Kinase Inhibitory Activities and Molecular Docking of a Novel Series of Anticancer Pyrazole Derivatives
Abstract
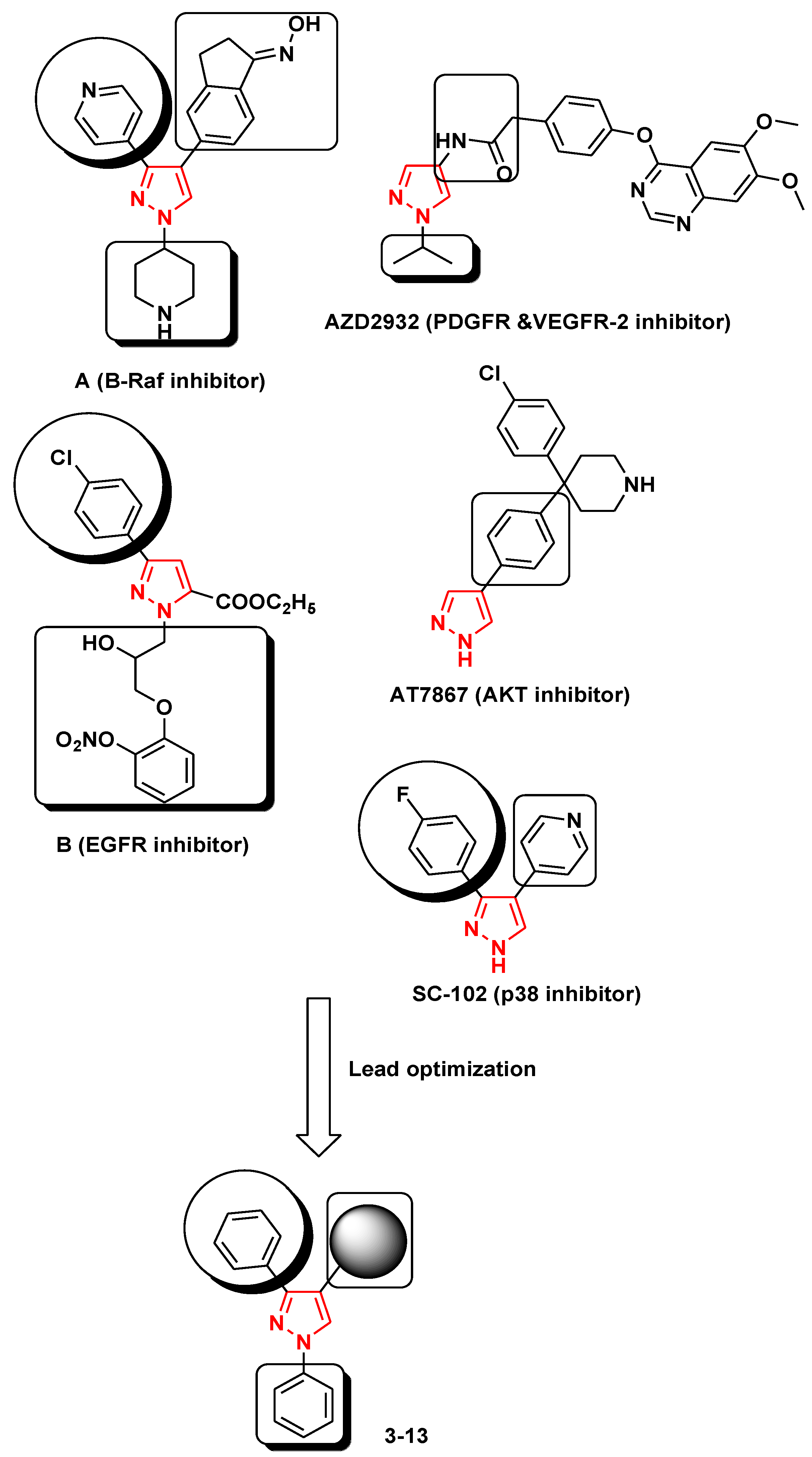
1. Introduction
2. Results and Discussion
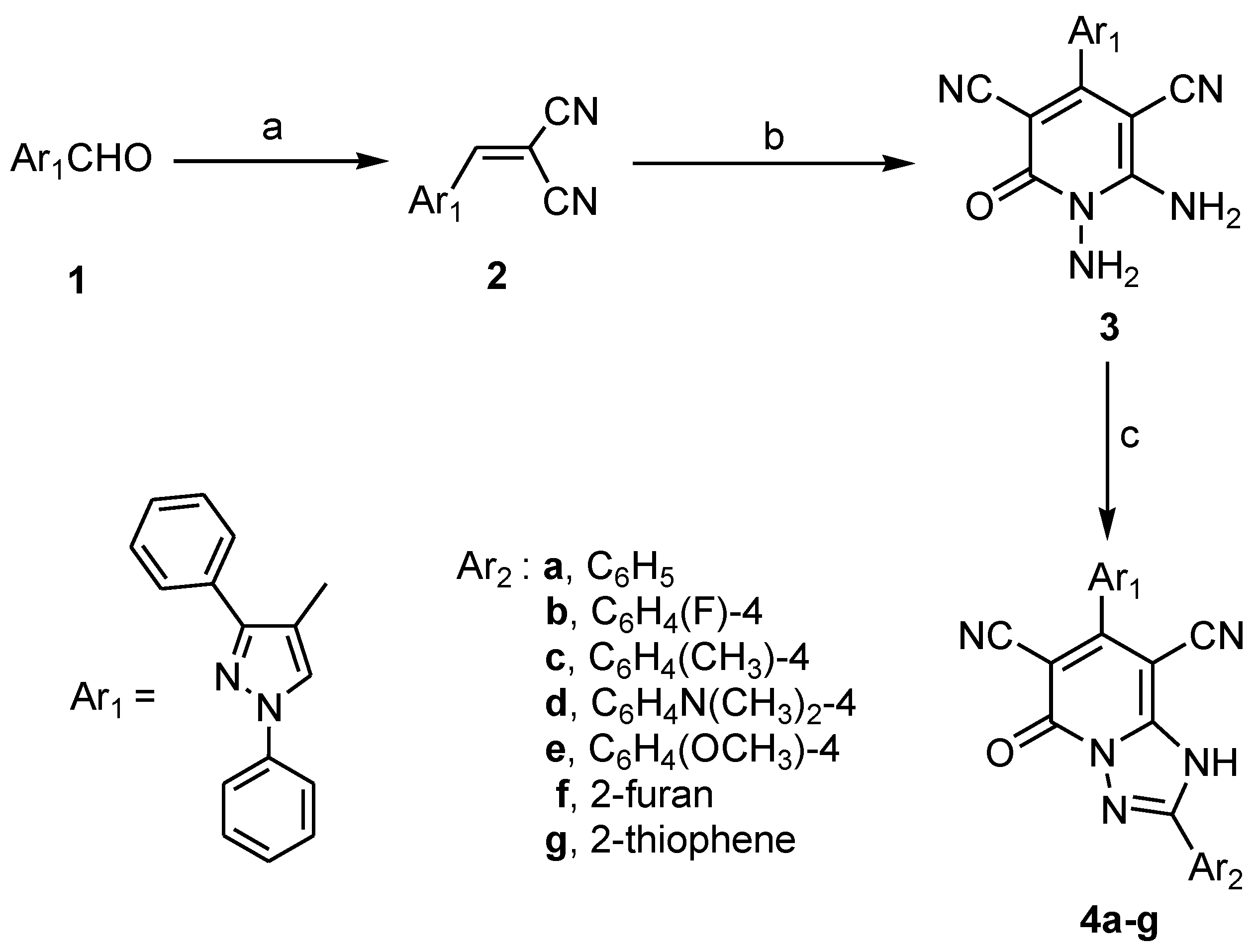
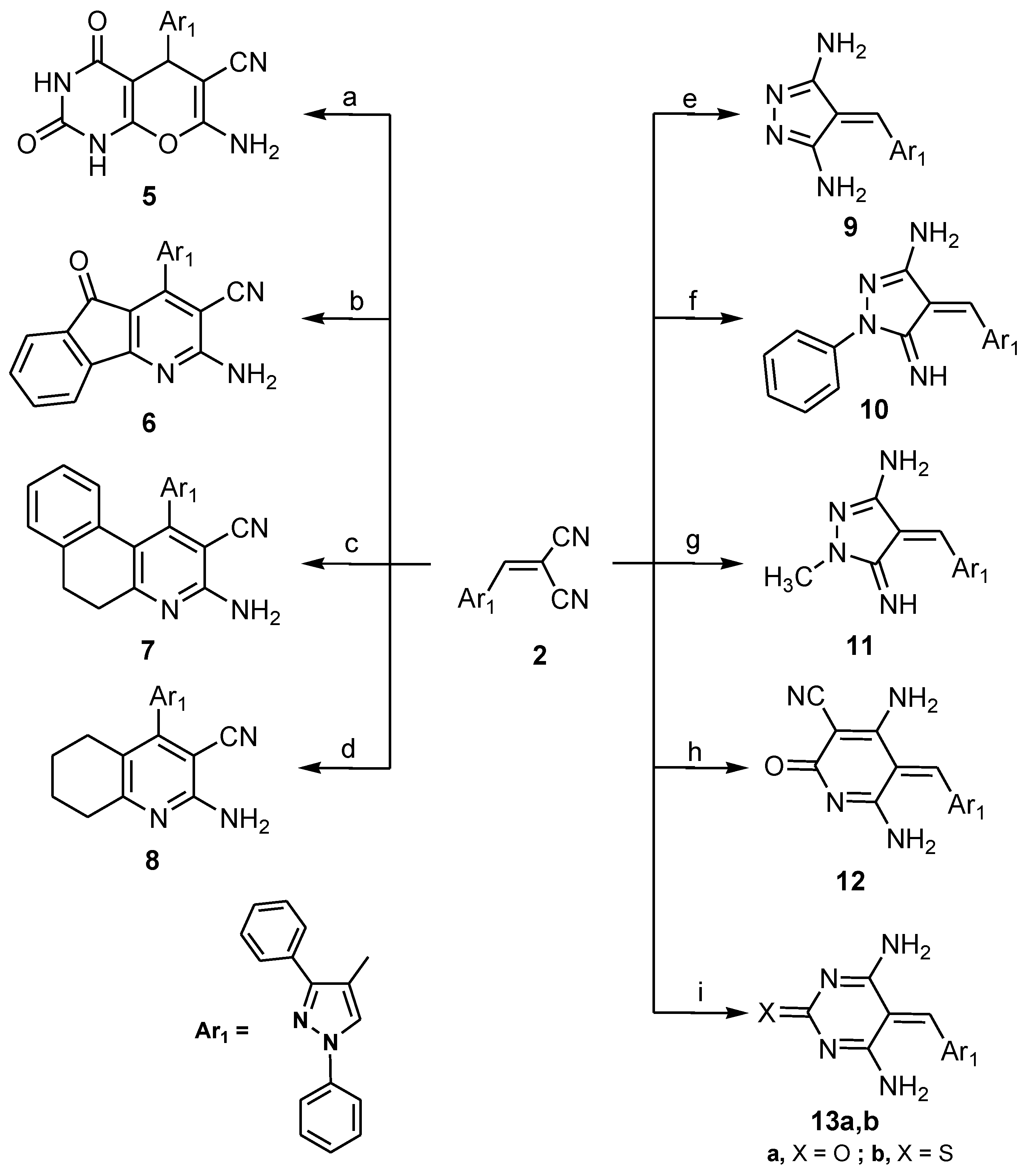
2.1. Chemistry
2.2. Biological Evaluation
2.2.1. In Vitro Cytotoxic Screening
2.2.2. Biochemical Assay (Kinase Inhibitor Activity)
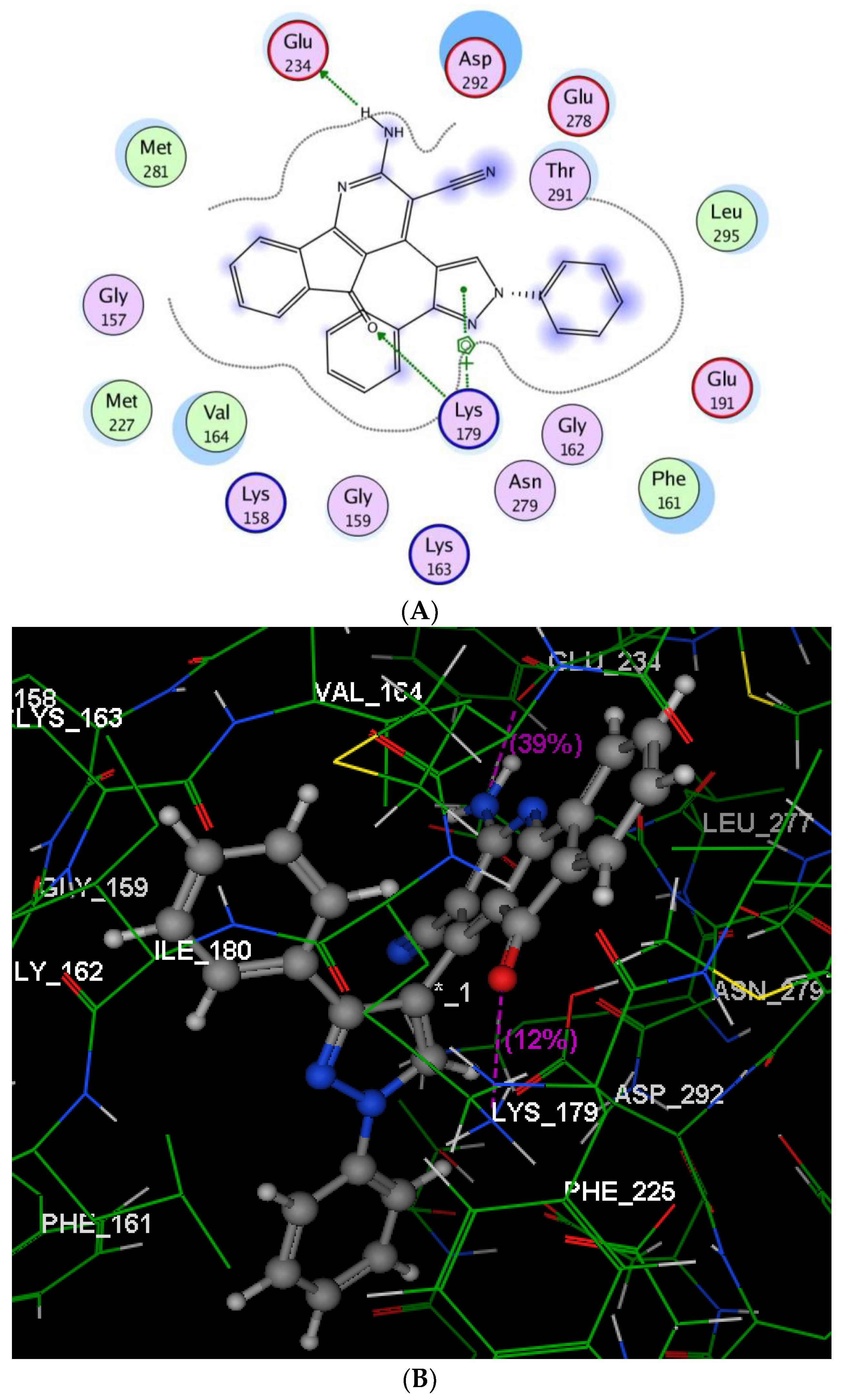
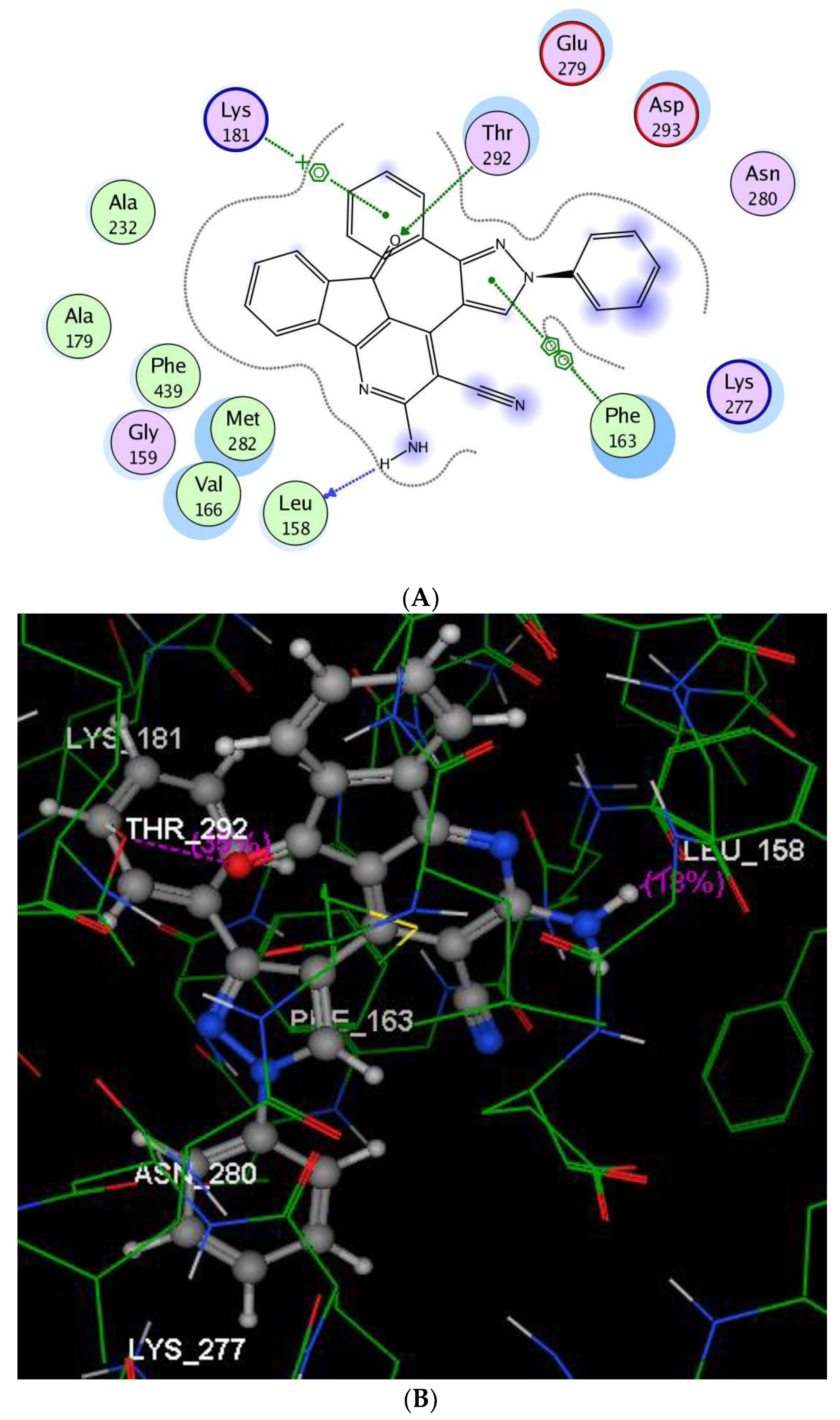
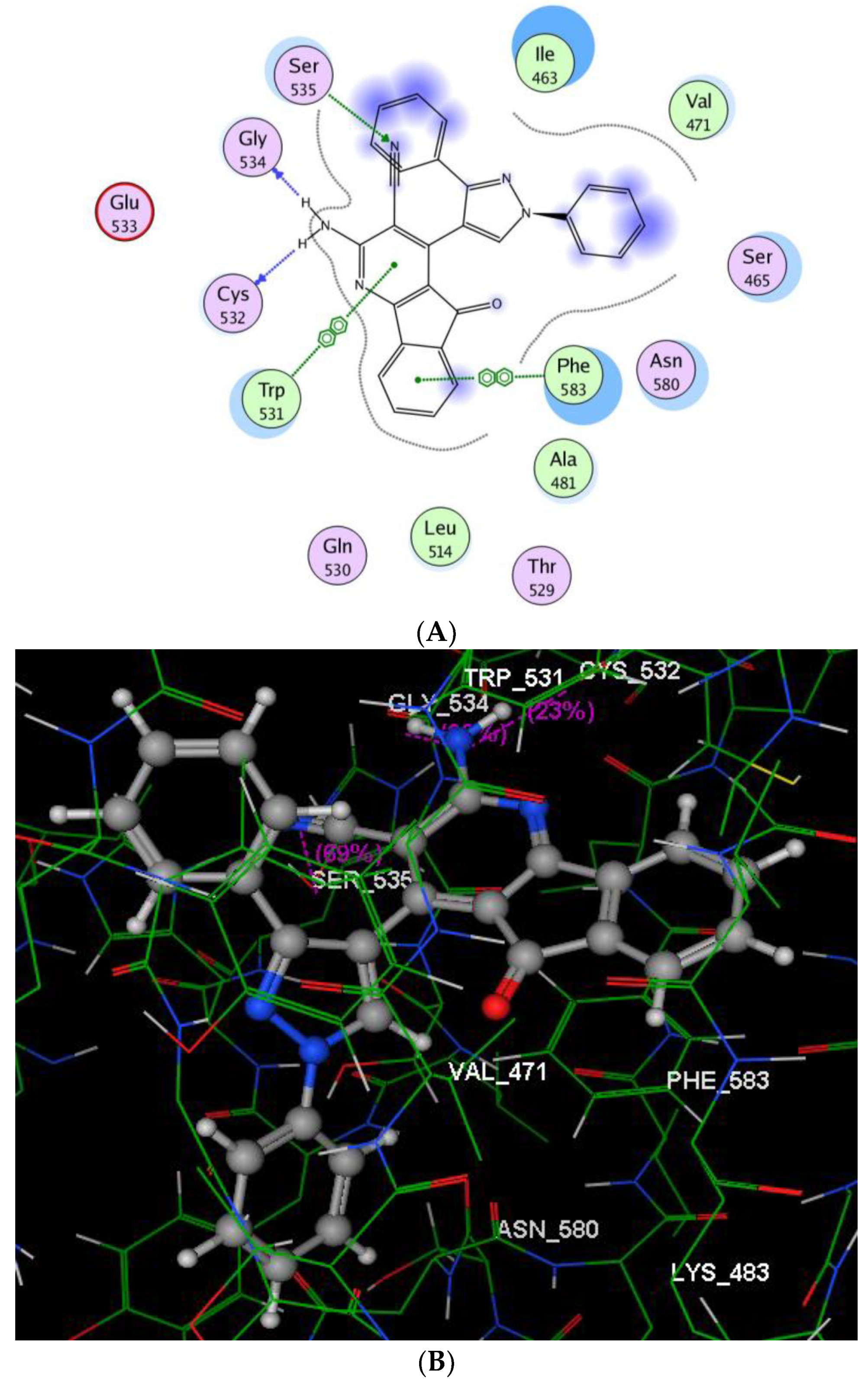
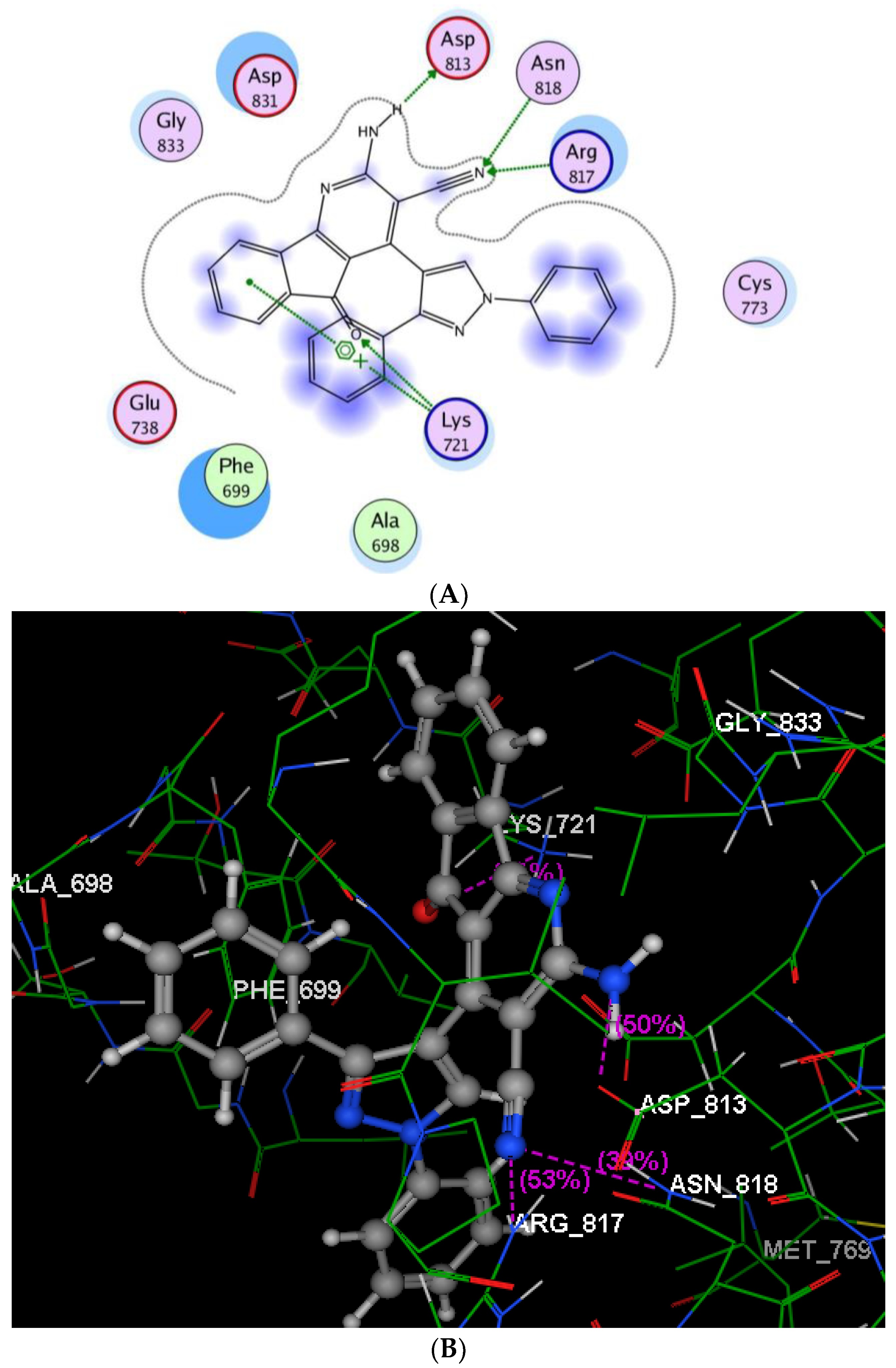
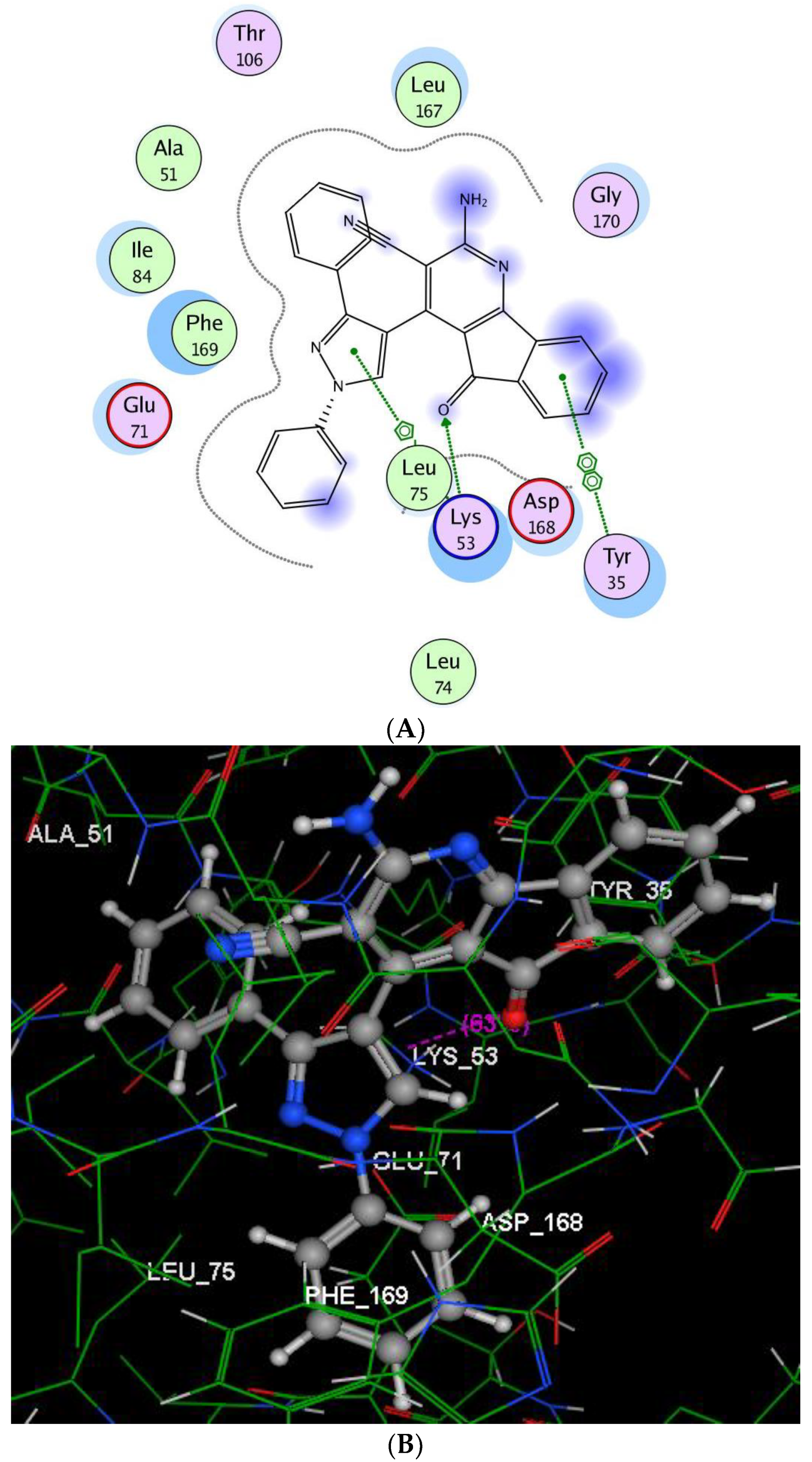
2.2.3. Molecular Modeling Studies
3. Experimental Section
3.1. General Information
3.1.1. 7-(1,3-Diphenyl-1H-pyrazol-4-yl)-5-oxo-2-substituted-1,5-dihydro-[1,2,4]triazolo[1,5-a]pyridine-6,8-Dicarbonitriles 4a–g
3.1.2. 7-Amino-5-(1,3-diphenyl-1H-pyrazol-4-yl)-2,4-dioxo-2,3,4,5-tetrahydro-1H-pyrano-[2,3-d]pyrimidine-6-carbonitrile (5)
