Selection of Specific Peptides for Coccidioides spp. Obtained from Antigenic Fractions through SDS-PAGE and Western Blot Methods by the Recognition of Sera from Patients with Coccidioidomycosis
Abstract
1. Introduction
2. Results
2.1. Species Identification for Isolates of Coccidioides spp.
2.2. Coccidioidins Characterization
2.3. Electrophoresis (SDS-PAGE)
2.4. Reactivity of Antigenic Fractions by WB
2.5. Analysis of Assembled Sequences
2.6. Peptide Analysis
3. Discussion
4. Materials and Methods
4.1. Isolates
4.2. Biological Samples
4.3. Obtaining Coccidioidins
4.4. Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE)
4.5. Recognition of Antigenic Fractions of Coccidioidins by Western Blot (WB)
4.6. Protein Identification of the Antigenic Fractions of C. immitis and C. posadasii by Mass Spectrometry
4.7. Analysis of Assembled Sequences
4.8. Peptide Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Fisher, M.G.; Koening, G.I.; White, T.J.; Taylor, J.W. Molecular and phenotypic description of Coccidioides posadasii sp. Nov., previously recognized as the non-California population of Coccidioides immitis. Mycologia 2002, 94, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Canteros, C.E.; Toranzo, A.; Ibarra-Camou, B.; David, V.; Carrizo, S.G.; Santillán-Iturres, A.; Serrano, J.; Fernández, N.; Capece, P.; Gorostiaga, J.; et al. Coccidioidomycosis in Argentina, 1892–2009. Rev. Argent. Microbiol. 2010, 42, 261–268. [Google Scholar] [PubMed]
- de Aguiar Cordeiro, R.; Bezerra Fechine, M.A.; Nogueira Brilhante, R.S.; Gadelha Rocha, M.F.; Freire da Costa, A.K.; Tiemi Dias Nagao, M.A.; Pires de Camargo, Z.; Costa Sidrim, J.J. Serologic detection of coccidioidomycosis antibodies in northeast Brazil. Mycopathologia 2009, 167, 187–190. [Google Scholar] [CrossRef] [PubMed]
- Hideo-Togashi, R.; Batista-Aguiar, F.M.; Barros-Ferreira, D.; Matos de Moura, C.; Montezuma-Sales, M.T.; Ximenes-Rios, N. Pulmonary and extrapulmonary coccidioidomycosis: Three cases in an endemic area in the state of Ceará, Brazil. J. Bras. Pneumol. 2009, 35, 275–279. [Google Scholar]
- Baptista-Rosas, R.C.; Hinojosa, A.; Riquelme, M. Ecological niche modeling of Coccidioides spp. in western North American deserts. Ann. N. Y. Acad. Sci. 2007, 1111, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Baptista-Rosas, R.C.; Riquelme, M. Epidemiología de la coccidioidomicosis en México. Rev. Iberoam. Micol. 2007, 24, 100–105. [Google Scholar] [CrossRef]
