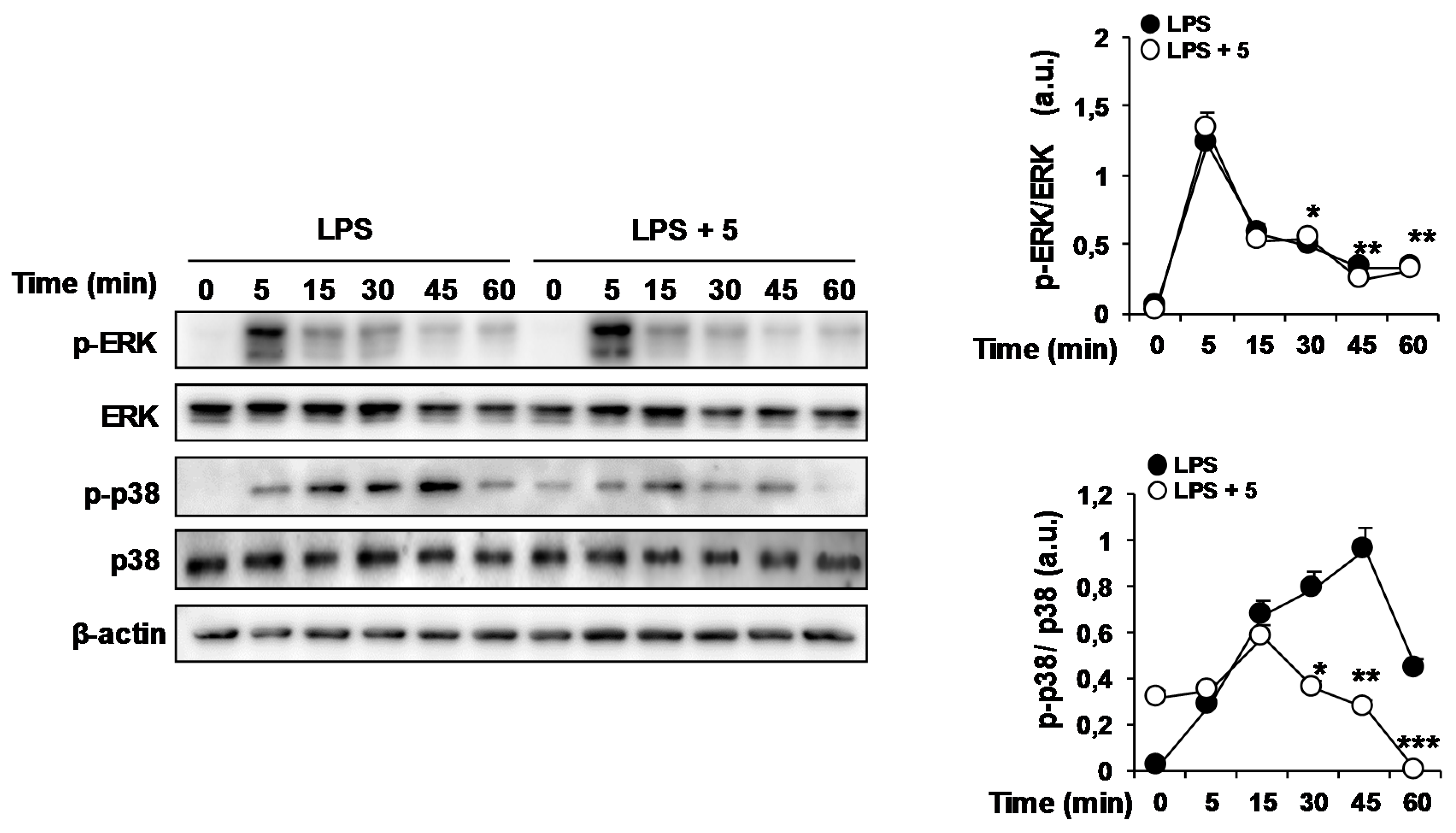
2.1. Effects of Solidagenone Derivatives on Inflammatory Responses
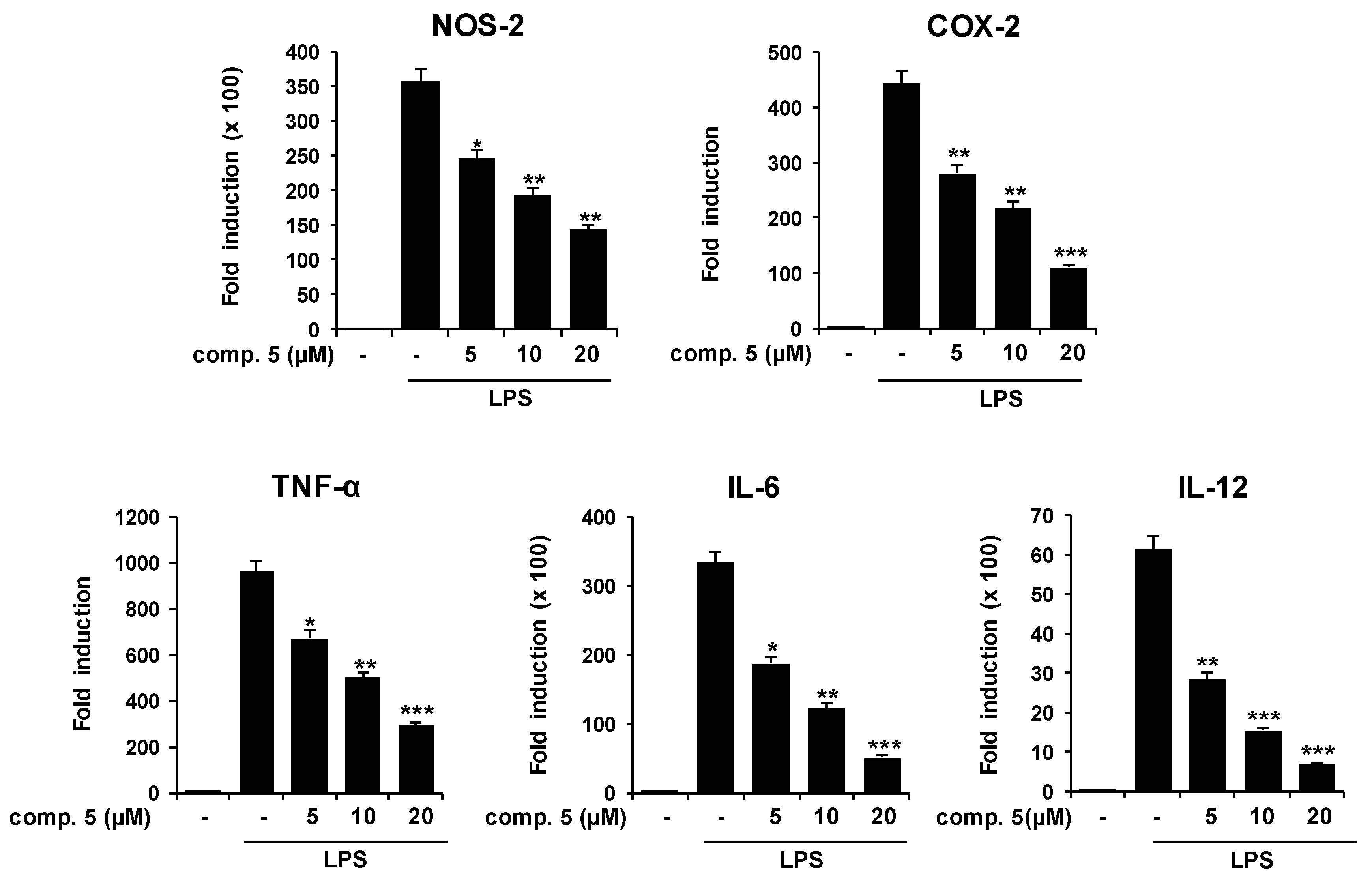
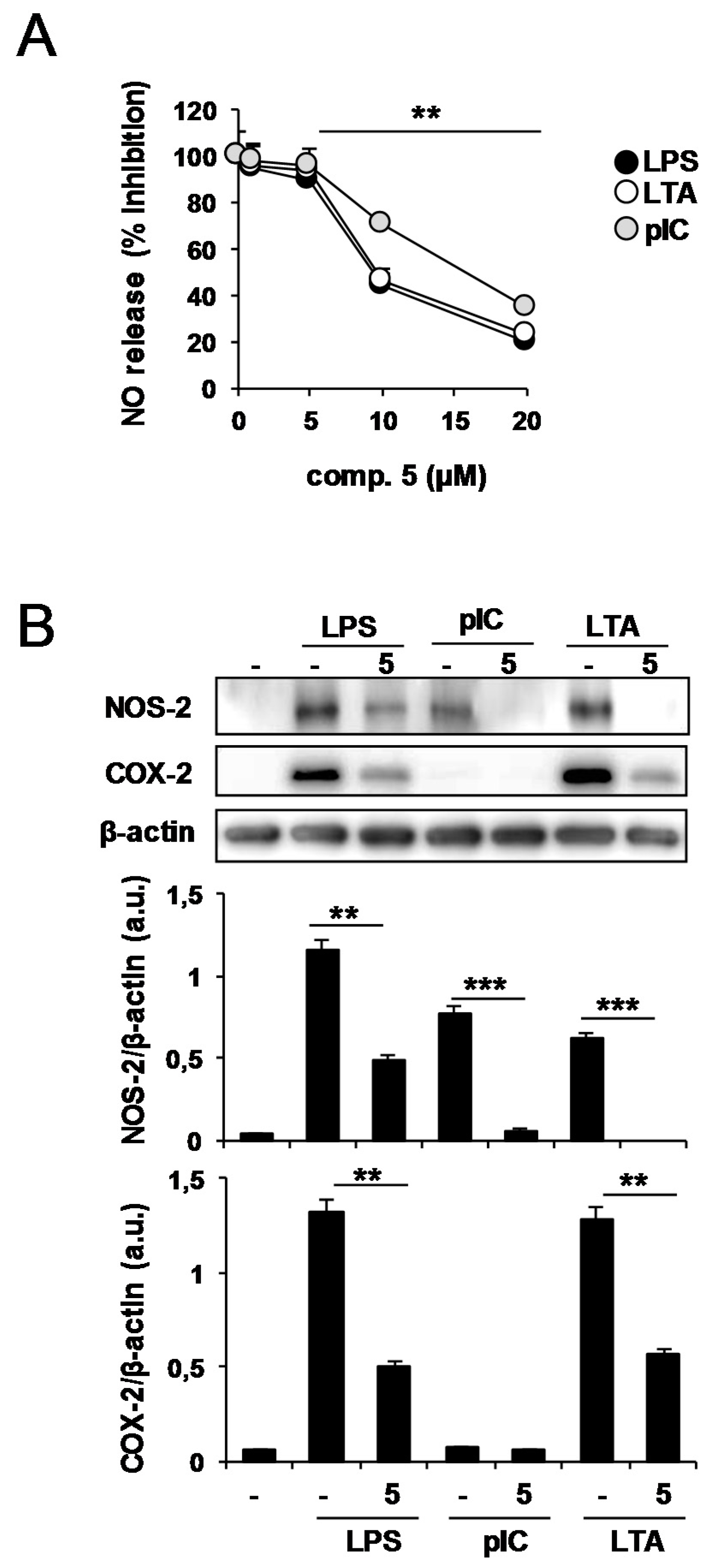
2.2. Effects of Compound 5 in Response to Different TLR Ligands
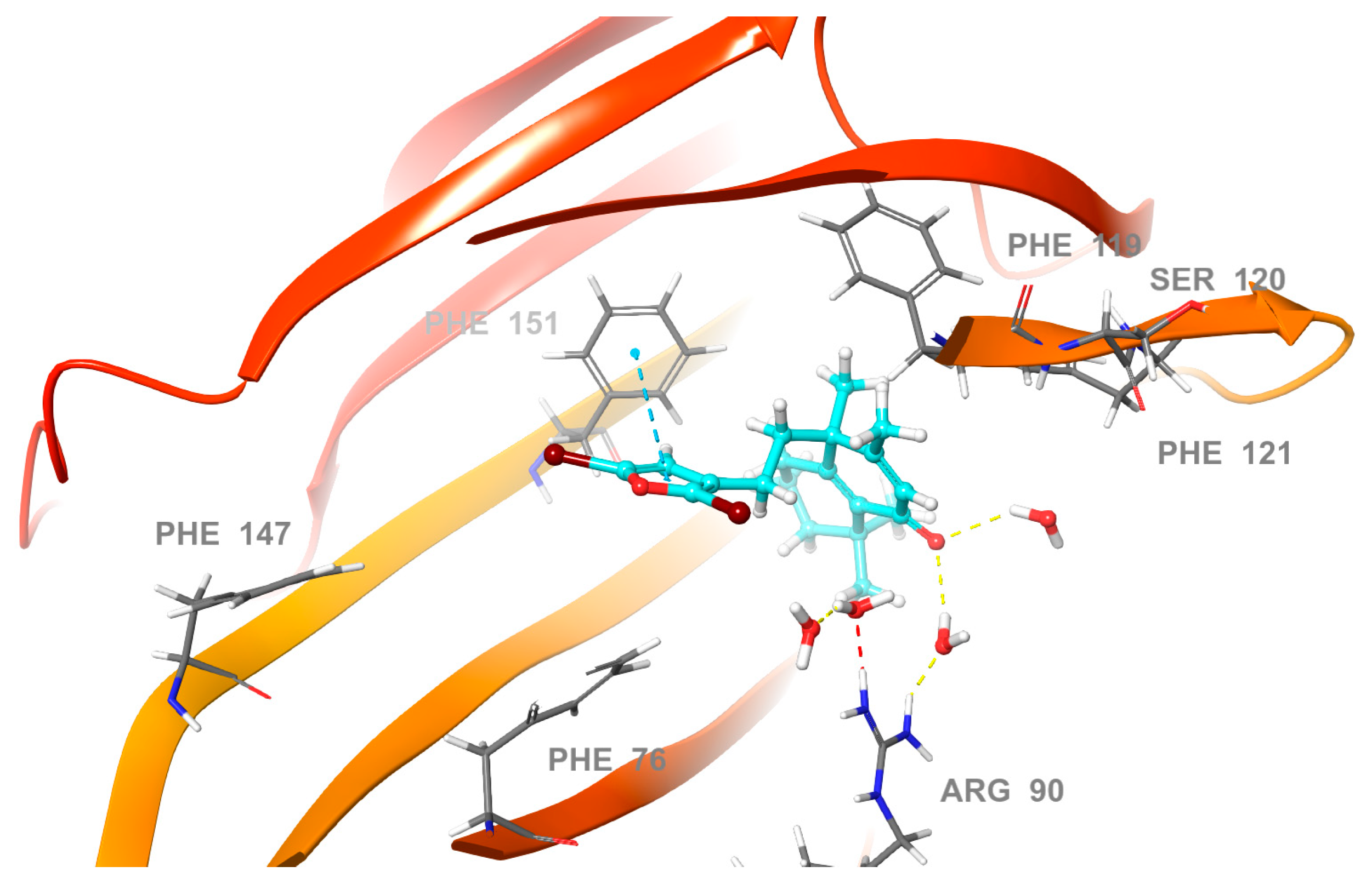
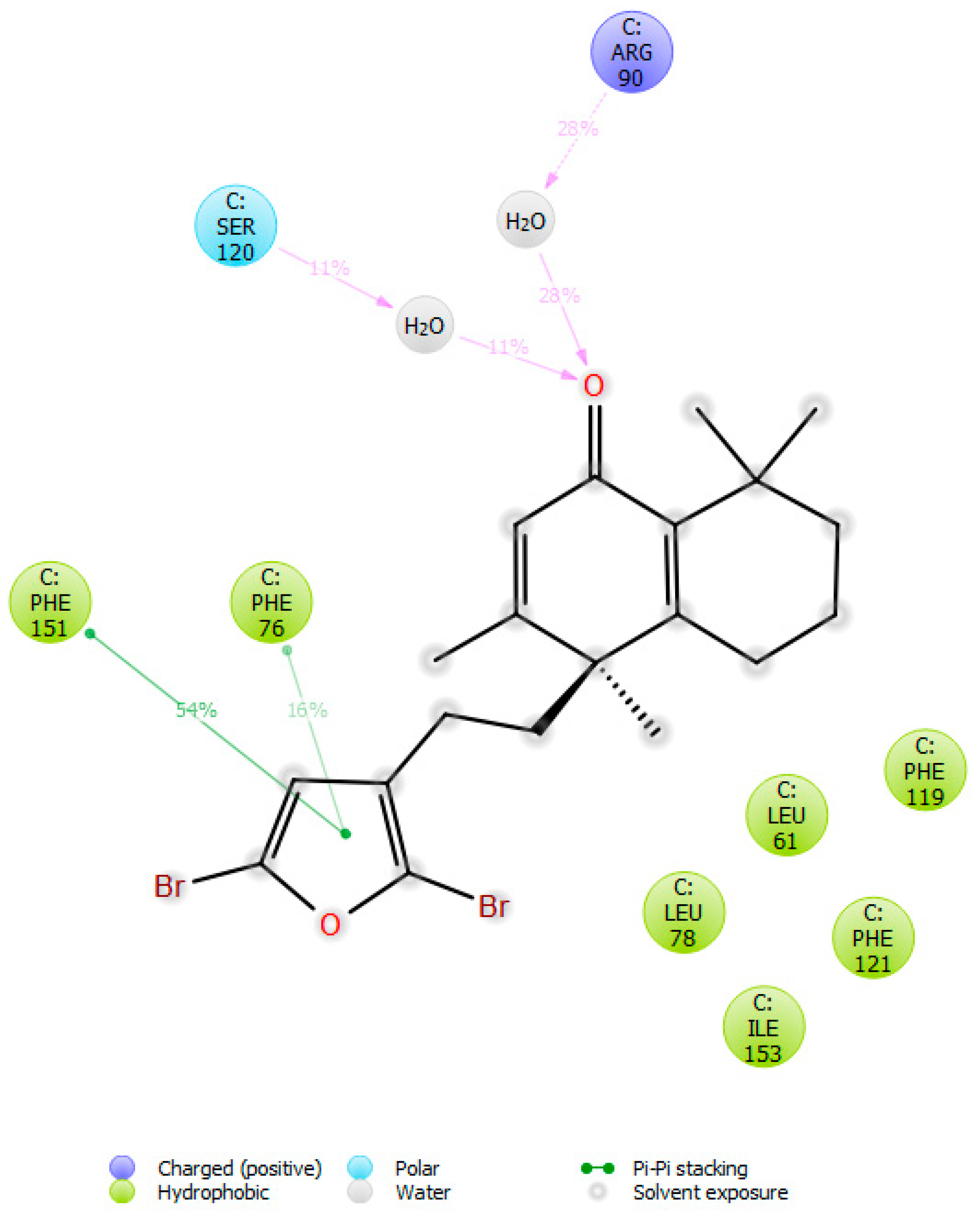
2.3. Molecular Docking Studies
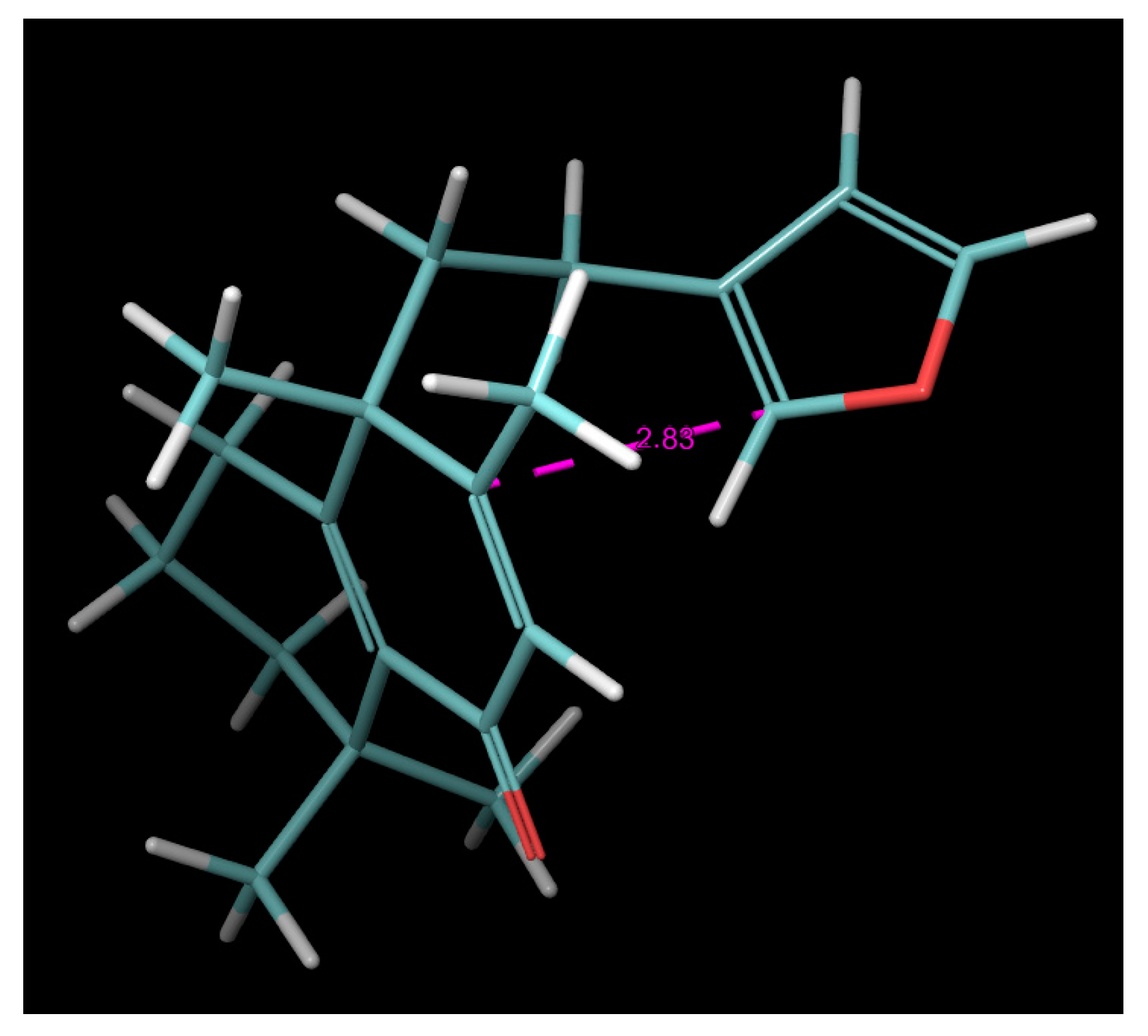
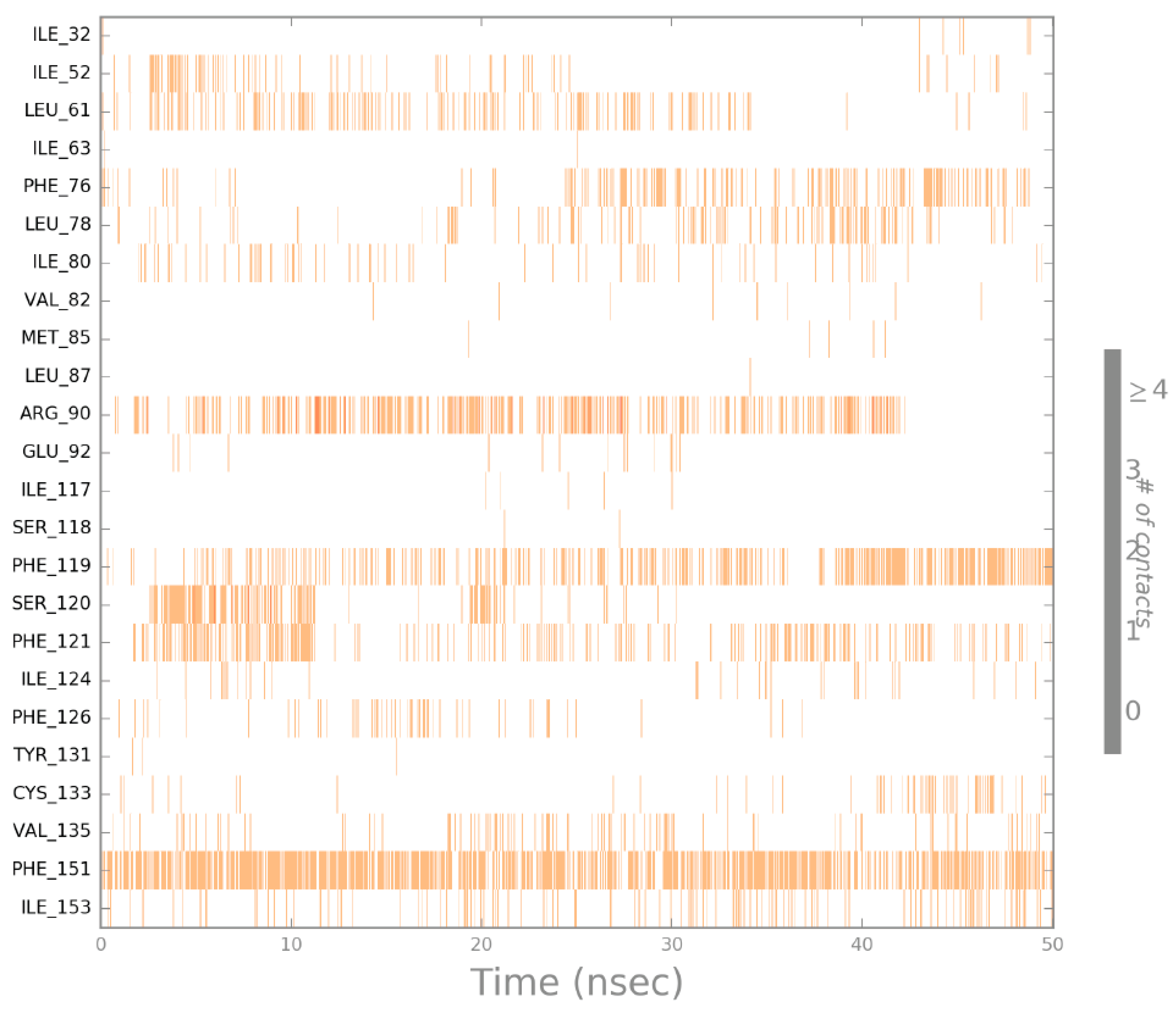
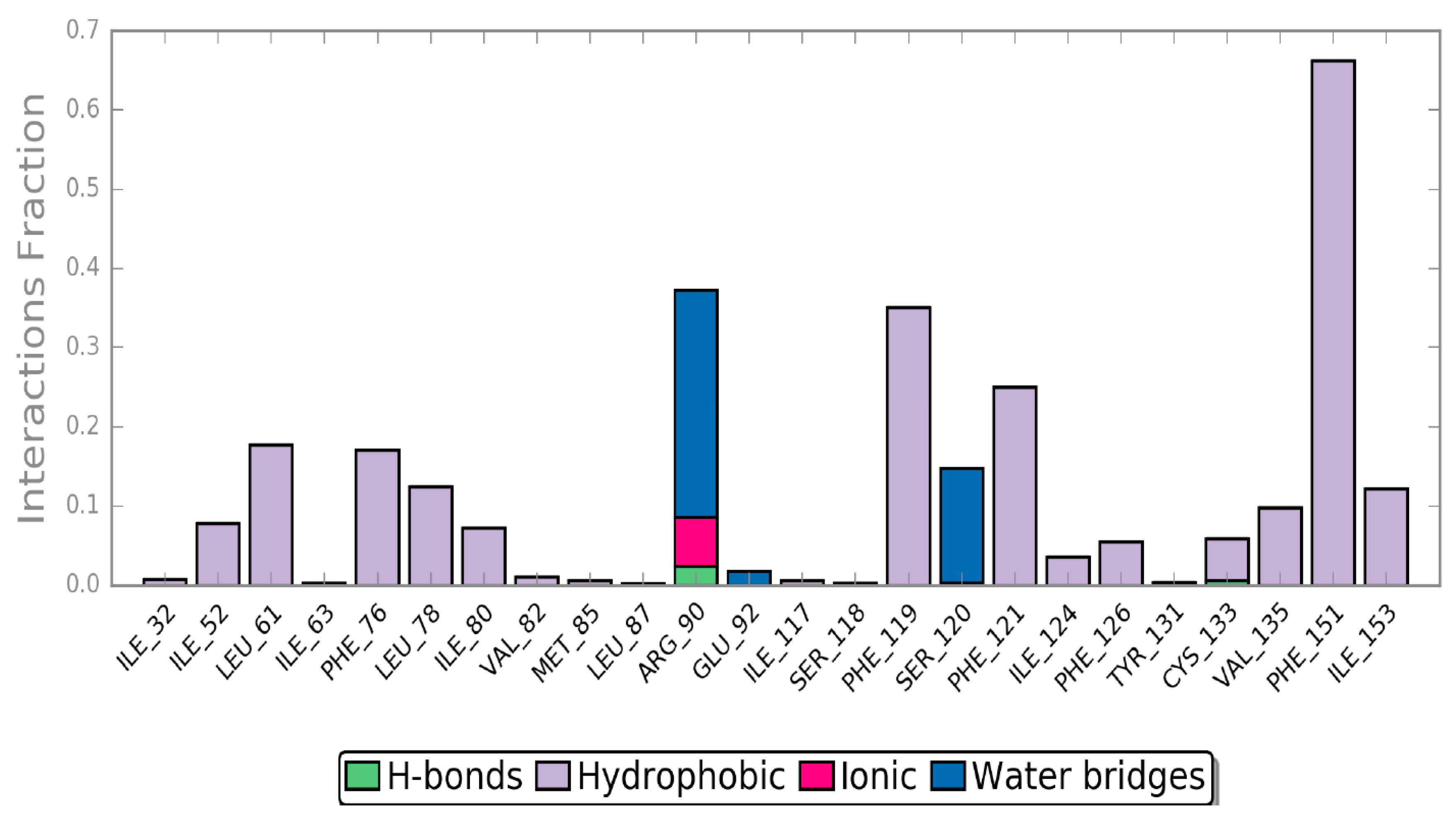
2.4. Molecular Dynamics (MD) Simulation
3. Materials and Methods
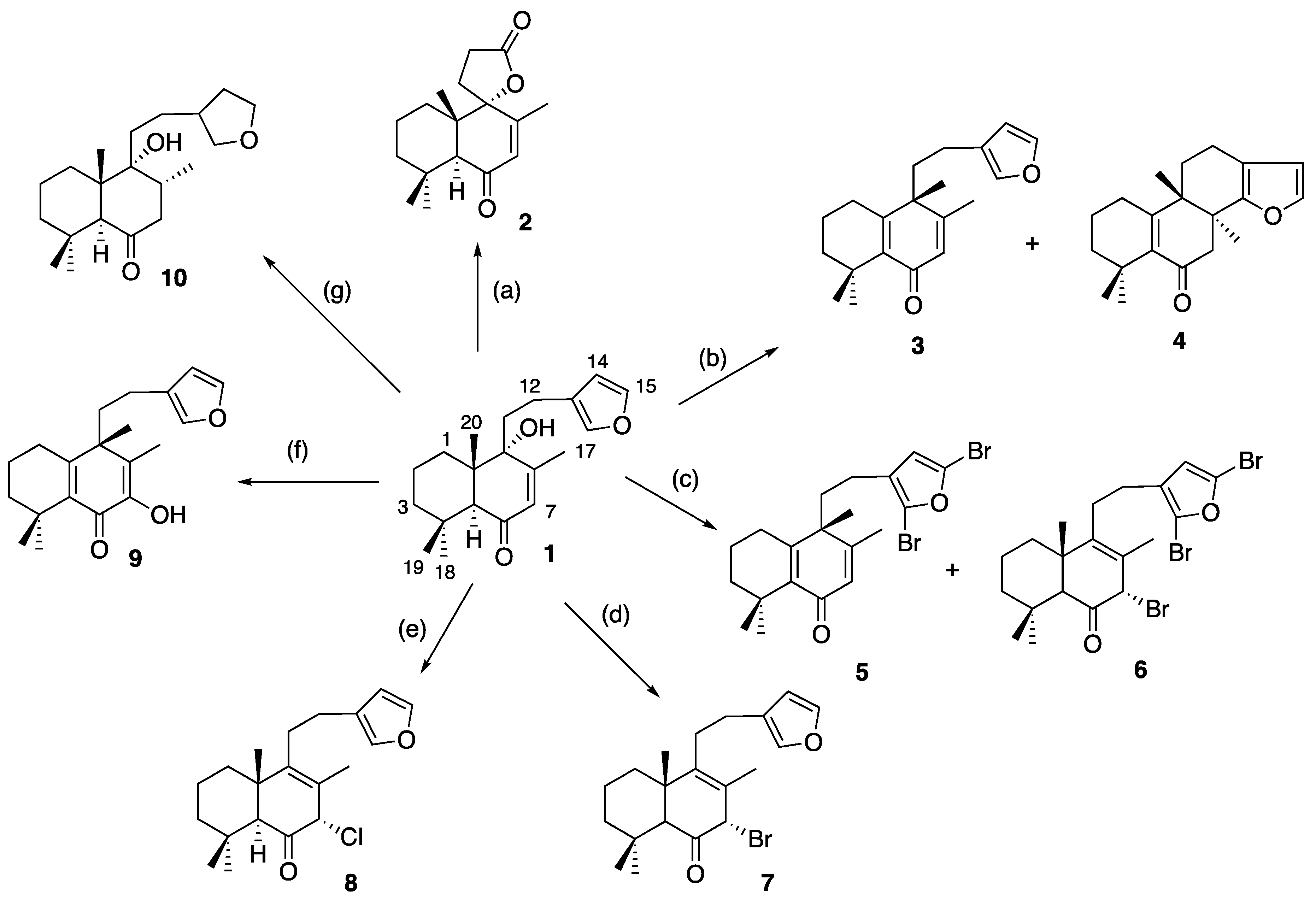
3.1. Chemistry
3.1.1. General Methods
3.1.2. Solidagenone (1)
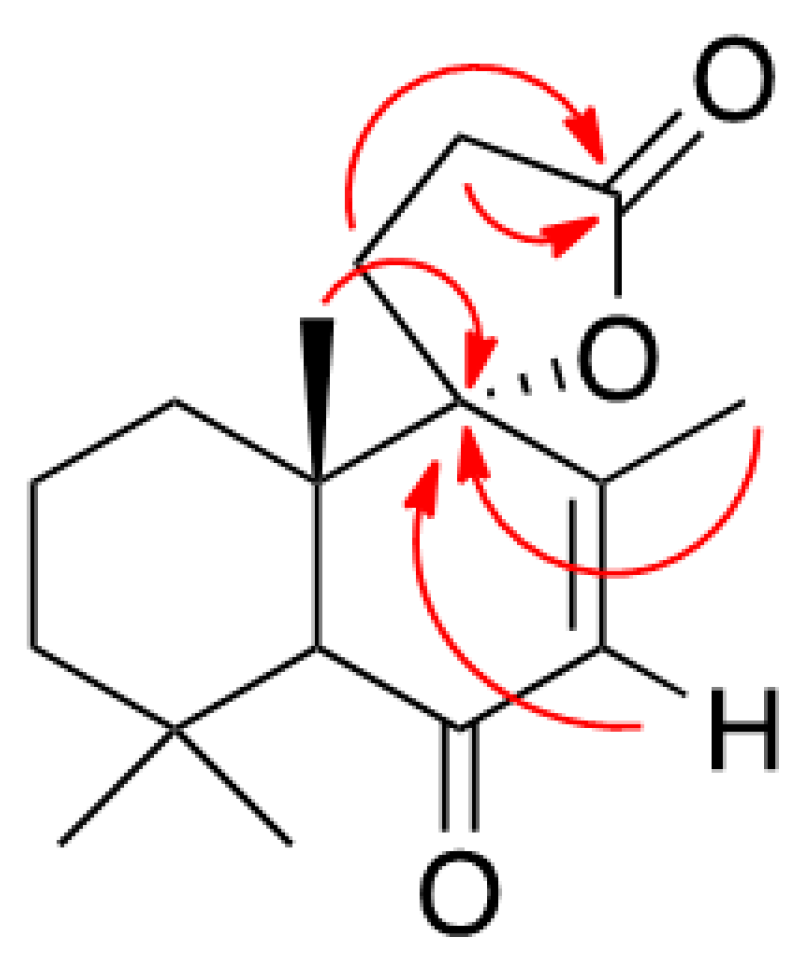
3.1.3. Preparation of Solidagenone Derivative (2)
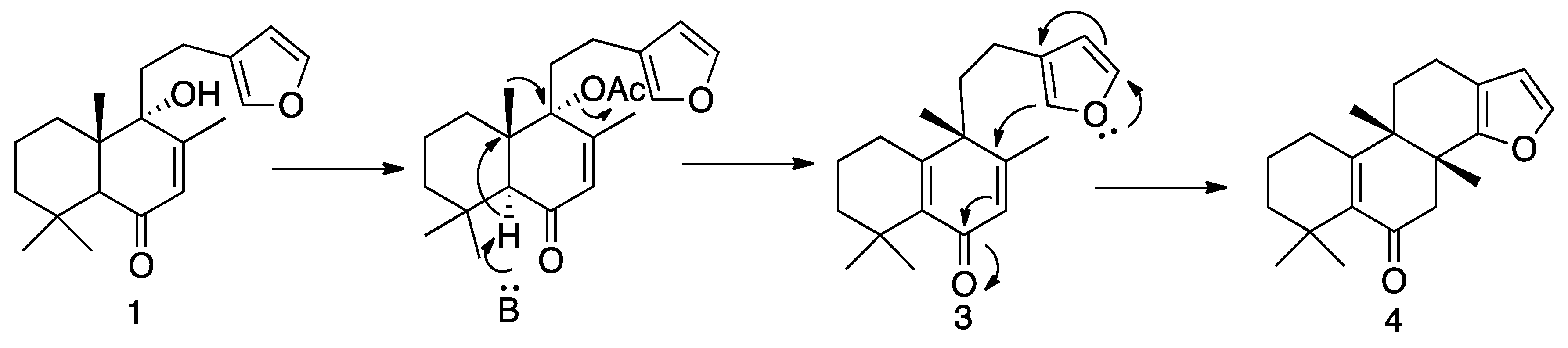
3.1.4. Preparation of Solidagenone Derivatives (3) and (4)
3.1.5. Preparation of Solidagenone Derivatives (5) and (6)
3.1.6. Preparation of Solidagenone Derivative (7)
3.1.7. Preparation of Solidagenone Derivative (8)
3.1.8. Preparation of Solidagenone Derivative (9)
3.1.9. Preparation of Solidagenone Derivative (10)
3.2. Biology
3.2.1. Cell Culture Conditions
3.2.2. Determination of NO Production and Cell Viability
3.2.3. Western Blot Analysis