3.1.3. 2-Amino-4-(1,3-diphenyl-1H-pyrazol-4-yl)-5-oxo-5H-indeno[1,2-b]pyridine-3-carbonitrile (6), 3-amino-1-(1,3-diphenyl-1H-pyrazol-4-yl)-5,6-dihydrobenzo[f] quinoline-2-carbonitrile (7) and 2-amino-4-(1,3-diphenyl-1H-pyrazol-4-yl)-5,6,7,8-tetrahydroquinoline-3-carbonitrile (8)
3.1.4. 4-((1,3-Diphenyl-1H-pyrazol-4-yl)methylene)-4H-pyrazole-3,5-diamine (9), 4-((1,3-diphenyl-1H-pyrazol-4-yl)methylene)-5-imino-1-phenyl-4,5-dihydro-1H-pyrazol-3-amine (10) and 4-((1,3-diphenyl-1H-pyrazol-4-yl)methylene)-5-imino-1-methyl-4,5-dihydro-1H-pyrazol-3-amine (11)
3.1.5. 4,6-Diamino-5-((1,3-diphenyl-1H-pyrazol-4-yl)methylene)-2-oxo-2,5-dihydropyridine-3-carbonitrile (12) and 4,6-Diamino-5-((1,3-diphenyl-1H-pyrazol-4-yl)methylene)pyrimidin-2(5H)-one/thione (13a,b)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [PubMed]
