Broccoli Fluorets: Split Aptamers as a User-Friendly Fluorescent Toolkit for Dynamic RNA Nanotechnology
Abstract
1. Introduction
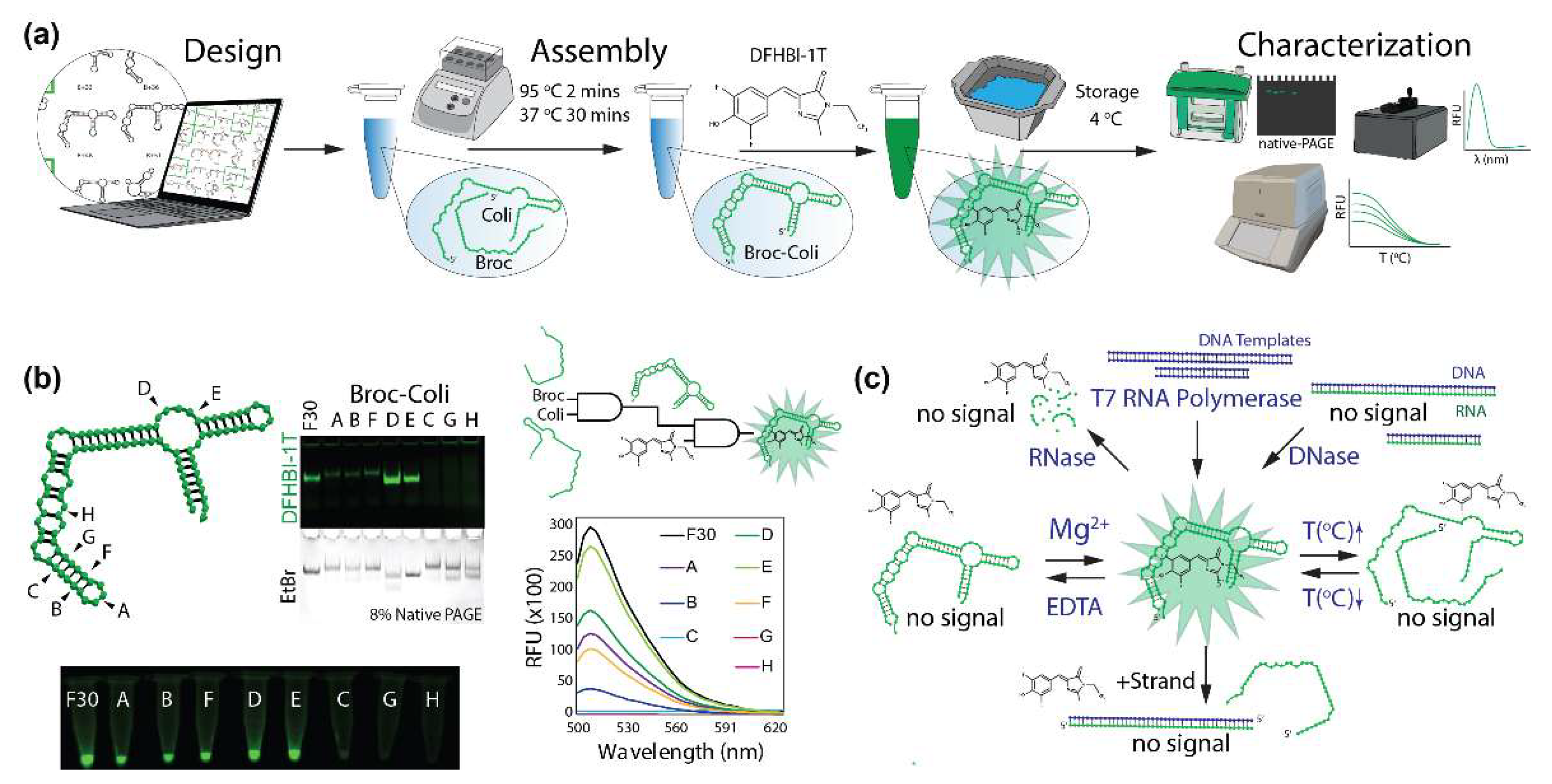
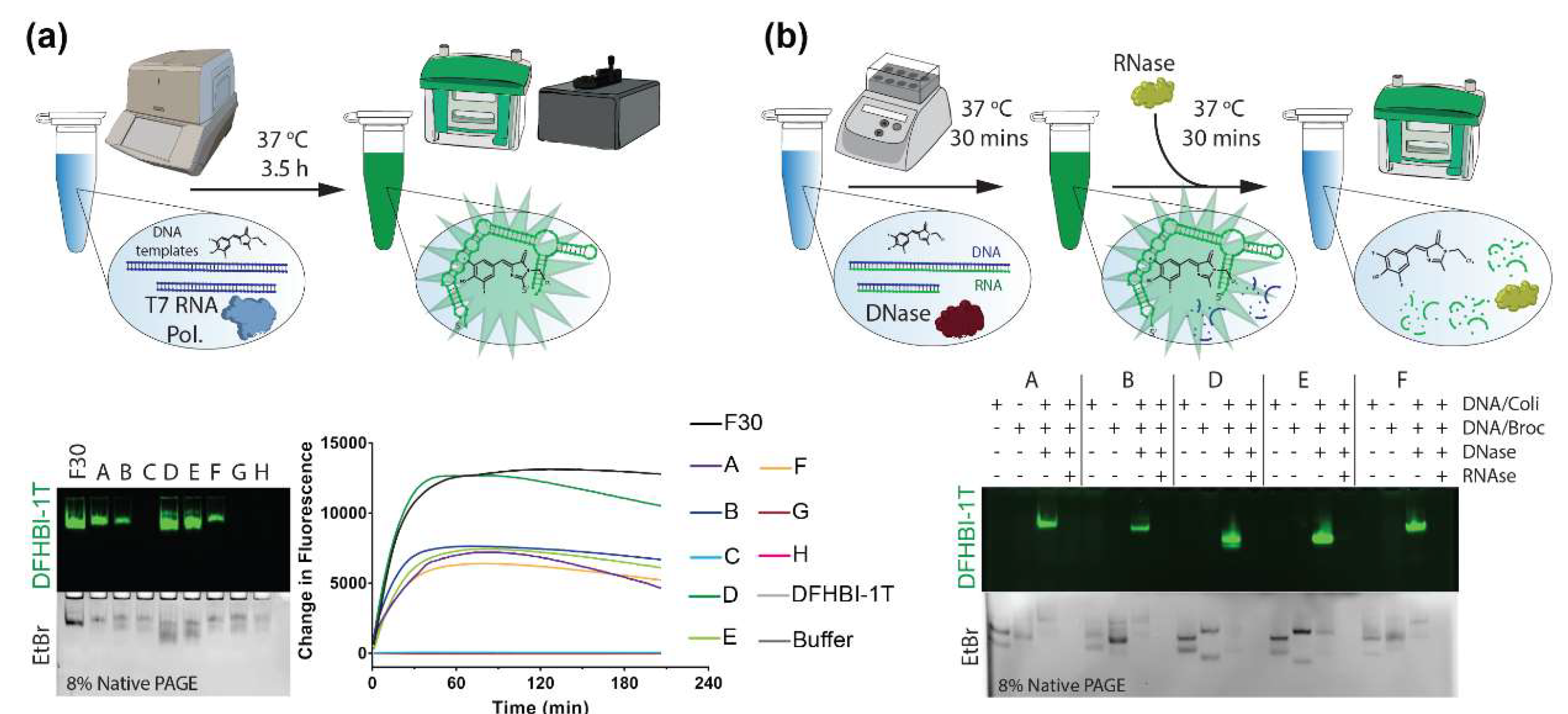
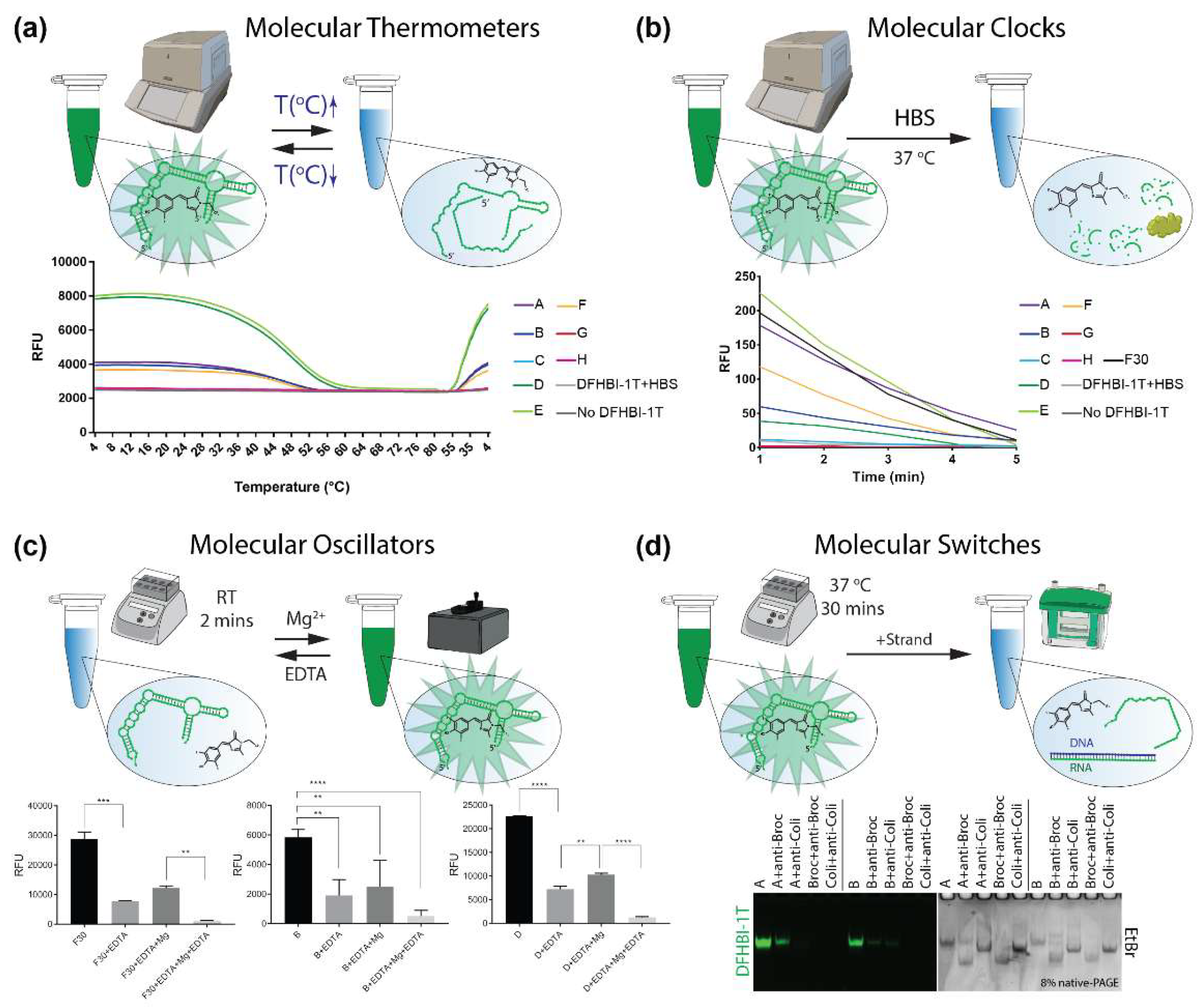
2. Results and Discussion
3. Materials and Methods
3.1. Design of Broccoli Fluorets
3.2. RNA Preparation
3.3. Broccoli Aptamer and Fluoret Assembly
3.4. Co-Transcriptional Assembly
3.5. Electrophoretic Mobility Shift Assays
3.6. EDTA Degradation and Mg2+ Formation
3.7. Nuclease-Driven Assembly/Degradation
3.8. Strand Displacement
3.9. Thermal Deactivation/Activation
3.10. Blood Stability
3.11. Statistics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shukla, G.C.; Haque, F.; Tor, Y.; Wilhelmsson, L.M.; Toulme, J.J.; Isambert, H.; Guo, P.; Rossi, J.J.; Tenenbaum, S.A.; Shapiro, B.A. A boost for the emerging field of RNA nanotechnology. ACS Nano 2011, 5, 3405–3418. [Google Scholar] [CrossRef] [PubMed]
- Afonin, K.A.; Kasprzak, W.K.; Bindewald, E.; Kireeva, M.; Viard, M.; Kashlev, M.; Shapiro, B.A. In silico design and enzymatic synthesis of functional RNA nanoparticles. Acc. Chem. Res. 2014, 47, 1731–1741. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, A.V.; Han, D.; Shih, W.M.; Yan, H. Challenges and opportunities for structural DNA nanotechnology. Nat. Nanotechnol. 2011, 6, 763–772. [Google Scholar] [CrossRef] [PubMed]
- Hong, E.; Halman, J.R.; Shah, A.B.; Khisamutdinov, E.F.; Dobrovolskaia, M.A.; Afonin, K.A. Structure and composition define immunorecognition of nucleic acid nanoparticles. Nano Lett. 2018, 18, 4309–4321. [Google Scholar] [CrossRef] [PubMed]
- Leontis, N.B.; Stombaugh, J.; Westhof, E. The non-watson-crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002, 30, 3497–3531. [Google Scholar] [CrossRef] [PubMed]
- Jasinski, D.; Haque, F.; Binzel, D.W.; Guo, P. Advancement of the emerging field of RNA nanotechnology. ACS Nano 2017, 11, 1142–1164. [Google Scholar] [CrossRef] [PubMed]