3.2.4. RNA Analysis and Quantitative PCR
3.2.5. Protein Preparation and Docking
3.2.6. Molecular Dynamics Simulation
3.2.7. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| NO | Nitric oxide |
| COX-2 | Cyclooxygenase-2 |
| NOS-2 | Nitric oxide synthase-2 |
| LPS | Lipopolysaccharide |
| TLRs | Toll-like receptors |
| NF-κB | Nuclear transcription factor-κB |
| MAPKs | Mitogen-activated protein kinases |
References
- Weiss, U. Inflammation. Nature 2008, 454, 427. [Google Scholar] [CrossRef]
- Achek, A.; Yesudhas, D.; Choi, S. Toll-like receptors: Promising therapeutic targets for inflammatory diseases. Arch. Pharm. Res. 2016, 39, 1032–1049. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.; Martinez, F.O. Alternative activation of macrophages: Mechanism and functions. Immunity 2010, 32, 593–604. [Google Scholar] [CrossRef]
- Nathan, C.; Ding, A. Nonresolving inflammation. Cell 2010, 140, 871–882. [Google Scholar] [CrossRef] [PubMed]
- Garantziotis, S.; Hollingsworth, J.W.; Zaas, A.K.; Schwartz, D.A. The effect of toll-like receptors and toll-like receptor genetics in human disease. Annu. Rev. Med. 2008, 59, 343–359. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Srivastava, M.; Saqib, U.; Liu, D.; Faisal, S.M.; Sugathan, S.; Bishnoi, S.; Baig, M.S. Potential therapeutic targets for inflammation in toll-like receptor 4 (TLR4)-mediated signaling pathways. Int. Immunopharmacol. 2016, 40, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Doyle, S.L.; O’Neill, L.A. Toll-like receptors: From the discovery of NFkappaB to new insights into transcriptional regulations in innate immunity. Biochem. Pharmacol. 2006, 72, 1102–1113. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, L.A.; Golenbock, D.; Bowie, A.G. The history of Toll-like receptors—redefining innate immunity. Nat. Rev. Immunol. 2013, 13, 453–460. [Google Scholar] [CrossRef] [PubMed]
- de las Heras, B.; Hortelano, S. Molecular basis of the anti-inflammatory effects of terpenoids. Inflamm. Allergy Drug Targets 2009, 8, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Chinou, I. Labdanes of natural origin-biological activities (1981-2004). Curr. Med. Chem. 2005, 12, 1295–1317. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, Y.; Torres-Mendoza, D.; Jones, G.E.; Fernandez, P.L. Marine diterpenoids as potential anti-inflammatory agents. Mediators Inflamm. 2015, 2015, 263543. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.C.; Chan, T.K.; Ng, D.S.; Sagineedu, S.R.; Stanslas, J.; Wong, W.S. Andrographolide and its analogues: Versatile bioactive molecules for combating inflammation and cancer. Clin. Exp. Pharmacol. Physiol. 2012, 39, 300–310. [Google Scholar] [CrossRef] [PubMed]
- Tran, Q.T.N.; Wong, W.S.F.; Chai, C.L.L. Labdane diterpenoids as potential anti-inflammatory agents. Pharmacol. Res. 2017, 124, 43–63. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.S.D.; Liao, W.; Zhou, S.; Wong, W.S.F. Is there a future for andrographolide to be an anti-inflammatory drug? Deciphering its major mechanisms of action. Biochem. Pharmacol. 2017, 139, 71–81. [Google Scholar] [CrossRef]
- Razmilic, I.; Schmeda-Hirschmann, G. Activity of solidagenone and their semisynthetic derivatives on the glucocorticoid-mediated signal transduction. Planta Med. 2000, 66, 86–88. [Google Scholar] [CrossRef] [PubMed]
- Karaman, R. Proximity vs. strain in intramolecular ring-closing reactions. Mol. Phys. 2010, 108, 1723–1730. [Google Scholar] [CrossRef]
- Lee, K.C.; Chang, H.H.; Chung, Y.H.; Lee, T.Y. Andrographolide acts as an anti-inflammatory agent in LPS-stimulated RAW264.7 macrophages by inhibiting STAT3-mediated suppression of the NF-kappaB pathway. J. Ethnopharmacol. 2011, 135, 678–684. [Google Scholar] [CrossRef]