- Collins, K.; Jacks, T.; Pavletich, N.P. The cell cycle and cancer. Proc. Natl. Acad. Sci. USA 1997, 94, 2776–2778. [Google Scholar] [CrossRef] [PubMed]
- Grant, S. Therapeutic protein kinase inhibitors. Cell. Mol. Life Sci. 2009, 66, 1163–1177. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Adjei, A. Novel agents on the horizon for cancer therapy. CA Cancer J. Clin. 2009, 59, 111–137. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yang, P.L.; Gray, N.S. Targeting cancer with small molecule kinase inhibitors. Nat. Rev. Cancer 2009, 9, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Cassinelli, G.; Zuco, V.; Gatti, L.; Lanzi, C.; Zaffaroni, N.; Colombo, D.; Perego, P. Targeting the Akt kinase to modulate survival, invasiveness and drug resistance of cancer cells. Curr. Med. Chem. 2013, 20, 1923–1945. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.J.; Reddy, E.P. Targeted inhibition of kinases in cancer therapy. Mt. Sinai J. Med. 2010, 77, 573–586. [Google Scholar] [CrossRef] [PubMed]
- Omar, H.A.; Sargeant, A.M.; Weng, J.R.; Wang, D.; Kulp, S.K.; Patel, T.; Chen, C.S. Targeting of the Akt-nuclear factor-kappa B signaling network by [1-(4-chloro-3-nitrobenzenesulfonyl)-1H-indol-3-yl]-methanol (OSU-A9), a novel indole-3-carbinol derivative, in a mouse model of hepatocellular carcinoma. Mol. Pharmacol. 2009, 76, 957–968. [Google Scholar] [CrossRef] [PubMed]
- Housman, G.; Byler, S.; Heerboth, S.; Lapinska, K.; Longacre, M.; Snyder, N.; Sarkar, S. Drug resistance in cancer: An overview. Cancers 2014, 6, 1769–1792. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, M.J.; Siddiqui, A.A.; Khan, A.A.; Ali, Z.; Dewangan, R.P.; Pasha, S.; Yar, M.S. Design, synthesis, docking and QSAR study of substituted benzimidazole linked oxadiazole as cytotoxic agents, EGFR and erbB2 receptor inhibitors. Eur. J. Med. Chem. 2017, 126, 853–869. [Google Scholar] [CrossRef] [PubMed]