- Afonin, K.A.; Viard, M.; Koyfman, A.Y.; Martins, A.N.; Kasprzak, W.K.; Panigaj, M.; Desai, R.; Santhanam, A.; Grabow, W.W.; Jaeger, L.; et al. Multifunctional RNA nanoparticles. Nano Lett. 2014, 14, 5662–5671. [Google Scholar] [CrossRef]
- Shibata, T.; Fujita, Y.; Ohno, H.; Suzuki, Y.; Hayashi, K.; Komatsu, K.R.; Kawasaki, S.; Hidaka, K.; Yonehara, S.; Sugiyama, H.; et al. Protein-driven RNA nanostructured devices that function in vitro and control mammalian cell fate. Nat. Commun. 2017, 8, 540. [Google Scholar] [CrossRef]
- Li, H.; Lee, T.; Dziubla, T.; Pi, F.; Guo, S.; Xu, J.; Li, C.; Haque, F.; Liang, X.J.; Guo, P. RNA as a stable polymer to build controllable and defined nanostructures for material and biomedical applications. Nano Today 2015, 10, 631–655. [Google Scholar] [CrossRef]
- Xie, Z.; Liu, S.J.; Bleris, L.; Benenson, Y. Logic integration of mRNA signals by an RNAi-based molecular computer. Nucleic Acids Res. 2010, 38, 2692–2701. [Google Scholar] [CrossRef]
- Xie, Z.; Wroblewska, L.; Prochazka, L.; Weiss, R.; Benenson, Y. Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science 2011, 333, 1307. [Google Scholar] [CrossRef] [PubMed]
- Rinaudo, K.; Bleris, L.; Maddamsetti, R.; Subramanian, S.; Weiss, R.; Benenson, Y. A universal RNAi-based logic evaluator that operates in mammalian cells. Nat. Biotechnol. 2007, 25, 795. [Google Scholar] [CrossRef] [PubMed]
- Penchovsky, R.; Breaker, R.R. Computational design and experimental validation of oligonucleotide-sensing allosteric ribozymes. Nat. Biotechnol. 2005, 23, 1424–1433. [Google Scholar] [CrossRef]
- Soukup, G.A.; Breaker, R.R. Nucleic acid molecular switches. Trends Biotechnol. 1999, 17, 469–476. [Google Scholar] [CrossRef]
- Roark, B.K.; Tan, L.A.; Ivanina, A.; Chandler, M.; Castaneda, J.; Kim, H.S.; Jawahar, S.; Viard, M.; Talic, S.; Wustholz, K.L.; et al. Fluorescence blinking as an output signal for biosensing. ACS Sens. 2016, 1, 1295–1300. [Google Scholar] [CrossRef] [PubMed]
- Zakrevsky, P.; Parlea, L.; Viard, M.; Bindewald, E.; Afonin, K.A.; Shapiro, B.A. Preparation of a conditional RNA switch. Methods Mol. Biol. 2017, 1632, 303–324. [Google Scholar] [PubMed]