- Laniado-Laborín, R. Expanding understanding of epidemiology of coccidioidomycosis in the western hemisphere. Ann. N. Y. Acad. Sci. 2007, 1111, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Laniado-Laborín, R. Coccidioidomycosis and other endemic mycoses in México. Rev. Iberoam. Micol. 2007, 24, 249–258. [Google Scholar] [CrossRef]
- Méndez-Tovar, L.J.; Ramos-Hernández, J.M.; Manzano-Gayosso, P.; García-González, A.; Hernández-Hernández, F.; Bazán-Mora, E.; López-Martínez, R. Micosis sistémicas: Experiencia de once años en el Hospital de Especialidades del Centro Médico Nacional Siglo XXI del IMSS, México. Rev. Mex. Mic. 2007, 25, 15–19. [Google Scholar]
- Herrera, L.E.; Gómez, V.; Morales-Blanhir, J.E. Coccidioidomicosis: Serie de casos. Neumol. Cir. Torax 2006, 65, 206–213. [Google Scholar]
- Torres-Nájera, M.; de la Garza-Galván, S.; Cerda-Flores, R.M.; Nocedal-Rustrián, F.C.; Calderón-Garcidueñas, A.L. Coccidioidomicosis osteoarticular: Estudio clinicopatológico de una serie de 36 pacientes mexicanos. Rev. Invest. Clin. 2006, 58, 211–216. [Google Scholar] [PubMed]
- Mondragón-González, R.; Méndez-Tovar, L.J.; Bernal-Vázquez, E.; Hernández-Hernández, F.; López-Martínez, R.; Manzano-Gayosso, P.; Ríos-Rosas, C.; Contreras-Pérez, C.; Anides-Fonseca, A.E. Detección de infección por Coccidioides immitis en Coahuila, México. Rev. Argent. Microbiol. 2005, 37, 135–138. [Google Scholar]
- Reviákina, V.; Panizo, M.; Dolande, M.; Maldonado, B. Micosis profundas sistémicas: Casuística del Departamento de Micología del Instituto Nacional de Higiene “Rafael Rangel” durante 5 años (1997–2001). Rev. Soc. Ven. Microbiol. 2002, 22, 164–168. [Google Scholar]
- Domínguez-Cota, G.; Zamora-Ângulo, L.E.; Osuna-Beltrán, E.; Vega-Rosas, C.E. Coccidioidomicosis cutánea primaria. Estudio epidemiológico y clínico. Serie de 16 casos. CIMEL 2001, 6, 49–53. [Google Scholar]
- Borelli, D.; Pérez, M.; Molina, T. Coccidioidomicosis: Un caso más en el bosque muy seco tropical. Dermatol. Venez. 1991, 29, 119–123. [Google Scholar]
- Laniado-Laborín, R.; Cárdenas-Moreno, R.P.; Álvarez-Cerro, M. Tijuana: Zona endémica de infección por Coccidioides immitis. Salud Pub. Méx. 1991, 33, 235–239. [Google Scholar]