- Lee, J.H.; Koo, T.H.; Hwang, B.Y.; Lee, J.J. Kaurane diterpene, kamebakaurin, inhibits NF-kappa B by directly targeting the DNA-binding activity of p50 and blocks the expression of antiapoptotic NF-kappa B target genes. J. Biol. Chem. 2002, 277, 18411–18420. [Google Scholar] [CrossRef]
- Zhu, T.; Wang, D.X.; Zhang, W.; Liao, X.Q.; Guan, X.; Bo, H.; Sun, J.Y.; Huang, N.W.; He, J.; Zhang, Y.K.; et al. Andrographolide protects against LPS-induced acute lung injury by inactivation of NF-kappaB. PLoS ONE 2013, 8, e56407. [Google Scholar]
- Kawai, T.; Akira, S. The role of pattern-recognition receptors in innate immunity: Update on toll-like receptors. Nat. Immunol. 2010, 11, 373–384. [Google Scholar] [CrossRef]
- Janssen-Heininger, Y.M.; Poynter, M.E.; Baeuerle, P.A. Recent advances towards understanding redox mechanisms in the activation of nuclear factor kappaB. Free Radic. Biol. Med. 2000, 28, 1317–1327. [Google Scholar] [CrossRef]
- Schulze-Osthoff, K.; Ferrari, D.; Riehemann, K.; Wesselborg, S. Regulation of NF-kappa B activation by MAP kinase cascades. Immunobiology 1997, 198, 35–49. [Google Scholar] [CrossRef]