- Bindewald, E.; Afonin, K.A.; Viard, M.; Zakrevsky, P.; Kim, T.; Shapiro, B.A. Multistrand structure prediction of nucleic acid assemblies and design of RNA switches. Nano Lett. 2016, 16, 1726–1735. [Google Scholar] [CrossRef] [PubMed]
- Halman, J.R.; Satterwhite, E.; Roark, B.; Chandler, M.; Viard, M.; Ivanina, A.; Bindewald, E.; Kasprzak, W.K.; Panigaj, M.; Bui, M.N.; et al. Functionally-interdependent shape-switching nanoparticles with controllable properties. Nucleic Acids Res. 2017, 45, 2210–2220. [Google Scholar] [CrossRef]
- Chandler, M.; Ke, W.; Halman, J.; Panigaj, M.; Afonin, K.A. Reconfigurable nucleic acid materials for cancer therapy. In Nanooncology; Gonçalves, G., Tobias, G., Eds.; Springer: New York, NY, USA, 2018; pp. 365–385. [Google Scholar]
- Douglas, S.M.; Bachelet, I.; Church, G.M. A logic-gated nanorobot for targeted transport of molecular payloads. Science 2012, 335, 831–834. [Google Scholar] [CrossRef]
- Afonin, K.A.; Viard, M.; Kagiampakis, I.; Case, C.L.; Dobrovolskaia, M.A.; Hofmann, J.; Vrzak, A.; Kireeva, M.; Kasprzak, W.K.; KewalRamani, V.N.; et al. Triggering of RNA interference with RNA-RNA, RNA-DNA, and DNA-RNA nanoparticles. ACS Nano 2015, 9, 251–259. [Google Scholar] [CrossRef]
- Afonin, K.A.; Viard, M.; Martins, A.N.; Lockett, S.J.; Maciag, A.E.; Freed, E.O.; Heldman, E.; Jaeger, L.; Blumenthal, R.; Shapiro, B.A. Activation of different split functionalities upon re-association of RNA-DNA hybrids. Nat. Nanotechnol. 2013, 8, 296–304. [Google Scholar] [CrossRef]
- Sajja, S.; Chandler, M.; Fedorov, D.; Kasprzak, W.K.; Lushnikov, A.; Viard, M.; Shah, A.; Dang, D.; Dahl, J.; Worku, B.; et al. Dynamic behavior of RNA nanoparticles analyzed by AFM on a mica/air interface. Langmuir 2018. [Google Scholar] [CrossRef] [PubMed]
- Rackley, L.; Stewart, J.M.; Salotti, J.; Krokhotin, A.; Shah, A.; Halman, J.; Juneja, R.; Smollett, J.; Roark, B.; Viard, M.; et al. RNA fibers as optimized nanoscaffolds for siRNA coordination and reduced immunological recognition. Adv. Funct. Mater. 2018. [Google Scholar] [CrossRef]