- Saubolle, M.A. Laboratory aspects in the diagnosis of coccidioidomycosis. Ann. N. Y. Acad. Sci. 2007, 1111, 301–314. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, L.; Clark, M.J. Value of the concomitant use of complement fixation and immunodiffusion tests in the diagnosis of coccidioidomycosis. Appl. Microbiol. 1974, 28, 641–643. [Google Scholar] [PubMed]
- Lindsley, M.D.; Warnock, D.W.; Morrison, C.J. Serological and molecular diagnosis of fungal infections. In Manual of Molecular and Clinical Laboratory Immunology, 7th ed.; Detrick, B.H., Hamilton, R.G., Folds, J.D., Eds.; ASM Press: Washington, DC, USA, 2006; pp. 569–605. [Google Scholar]
- Pappagianis, D. Serologic studies in coccidioidomycosis. Semin. Respir. Infect. 2001, 16, 242–250. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, C.R.; Johnson, S.M.; Martens, G.W.; White, A.G.; Pappagianis, D. Cloning and characterization of the complement fixation antigen-chitinase of Coccidioides immitis. Infect. Immun. 1996, 64, 4967–4975. [Google Scholar]
- Johnson, S.M.; Zimmermann, R.; Pappagianis, D. Use of a recombinant Coccidioides immitis complement fixation antigen-chitinase in conventional serological assays. J. Clin. Microbiol. 1996, 34, 3160–3164. [Google Scholar] [PubMed]
- Yang, M.C.; Magee, M.; Kaufman, L.; Zhu, Y.; Cox, R.A. Recombinant Coccidioides immitis complement-fixing antigen: Detection of an epitope shared by Coccidiodes immitis, Histoplasma capsulatum, and Blastomyces dermatitidis. Clin. Diag. Lab. Immunol. 1997, 4, 19–22. [Google Scholar]
- Peng, T.; Orsborn, K.I.; Orbach, M.J.; Galgiani, J.N. Proline-rich vaccine candidate antigen of Coccidioides immitis: Conservation among isolates and differential expression with spherule maturation. J. Infect. Dis. 1999, 179, 518–521. [Google Scholar] [CrossRef] [PubMed]
- Orsborn, K.I.; Galgiani, J.N. Detecting serum antibodies to a purified recombinant proline-rich antigen of Coccidioides immitis in patients with coccidioidomycosis. Clin. Infect. Dis. 1998, 27, 1475–1478. [Google Scholar] [CrossRef] [PubMed]
- Filho, R.E.; Bandeira, S.P.; Brillhante, R.S.; Rocha, M.F.; Vasconcelos, I.M.; Pereira, M.L.; Castelo-Branco, D.D.S.C.; Rocha, F.A.; Camargo, Z.P.; Ramos, M.V.; et al. Biochemical characterization of an in-house Coccidioides antigen: Perspectives for the immunodiagnosis of coccidioidomycosis. Molecules 2012, 17, 7854–7863. [Google Scholar] [CrossRef] [PubMed]