- Barrat, F.J.; Coffman, R.L. Development of TLR inhibitors for the treatment of autoimmune diseases. Immunol. Rev. 2008, 223, 271–283. [Google Scholar] [CrossRef]
- Gao, W.; Xiong, Y.; Li, Q.; Yang, H. Inhibition of toll-like receptor signaling as a promising therapy for inflammatory diseases: A journey from molecular to nano therapeutics. Front. Physiol. 2017, 8, 508. [Google Scholar] [CrossRef]
- Hennessy, E.J.; Parker, A.E.; O’Neill, L.A. Targeting toll-like receptors: Emerging therapeutics? Nat. Rev. Drug Discov. 2010, 9, 293–307. [Google Scholar] [CrossRef]
- Schrodinger: Glide Software, Glide, Version 7; Ed LLC: New York, NY, USA, 2018.
- Perrin, D.D.; Amarego, W.L.F. Purification of Laboratory Chemicals; Pergamon Press: Oxford, UK, 1988. [Google Scholar]
- Hirschmann, G.S. A labdan diterpene from Solidago chilensis roots. Planta Med. 1988, 54, 179–180. [Google Scholar] [CrossRef]
- Zeini, M.; Traves, P.G.; Lopez-Fontal, R.; Pantoja, C.; Matheu, A.; Serrano, M.; Bosca, L.; Hortelano, S. Specific contribution of p19(ARF) to nitric oxide-dependent apoptosis. J. Immunol. 2006, 177, 3327–3336. [Google Scholar] [CrossRef]