- Ouellet, J. RNA fluorescence with light-up aptamers. Front. Chem. 2016, 4. [Google Scholar] [CrossRef] [PubMed]
- Marras, S.A.; Kramer, F.R.; Tyagi, S. Multiplex detection of single-nucleotide variations using molecular beacons. Genet. Anal. 1999, 14, 151–156. [Google Scholar] [CrossRef]
- Feng, S.; Shang, Y.; Wu, F.; Ding, F.; Li, B.; Xu, J.; Xu, L.; Zhou, X. DNA nanomachines as evolved molecular beacons for in vitro and in vivo detection. Talanta 2014, 120, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, E.; Chartrand, P.; Schaefer, M.; Shenoy, S.M.; Singer, R.H.; Long, R.M. Localization of ASH1 mRNA particles in living yeast. Mol. Cell. 1998, 2, 437–445. [Google Scholar] [CrossRef]
- Valencia-Burton, M.; McCullough, R.M.; Cantor, C.R.; Broude, N.E. RNA visualization in live bacterial cells using fluorescent protein complementation. Nat. Methods 2007, 4, 421–427. [Google Scholar] [CrossRef]
- Demidov, V.V.; Dokholyan, N.V.; Witte-Hoffmann, C.; Chalasani, P.; Yiu, H.-W.; Ding, F.; Yu, Y.; Cantor, C.R.; Broude, N.E. Fast complementation of split fluorescent protein triggered by DNA hybridization. Proc. Nat. Acad. Sci. USA 2006, 103, 2052–2056. [Google Scholar] [CrossRef]
- Schwarz-Schilling, M.; Dupin, A.; Chizzolini, F.; Krishnan, S.; Mansy, S.S.; Simmel, F.C. Optimized assembly of a multifunctional RNA-protein nanostructure in a cell-free gene expression system. Nano Lett. 2018, 18, 2650–2657. [Google Scholar] [CrossRef]
- Grate, D.; Wilson, C. Laser-mediated, site-specific inactivation of RNA transcripts. Proc. Nat. Acad. Sci. USA 1999, 96, 6131–6136. [Google Scholar] [CrossRef] [PubMed]
- Stojanovic, M.N.; Kolpashchikov, D.M. Modular aptameric sensors. J. Am. Chem. Soc. 2004, 126, 9266–9270. [Google Scholar] [CrossRef] [PubMed]
- Kolpashchikov, D.M. Binary malachite green aptamer for fluorescent detection of nucleic acids. J. Am. Chem. Soc. 2005, 127, 12442–12443. [Google Scholar] [CrossRef] [PubMed]