- de Aguiar Cordeiro, R.; Patoilo, K.R.; Praciano, S.B.; Medrano, D.J.; de Farias Marques, F.J.; Martins, L.M.; Eulalio, K.D.; de Deus Filho, A.; Cavalvanti Mdo, A.; Fechine, M.A.; et al. Antigens of Coccidioides posadasii as an important tool for the immunodiagnosis of coccidioidomycosis. Mycopathologia 2013, 175, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Navalkar, K.A.; Johnston, S.A.; Stafford, P. Peptide based diagnostics: Are random-sequence peptides more useful than tiling proteome sequences? J. Immunol. Methods 2015, 417, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Meloen, R.H.; Puijk, W.C.; Slootstra, J.W. Mimotopes: Realization of an unlikely concept. J. Mol. Recognit. 2000, 13, 352–359. [Google Scholar] [CrossRef]
- Ampel, N.M. The diagnosis of coccidioidomycosis. F1000 Med. Rep. 2010, 2, 2. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.E.; Jeffery, B.; Huppert, M. Evaluation of five commercially available immunodiffusion kits for detection of Coccidioides immitis and Histoplasma capsulatum antibodies. J. Clin. Microbiol. 1984, 20, 530–532. [Google Scholar] [PubMed]
- Pappagianis, D.; Zimmer, B.L. Serology of coccidioidomycosis. Clin. Microbiol. Rev. 1990, 3, 247–268. [Google Scholar] [CrossRef] [PubMed]
- Stevens, D.A.; Levine, H.B.; TenEyck, D.R. Dermal sensitivity to different doses of spherulin and coccidioidin. Chest 1974, 65, 530–533. [Google Scholar] [CrossRef] [PubMed]
- Brilhante, R.S.; Cordeiro, R.A.; Rocha, M.F.; Fechine, M.A.; Furtado, F.M.; Nagao-Dias, A.T.; Camargo, Z.P.; Sidrim, J.J. Coccidioidal pericarditis: A rapid presumptive diagnosis by an in-house antigen confirmed by mycological and molecular methods. J. Med. Microbiol. 2008, 57, 1288–1292. [Google Scholar] [CrossRef] [PubMed]
- Welsh, O.; Vera-Cabrera, L.; Rendon, A.; González, G.; Bonifaz, A. Coccidioidomycosis. Clin. Dermatol. 2012, 30, 573–591. [Google Scholar] [CrossRef] [PubMed]
- Huppert, M.; Krasnow, I.; Vukovich, K.R.; Sun, S.H.; Rice, E.H.; Kutner, L.J. Comparison of coccidioidin and spherulin in complement fixation tests for coccidioidomycosis. J. Clin. Microbiol. 1977, 6, 33–41. [Google Scholar] [PubMed]
- Mitchell, N.M.; Sherrard, A.L.; Dasari, S.; Magee, D.M.; Grys, T.E.; Lake, D.F. Proteogenomic re-annotation of Coccidioides posadasii strain silveira. Proteomics 2018, 18, 1700173. [Google Scholar] [CrossRef] [PubMed]