- Giron, N.; Traves, P.G.; Rodriguez, B.; Lopez-Fontal, R.; Bosca, L.; Hortelano, S.; de las Heras, B. Suppression of inflammatory responses by labdane-type diterpenoids. Toxicol. Appl. Pharmacol. 2008, 228, 179–189. [Google Scholar] [CrossRef]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef]
- Eldridge, M.D.; Murray, C.W.; Auton, T.R.; Paolini, G.V.; Mee, R.P. Empirical scoring functions: I. The development of a fast empirical scoring function to estimate the binding affinity of ligands in receptor complexes. J. Comput. Aided Mol. Des. 1997, 11, 425–445. [Google Scholar] [CrossRef]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A new approach for rapid, accurate docking and scoring. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef]
- Banks, J.L.; Beard, H.S.; Cao, Y.; Cho, A.E.; Damm, W.; Farid, R.; Felts, A.K.; Halgren, T.A.; Mainz, D.T.; Maple, J.R.; et al. Integrated Modeling Program, Applied Chemical Theory (IMPACT). J. Comput. Chem. 2005, 26, 1752–1780. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Bowers, K.J.; Dror, R.O.; Shaw, D.E. The midpoint method for parallelization of particle simulations. J. Chem. Phys. 2006, 124, 184109. [Google Scholar] [CrossRef]