- Afonin, K.A.; Bindewald, E.; Yaghoubian, A.J.; Voss, N.; Jacovetty, E.; Shapiro, B.A.; Jaeger, L. In vitro assembly of cubic RNA-based scaffolds designed in silico. Nat. Nanotechnol. 2010, 5, 676–682. [Google Scholar] [CrossRef]
- Afonin, K.A.; Desai, R.; Viard, M.; Kireeva, M.L.; Bindewald, E.; Case, C.L.; Maciag, A.E.; Kasprzak, W.K.; Kim, T.; Sappe, A.; et al. Co-transcriptional production of RNA-DNA hybrids for simultaneous release of multiple split functionalities. Nucleic Acids Res. 2014, 42, 2085–2097. [Google Scholar] [CrossRef] [PubMed]
- Paige, J.S.; Wu, K.Y.; Jaffrey, S.R. RNA mimics of green fluorescent protein. Science 2012, 333, 642–646. [Google Scholar] [CrossRef] [PubMed]
- Paige, J.S.; Nguyen-Duc, T.; Song, W.; Jaffrey, S.R. Fluorescence imaging of cellular metabolites with RNA. Science 2012, 335, 1194. [Google Scholar] [CrossRef]
- Strack, R.L.; Disney, M.D.; Jaffrey, S.R. A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat–containing RNA. Nat. Methods 2013, 10, 1219. [Google Scholar] [CrossRef]
- Filonov, G.S.; Kam, C.W.; Song, W.; Jaffrey, S.R. In-gel imaging of RNA processing using broccoli reveals optimal aptamer expression strategies. Chem. Biol. 2015, 22, 649–660. [Google Scholar] [CrossRef]
- Filonov Grigory, S.; Jaffrey, S.R. RNA imaging with dimeric broccoli in live bacterial and mammalian cells. Curr. Protoc. Chem. Biol. 2016, 8, 1–28. [Google Scholar]
- Filonov, G.S.; Moon, J.D.; Svensen, N.; Jaffrey, S.R. Broccoli: Rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J. Am. Chem. Soc. 2014, 136, 16299–16308. [Google Scholar] [CrossRef] [PubMed]
- Svensen, N.; Jaffrey, S.R. Fluorescent RNA aptamers as a tool to study RNA-modifying enzymes. Cell Chem. Biol. 2016, 23, 415–425. [Google Scholar] [PubMed]
- Autour, A.C.Y.; Jeng, S.D.; Cawte, A.; Abdolahzadeh, A.; Galli, A.; Panchapakesan, S.S.S.; Rueda, D.; Ryckelynck, M.; Unrau, P.J. Fluorogenic RNA mango aptamers for imaging small non-coding RNAs in mammalian cells. Nat. Commun. 2018, 9, 656. [Google Scholar] [CrossRef]