- Antoni, G.; Presentini, R.; Perin, F.; Tagliabue, A.; Ghiara, P.; Censini, S.; Volpini, G.; Villa, L.; Boraschi, D. A short synthetic peptide fragment of human interleukin 1 with immunostimulatory but not inflammatory activity. J. Immunol. 1986, 137, 3201–3204. [Google Scholar] [PubMed]
- Schotanus, K.; Meloen, R.H.; Puijk, W.C.; Berkenbosch, F.; Binnekade, R.; Tilders, F.J.H. Effects of monoclonal antibodies to specific epitopes of rat interleukin-1 beta (IL-β) on IL-1β-induced ACTH, corticosterone and IL-6 responses in rats. J. Neuroendocrinol. 1995, 7, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Meloen, R.H.; Casal, J.I.; Dalsgaard, K.; Langeveld, J.P.M. Synthetic peptide vaccines: Success at last. Vaccine 1995, 13, 885–886. [Google Scholar] [CrossRef]
- Meloen, R.H.; Langedijk, J.P.M.; Langeveld, J.P.M. Synthetic peptides for diagnostic use. Vet. Q. 1997, 19, 122–126. [Google Scholar] [CrossRef] [PubMed]
- Langedijk, J.P.M.; Brandenburg, A.H.; Middel, W.G.J.; Osterhaus, A.; Meloen, R.H.; van Oirschot, J.T. A subtype-specifc peptide-based enzyme immunoassay for detection of antibodies to the G protein of human respiratory syncytial virus is more sensitive than routine serological tests. J. Clin. Microbiol. 1997, 55, 1656–1660. [Google Scholar]
- Grunsven, W.M.; Spaan, W.J.; Middeldorp, J.M. Localisation and diagnostic application of immunodominant domains of BFRF3 encoded EBV capsid protein. J. Infect. Dis. 1994, 170, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Brusic, V.; Petrovsky, N. Immunoinformatics and its relevance to understanding human immune disease. Expert Rev. Clin. Immunol. 2005, 1, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Tomar, N.; De, R.K. Immunoinformatics: An integrated scenario. Immunology 2010, 131, 153–168. [Google Scholar] [CrossRef] [PubMed]
- Duarte-Escalante, E.; Zúñiga, G.; Frías-De-León, M.G.; Canteros, C.; Castañón-Olivares, L.R.; Reyes-Montes, M.R. AFLP analysis reveals high genetic diversity but low population structure in Coccidioides posadasii isolates from Mexico and Argentina. BMC Infect. Dis. 2013, 13, 411. [Google Scholar] [CrossRef] [PubMed]