- Gibson, D.A.; Carter, E.A. Time-reversible multiple time-scale ab-initio molecular-dynamics. J. Phys. Chem. 1993, 97, 13429–13434. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cuadrado, I.; Amesty, Á.; Cedrón, J.C.; Oberti, J.C.; Estévez-Braun, A.; Hortelano, S.; De las Heras, B. Semisynthesis and Inhibitory Effects of Solidagenone Derivatives on TLR-Mediated Inflammatory Responses. Molecules 2018, 23, 3197. https://doi.org/10.3390/molecules23123197
Cuadrado I, Amesty Á, Cedrón JC, Oberti JC, Estévez-Braun A, Hortelano S, De las Heras B. Semisynthesis and Inhibitory Effects of Solidagenone Derivatives on TLR-Mediated Inflammatory Responses. Molecules. 2018; 23(12):3197. https://doi.org/10.3390/molecules23123197
Chicago/Turabian StyleCuadrado, Irene, Ángel Amesty, Juan Carlos Cedrón, Juan Carlos Oberti, Ana Estévez-Braun, Sonsoles Hortelano, and Beatriz De las Heras. 2018. "Semisynthesis and Inhibitory Effects of Solidagenone Derivatives on TLR-Mediated Inflammatory Responses" Molecules 23, no. 12: 3197. https://doi.org/10.3390/molecules23123197
APA StyleCuadrado, I., Amesty, Á., Cedrón, J. C., Oberti, J. C., Estévez-Braun, A., Hortelano, S., & De las Heras, B. (2018). Semisynthesis and Inhibitory Effects of Solidagenone Derivatives on TLR-Mediated Inflammatory Responses. Molecules, 23(12), 3197. https://doi.org/10.3390/molecules23123197