- Jepsen, M.D.E.; Sparvath, S.M.; Nielsen, T.B.; Langvad, A.H.; Grossi, G.; Gothelf, K.V.; Andersen, E.S. Development of a genetically encodable FRET system using fluorescent RNA aptamers. Nat. Commun. 2018, 9, 18. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Filonov, G.S.; Kim, H.; Hirsch, M.; Li, X.; Moon, J.D.; Jaffrey, S.R. Imaging RNA polymerase III transcription using a photostable RNA–fluorophore complex. Nat. Chem. Biol. 2017, 13, 1187. [Google Scholar] [CrossRef]
- Rogers, T.A.; Andrews, G.E.; Jaeger, L.; Grabow, W.W. Fluorescent monitoring of RNA assembly and processing using the split-spinach aptamer. ACS Synth. Biol. 2015, 4, 162–166. [Google Scholar] [CrossRef]
- Warner, K.D.; Chen, M.C.; Song, W.; Strack, R.L.; Thorn, A.; Jaffrey, S.R.; Ferre-D‘Amare, A.R. Structural basis for activity of highly efficient RNA mimics of green fluorescent protein. Nat. Struct. Mol. Biol. 2014, 21, 658–663. [Google Scholar] [CrossRef]
- Zadeh, J.N.; Steenberg, C.D.; Bois, J.S.; Wolfe, B.R.; Pierce, M.B.; Khan, A.R.; Dirks, R.M.; Pierce, N.A. NUPACK: Analysis and design of nucleic acid systems. J. Comput. Chem. 2011, 32, 170–173. [Google Scholar] [CrossRef]
- Afonin, K.A.; Kireeva, M.; Grabow, W.W.; Kashlev, M.; Jaeger, L.; Shapiro, B.A. Co-transcriptional assembly of chemically modified RNA nanoparticles functionalized with siRNAs. Nano Lett. 2012, 12, 5192–5195. [Google Scholar] [CrossRef]
- Britton, P.; Green, P.; Kottier, S.; Mawditt, K.L.; Penzes, Z.; Cavanagh, D.; Skinner, M.A. Expression of bacteriophage T7 RNA polymerase in avian and mammalian cells by a recombinant fowlpox virus. J. Gen. Virol. 1996, 77, 963–967. [Google Scholar] [CrossRef]
- Elroy-Stein, O.; Moss, B. Cytoplasmic expression system based on constitutive synthesis of bacteriophage T7 RNA polymerase in mammalian cells. Proc. Nat. Acad. Sci. USA 1990, 87, 6743–6747. [Google Scholar] [CrossRef] [PubMed]