- Bialek, R.; Kern, J.; Herrmann, T.; Tijerina, R.; Ceceñas, L.; Reischl, U.; González, G.M. PCR assays for identification of Coccidioides posadasii based on the nucleotide sequence of the antigen 2/proline-rich antigen. J. Clin. Microbiol. 2004, 42, 778–783. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.E.; Saito, M.T.; Beard, R.R.; Kepp, R.M.; Clark, R.W.; Eddie, B.U. Serological tests in the diagnosis and prognosis of coccidioidomycosis. Am. J. Hyg. 1950, 52, 1–20. [Google Scholar] [PubMed]
- Ouchterlony, O.; Nilsson, L.A. Immunodiffusion and immunoelectrophoresis. In Handbook of Experimental Immunology, 1st ed.; Weir, D.M., Ed.; Blackwell Scientific Publications: Oxford, UK, 1978; Volume 1, pp. 19.1–19.44. ISBN 0632014997. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the bacteriophage T4. Nature 1970, 227, 679–685. [Google Scholar] [CrossRef]
- Towbin, H.; Staehelin, T.; Gordon, J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. USA 1979, 76, 4350–4354. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |

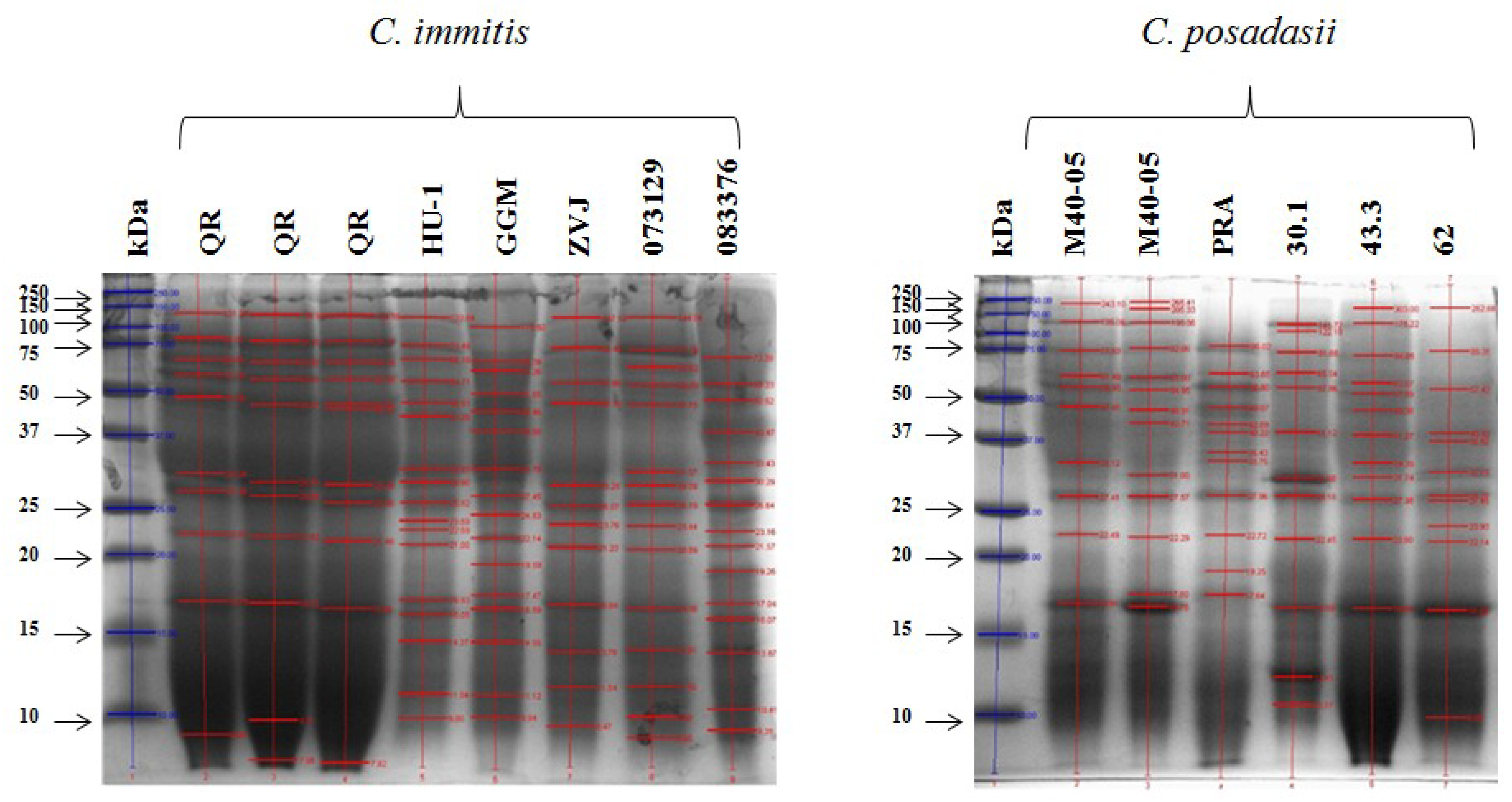

| Coccidioidins | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| kDa | QR | HU-1 | GGM | ZVJ | 073329 | 083376 | M40-05 | PRA | 30.1 | 43.3 | 62 |
| 100 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 |
| 75 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 65 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 |
| 55 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 50 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 37 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | |
| 35 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 30 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 28 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 23 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 |
| 21 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| 20 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 19 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 |
| 18 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| 16 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| kDa | Frequency (%) Anti-C. immitis | Frequency (%) Anti-C. posadasii |
|---|---|---|
| 250 | 21.42 (6/28) | 10.71 (3/28) |
| 150 | 10.71 (3/28) | 10.71 (3/28) |
| 100 | 46.42 (13/28) | 46.42 (13/28) |
| 75 | 10.71 (3/28) | 10.71 (3/28) |
| 50 | 42.85 (12/28) | 42.85 (12/28) |
| 37 | 28.57 (8/28) | 28.57 (8/28) |
| 28 | 20.0 (5/28) | 25.0 (7/28) |