- Regad, T.; Targeting, R.T.K. Signaling Pathways in Cancer. Cancers 2015, 7, 1758–1784. [Google Scholar] [CrossRef] [PubMed]
- Hansen, J.D.; Grina, J.; Newhouse, B.; Welch, M.; Topalov, G.; Littman, N.; Callejo, M.; Gloor, S.; Martinson, M.; Laird, E.; et al. Potent and selective pyrazole-based inhibitors of B-Raf kinase. Bioorg. Med. Chem. Lett. 2008, 18, 4692–4695. [Google Scholar] [CrossRef] [PubMed]
- Plé, P.A.; Jung, F.; Ashton, S.; Hennequin, L.; Laine, R.; Morgentin, R.; Pasquet, G.; Taylor, S. Discovery of AZD2932, a new Quinazoline Ether Inhibitor with high affinity for VEGFR-2 and PDGFR tyrosine kinases. Bioorg. Med. Chem. Lett. 2011, 22, 262–266. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Xing, M.; Zhao, T.; Ren, Y.; Yang, X.; Yang, Y.; Lv, P.; Zhu, H. Synthesis, molecular modeling and biological evaluation of cinnamic acid derivatives with pyrazole moieties as novel anticancer agents. RSC Adv. 2014, 4, 37197–37207. [Google Scholar] [CrossRef]