- Goldsworthy, V.; LaForce, G.; Abels, S.; Khisamutdinov, E. Fluorogenic RNA aptamers: A nano-platform for fabrication of simple and combinatorial logic gates. Nanomaterials 2018, 8, 984. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, J.; Tran, C.H.; Wadhwani, K.; Cuba Samaniego, C.; Subramanian, H.K.K.; Franco, E. Dynamic control of aptamer-ligand activity using strand displacement reactions. ACS Synth. Biol. 2018, 7, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Ke, W.; Hong, E.; Saito, R.F.; Rangel, M.C.; Wang, J.; Viard, M.; Richardson, M.; Khisamutdinov, E.F.; Panigaj, M.; Dokholyan, N.V.; et al. RNA-DNA fibers and polygons with controlled immunorecognition activate RNAi, FRET, and transcriptional regulation of NF-κB in human cells. Nucleic Acids Res. 2018. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chandler, M.; Lyalina, T.; Halman, J.; Rackley, L.; Lee, L.; Dang, D.; Ke, W.; Sajja, S.; Woods, S.; Acharya, S.; et al. Broccoli Fluorets: Split Aptamers as a User-Friendly Fluorescent Toolkit for Dynamic RNA Nanotechnology. Molecules 2018, 23, 3178. https://doi.org/10.3390/molecules23123178
Chandler M, Lyalina T, Halman J, Rackley L, Lee L, Dang D, Ke W, Sajja S, Woods S, Acharya S, et al. Broccoli Fluorets: Split Aptamers as a User-Friendly Fluorescent Toolkit for Dynamic RNA Nanotechnology. Molecules. 2018; 23(12):3178. https://doi.org/10.3390/molecules23123178
Chicago/Turabian StyleChandler, Morgan, Tatiana Lyalina, Justin Halman, Lauren Rackley, Lauren Lee, Dylan Dang, Weina Ke, Sameer Sajja, Steven Woods, Shrija Acharya, and et al. 2018. "Broccoli Fluorets: Split Aptamers as a User-Friendly Fluorescent Toolkit for Dynamic RNA Nanotechnology" Molecules 23, no. 12: 3178. https://doi.org/10.3390/molecules23123178
APA StyleChandler, M., Lyalina, T., Halman, J., Rackley, L., Lee, L., Dang, D., Ke, W., Sajja, S., Woods, S., Acharya, S., Baumgarten, E., Christopher, J., Elshalia, E., Hrebien, G., Kublank, K., Saleh, S., Stallings, B., Tafere, M., Striplin, C., & Afonin, K. A. (2018). Broccoli Fluorets: Split Aptamers as a User-Friendly Fluorescent Toolkit for Dynamic RNA Nanotechnology. Molecules, 23(12), 3178. https://doi.org/10.3390/molecules23123178