| Access Number | Description | Alignment | Query Cover | E-Value | Identity |
|---|---|---|---|---|---|
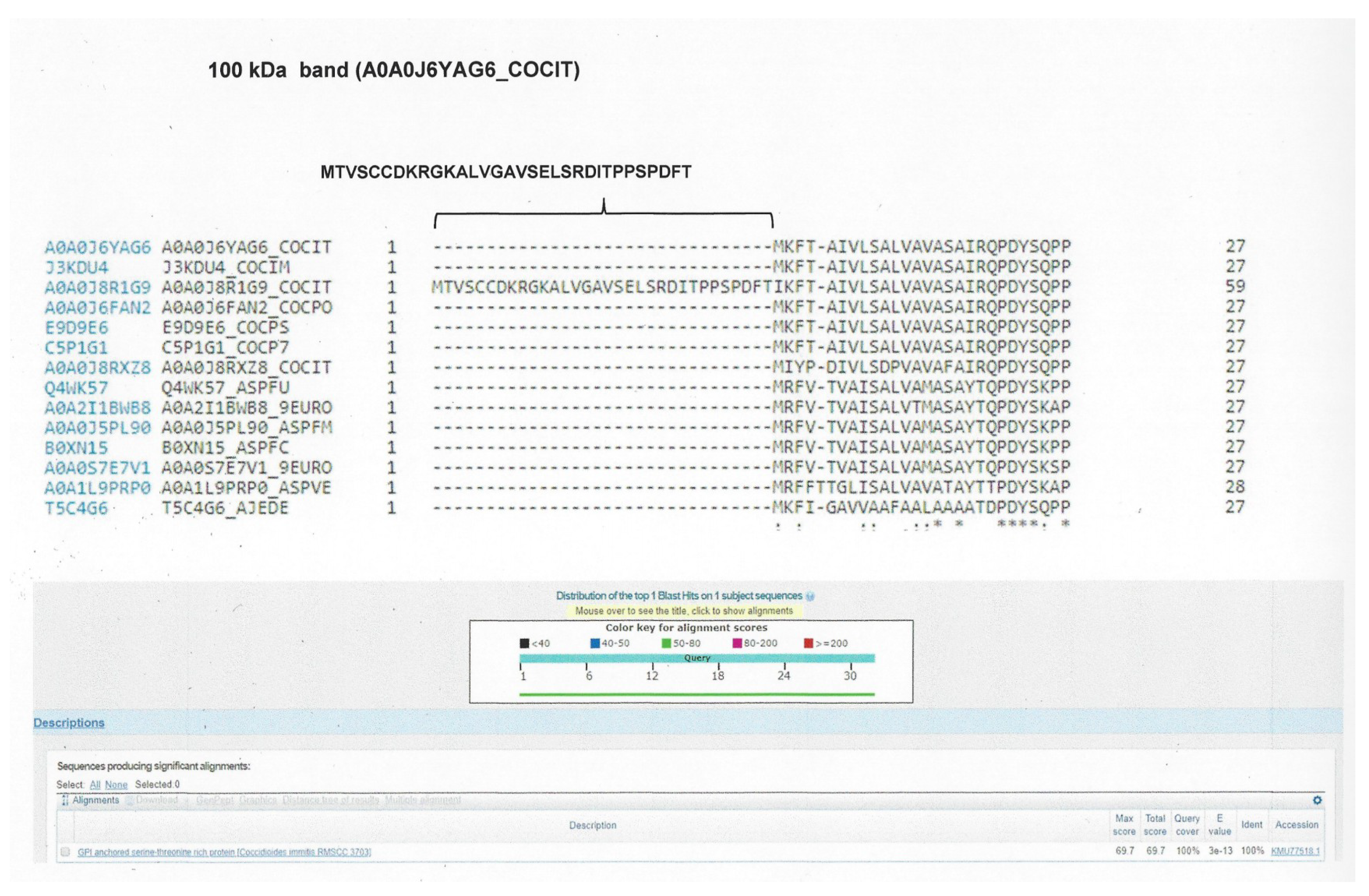
| 100 kDa band | |||||
| A0A0J6YAG6_COCIT | GPI anchored serine threonine-rich protein OS C. immitis | C. immitis | 93% | 7 × 10−96 | 75% |
| C. posadasii | 93% | 1 × 10−94 | 74% | ||
| H. capsulatum | 68% | 8 × 10−38 | 52% | ||
| A. fumigatus | 63% | 6 × 10−45 | 57% | ||
| P. brasiliensis | 58% | 2 × 10−30 | 49% | ||
| A0A0J8R1I4_COCIT | Uncharacterized protein OS C. immitis | C. immitis | 100% | 0 | 90% |
| C. posadasii | 100% | 0 | 89% | ||
| P. lutzi | 49% | 1 × 10−67 | 60% | ||
| 50 kDa band | H. capsulatum | 47% | 1 × 10−64 | 58% | |
| A0A0J6F383_COCPO | Fatty acid amide hydrolase 1 OS C. posadasii | C. posadasii | 100% | 0 | 97% |
| C. immitis | 100% | 0 | 96% | ||
| P. brasiliensis | 91% | 0 | 54% | ||
| P. lutzi | 91% | 0 | 54% | ||
| H. capsulatum | 88% | 0 | 55% |
| Isolate | Specie | Source | Geographical Origin |
|---|---|---|---|
| HU1 | C. posadasii | ND | Monterrey, MX |
| 073129 | C. posadasii | Biopsy | Salta, AR |
| 083376 | C. posadasii | BAL | Catamarca, AR |
| 30.1 | C. immitis | Sputum | Tijuana, MX |
| 43.3 | C. immitis | Sputum | Guerrero, MX |
| 62 | C. immitis | Sputum | ND |
| GGM | C. posadasii | Sputum | Baja California, MX |
| ZVJ | C. posadasii | Pulmonary nodule | Michoacán, MX |
| QR | C. posadasii | Knee puncture | ND |
| PRA | C. immitis | BAL | Tijuana, MX |
| M40-05 | C. immitis | Wound secretion | Guadalajara, MX |
| Sera | Gender | Age | IDG | PTC | ELISA | Culture |
|---|---|---|---|---|---|---|
| 1 | F | 31 | Positive | 1:32 | ND | Positive |
| 2 | F | 39 | Negative | 1:32 | ND | Negative |
| 3 | M | 75 | Positive | 1:16 | 1:256 | Positive |
| 5 | F | 53 | Positive | 1:16 | ND | Negative |
| 9 | F | 37 | Negative | 1:32 | ND | Negative |
| 10 | F | 38 | Negative | 1:2 | ND | Negative |
| 14 | M | 41 | Positive | 1:8 | ND | Negative |
| 17 | F | 38 | Negative | 1:16 | ND | Negative |
| 18 | F | 46 | Negative | 1:64 | ND | Negative |
| 23 | M | 79 | Negative | 1:32 | ND | Negative |
| 29 | M | 42 | Positive | 1:32 | 1:128 | Positive |
| 31 | M | 19 | Positive | 1:128 | ND | Positive |
| 32 | M | 32 | Positive | 1:2 | 1:256 | Positive |
| 35 | M | 44 | Positive | 1:2 | ND | Positive |
| 45 | F | 47 | Negative | 1:8 | ND | Negative |
| 52 | M | 42 | Negative | 1:256 | ND | Negative |