- Grimshaw, K.M.; Hunter, L.K.; Yap, T.A.; Heaton, S.P.; Walton, M.I.; Woodhead, S.; Fazal, L.; Reule, M.; Davies, T.G.; Seavers, L.C.; et al. AT7867 Is a potent and oral inhibitor of AKT and p70 S6 kinase that induces pharmacodynamicchanges and inhibits human tumor xenograftgrowth. Mol. Cancer Ther. 2010, 9, 1100–1110. [Google Scholar] [CrossRef] [PubMed]
- Graneto, M.J.; Kurumbail, R.G.; Vazquez, M.L.; Shieh, H.-S.; Pawlitz, J.L.; Williams, J.M.; Stallings, W.C.; Geng, L.; Naraian, A.S.; Koszyk, F.J.; et al. Synthesis, crystal structure, and activity of pyrazole-based inhibitors of p38 kinase. J. Med. Chem. 2007, 50, 5712–5719. [Google Scholar] [CrossRef] [PubMed]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Nossier, E.S.; El-Hallouty, S.M.; Zaki, E.R. Synthesis, anticancer evaluation and molecular modeling of some substituted thiazolidinonyl and thiazolyl pyrazole derivatives. Int. J. Pharm. Sci. 2015, 7, 353–359. [Google Scholar]
- El-Serwy, W.S.; Mohamed, N.A.; Nossier, E.S.; Mahmoud, K. Synthesis, molecular modeling studies and biological evaluation of novel pyrazole as antitumor and EGFR inhibitors. Int. J. Pharm. Technol. 2016, 8, 25192–25209. [Google Scholar]
- Elzahabi, H.S.A.; Nossier, E.S.; Khalifa, N.M.; Alasfoury, R.A.; El-Manawaty, M.A. Anticancer evaluation and molecular modeling of multi-targeted kinase inhibitors based pyrido[2,3-d]pyrimidine scaffold. J. Enzyme Inhib. Med. Chem. 2018, 33, 546–557. [Google Scholar] [CrossRef] [PubMed]
- Molecular Operating Environment (MOE). Chemical Computing Group ULC, 1010 Sherbrooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7. 2008. Available online: https://www.chemcomp.com/MOEMolecular_Operating_Environment.htm.