| 53 | F | 19 | Negative | 1:64 | ND | Negative |
| 55 | M | 45 | Positive | 1:32 | ND | Negative |
| 59 | M | 11 | Negative | 1:16 | ND | Negative |
| 61 | F | 26 | Negative | Negative | ND | Negative |
| 64 | F | 63 | Positive | Negative | 1:128 | Negative |
| 69 | M | 35 | Negative | 1:8 | ND | Positive |
| 74 | F | 35 | Negative | 1:8 | ND | Negative |
| 75 | F | 57 | Positive | 1:64 | ND | Negative |
| 81 | M | 45 | Positive | 1:32 | 1:256 | Positive |
| 82 | M | 67 | Negative | 1:64 | ND | Negative |
| 85 | F | 77 | Positive | 1:64 | ND | Negative |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duarte Escalante, E.; Frías De León, M.G.; Martínez García, L.G.; Herrera, J.; Acosta Altamirano, G.; Cabello, C.; Palma, G.; Reyes Montes, M.d.R. Selection of Specific Peptides for Coccidioides spp. Obtained from Antigenic Fractions through SDS-PAGE and Western Blot Methods by the Recognition of Sera from Patients with Coccidioidomycosis. Molecules 2018, 23, 3145. https://doi.org/10.3390/molecules23123145
Duarte Escalante E, Frías De León MG, Martínez García LG, Herrera J, Acosta Altamirano G, Cabello C, Palma G, Reyes Montes MdR. Selection of Specific Peptides for Coccidioides spp. Obtained from Antigenic Fractions through SDS-PAGE and Western Blot Methods by the Recognition of Sera from Patients with Coccidioidomycosis. Molecules. 2018; 23(12):3145. https://doi.org/10.3390/molecules23123145
Chicago/Turabian StyleDuarte Escalante, Esperanza, María Guadalupe Frías De León, Luz Gisela Martínez García, Jorge Herrera, Gustavo Acosta Altamirano, Carlos Cabello, Gabriel Palma, and María del Rocío Reyes Montes. 2018. "Selection of Specific Peptides for Coccidioides spp. Obtained from Antigenic Fractions through SDS-PAGE and Western Blot Methods by the Recognition of Sera from Patients with Coccidioidomycosis" Molecules 23, no. 12: 3145. https://doi.org/10.3390/molecules23123145
APA StyleDuarte Escalante, E., Frías De León, M. G., Martínez García, L. G., Herrera, J., Acosta Altamirano, G., Cabello, C., Palma, G., & Reyes Montes, M. d. R. (2018). Selection of Specific Peptides for Coccidioides spp. Obtained from Antigenic Fractions through SDS-PAGE and Western Blot Methods by the Recognition of Sera from Patients with Coccidioidomycosis. Molecules, 23(12), 3145. https://doi.org/10.3390/molecules23123145