- Addie, M.; Ballard, P.; Buttar, D.; Crafter, C.; Currie, G.; Davies, B.R.; Debreczeni, J.; Dry, H.; Dudley, P.; Greenwood, R.; et al. Discovery of 4-Amino-N-[(1S)-1-(4-chlorophenyl)-3-hydroxypropyl]-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidine-4-carboxamide (AZD5363), an Orally Bioavailable, Potent Inhibitor of Akt Kinases. J. Med. Chem. 2013, 56, 2059–2073. [Google Scholar] [CrossRef] [PubMed]
- Davies, T.G.; Verdonk, M.L.; Graham, B.; Saalau-Bethell, S.; Hamlett, C.C.F.; Mchardy, T.; Collins, I.; Garrett, M.D.; Workman, P.; Woodhead, S.J.; et al. A structural comparison of inhibitor binding to Pkb, Pka and Pka-Pkb chimera. J. Mol. Biol. 2007, 367, 882–894. [Google Scholar] [CrossRef] [PubMed]
- Stamos, J.; Sliwkowski, M.X.; Eigenbrot, C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J. Biol. Chem. 2002, 277, 46265–46272. [Google Scholar] [CrossRef] [PubMed]
- Vogtherr, M.; Saxena, K.; Hoelder, S.; Grimme, S.; Betz, M.; Schieborr, U.; Pescatore, B.; Robin, M.; Delarbre, L.; Langer, T.; et al. NMR characterization of kinase p38 dynamics in free and ligand-bound forms. Angew. Chem. Int. Ed. Engl. 2006, 45, 993–997. [Google Scholar] [CrossRef] [PubMed]
- Conconi, M.T.; Marzaro, G.; Urbani, L.; Zanusso, I.; Di Liddo, R.; Castagliuolo, I.; Brun, P.; Tonus, F.; Ferrarese, A.; Guiotto, A.; et al. Quinazoline-based multi-tyrosine kinase inhibitors: Synthesis, modeling, antitumor and antiangiogenic properties. Eur. J. Med. Chem. 2013, 67, 373–383. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: All the compounds are available from the authors. |
| Compound | Growth Inhibition (%) | ||||
|---|---|---|---|---|---|
| HepG-2 | MCF-7 | PC-3 | A-549 | HCT-116 | |
| 3 | 95.7 | 70.4 | 98.2 | 93.7 | 82.1 |
| 4a | 98.2 | 99.1 | 98.8 | 95.6 | 86.9 |
| 4b | 7.7 | 28.3 | 38.2 | 0 | 0 |
| 4c | 0 | 0 | 32.2 | 0 | 47.9 |
| 4d | 0 | 0 | 25.4 | 0 | 35.4 |
| 4e | 0 | 0 | 29.8 | 11.9 | 6.79 |
| 4f | 32.4 | 33.7 | 89.4 | 64.3 | 89.9 |
| 4g | 0 | 0 | 27 | 0 | 0 |
| 5 | 2.9 | 8.6 | 91.2 | 80 | 94.8 |
| 6 | 15.01 | 95.9 | 10.7 | 90.7 | 92.2 |
| 7 | 4.8 | 0 | 33.7 | 0 | 0 |
| 8 | 46.4 | 70.6 | 66.7 | 34.5 | 0 |
| 9 | 62.8 | 92.6 | 89.6 | 65.8 | 67.8 |
| 10 | - | - | 53.8 | - | - |
| 11 | 97.8 | 98.03 | 99.5 | 0 | 92.8 |
| 12a | 95.9 | 95 | 99.1 | 79.3 | 80.4 |
| 12b | - | - | 96 | - | - |
| 13a | 99.2 | 99.3 | 94.5 | 82.8 | 95.9 |
| 13b | 82.6 | 97 | 98.3 | 83.6 | 84 |
| Negative control | 0 | 0 | 0 | 0 | 0 |
| Doxorubicin | 100 | 100 | 100 | 100 | 100 |
| Compounds | IC50 (μM) | ||||
|---|---|---|---|---|---|
| HepG-2 | MCF-7 | PC-3 | A-549 | HCT-116 | |
| 3 | 29.23 | - | 18.81 | 33.07 | - |
| 4a | 25.13 | 12.00 | 98.23 | 108.20 | - |
| 4f | - | - | 105.41 | - | - |
| 5 | - | - | 55.61 | - | 112.86 |
| 6 | - | 6.53 | - | 26.40 | 59.84 |
| 8 | - | 105.14 | - | - | - |
| 9 | - | 87.65 | - | - | - |
| 11 | 104.56 | 18.22 | 104.56 | 97.84 | 141.65 |
| 12a | 39.96 | 36.67 | 78.34 | - | - |
| 12b | - | - | 26.81 | - | - |
| 13a | 28.40 | 48.49 | 67.06 | - | 154.89 |
| 13b | - | 65.38 | 73.57 | - | - |
| Doxorubicin | 37.8 ± 1.50 | 45.0 ± 2.20 | 41.1 ± 2.01 | 48.8 ± 1.30 | 65.1 ± 1.00 |
| Kinase | Compound 6 |
|---|---|
| % Inhibition | |
| AKT1 | −97 |
| AKT2 | −94 |
| BRAF (V600E) | −95 |
| CDK2/Cyclin A1 | −73 |
| CHK1 | −8 |
| EGFR | −99 |
| KDR (VEGFR) | −76 |
| p38α | −94 |
| PDGFRβ | −96 |
| PI3K (p110a/p85a) * | −47 |
| PI3K (p110b/p85a) * | −63 |
| c-RAF | 43 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nossier, E.S.; Abd El-Karim, S.S.; Khalifa, N.M.; El-Sayed, A.S.; Hassan, E.S.I.; El-Hallouty, S.M. Kinase Inhibitory Activities and Molecular Docking of a Novel Series of Anticancer Pyrazole Derivatives. Molecules 2018, 23, 3074. https://doi.org/10.3390/molecules23123074
Nossier ES, Abd El-Karim SS, Khalifa NM, El-Sayed AS, Hassan ESI, El-Hallouty SM. Kinase Inhibitory Activities and Molecular Docking of a Novel Series of Anticancer Pyrazole Derivatives. Molecules. 2018; 23(12):3074. https://doi.org/10.3390/molecules23123074
Chicago/Turabian StyleNossier, Eman S., Somaia S. Abd El-Karim, Nagy M. Khalifa, Ali S. El-Sayed, Emad S. I. Hassan, and Salwa M. El-Hallouty. 2018. "Kinase Inhibitory Activities and Molecular Docking of a Novel Series of Anticancer Pyrazole Derivatives" Molecules 23, no. 12: 3074. https://doi.org/10.3390/molecules23123074
APA StyleNossier, E. S., Abd El-Karim, S. S., Khalifa, N. M., El-Sayed, A. S., Hassan, E. S. I., & El-Hallouty, S. M. (2018). Kinase Inhibitory Activities and Molecular Docking of a Novel Series of Anticancer Pyrazole Derivatives. Molecules, 23(12), 3074. https://doi.org/10.3390/molecules23123074





