Natalenamides A–C, Cyclic Tripeptides from the Termite-Associated Actinomadura sp. RB99
Abstract
1. Introduction
2. Results and Discussion
2.1. Liquid chromatography (LC)/Ultraviolet (UV)/Mass Spectrometry (MS)-Based Isolation of Compounds 1–3
2.2. Structural Elucidation of the Compounds
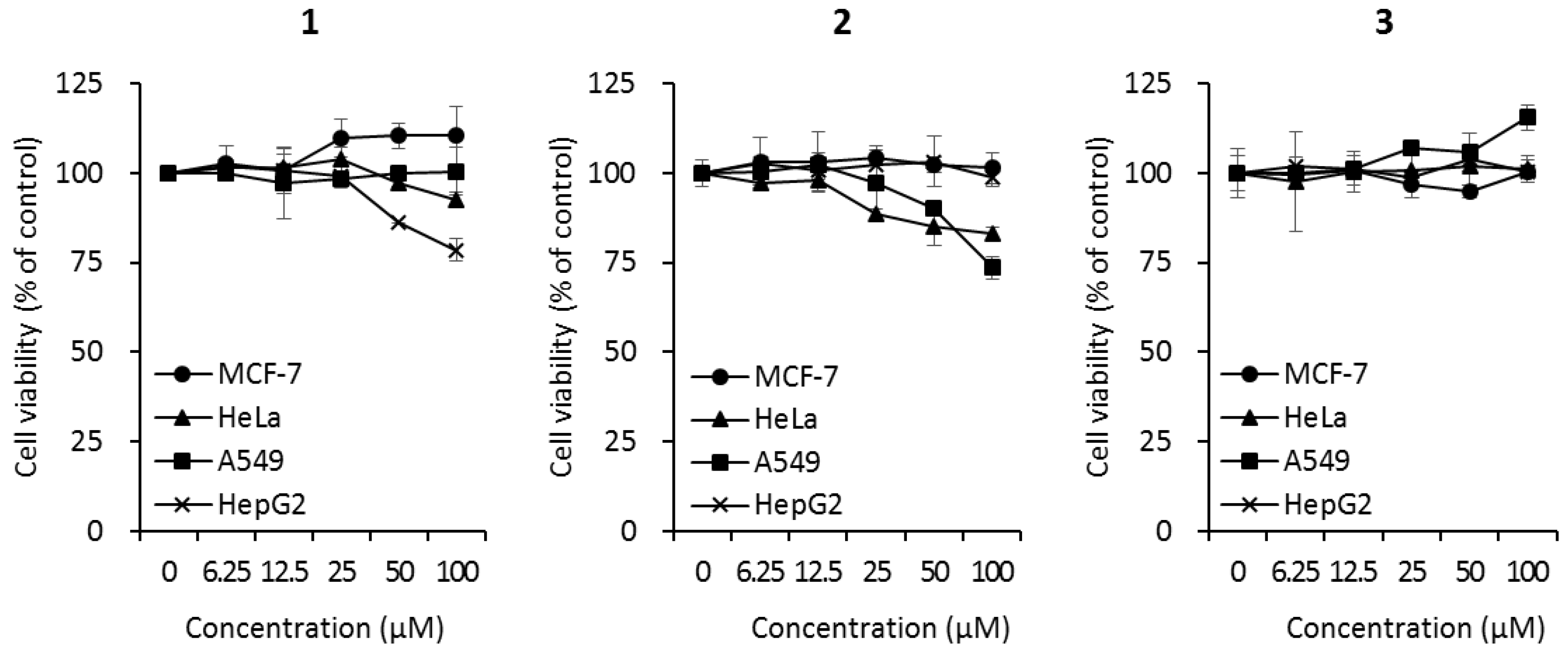
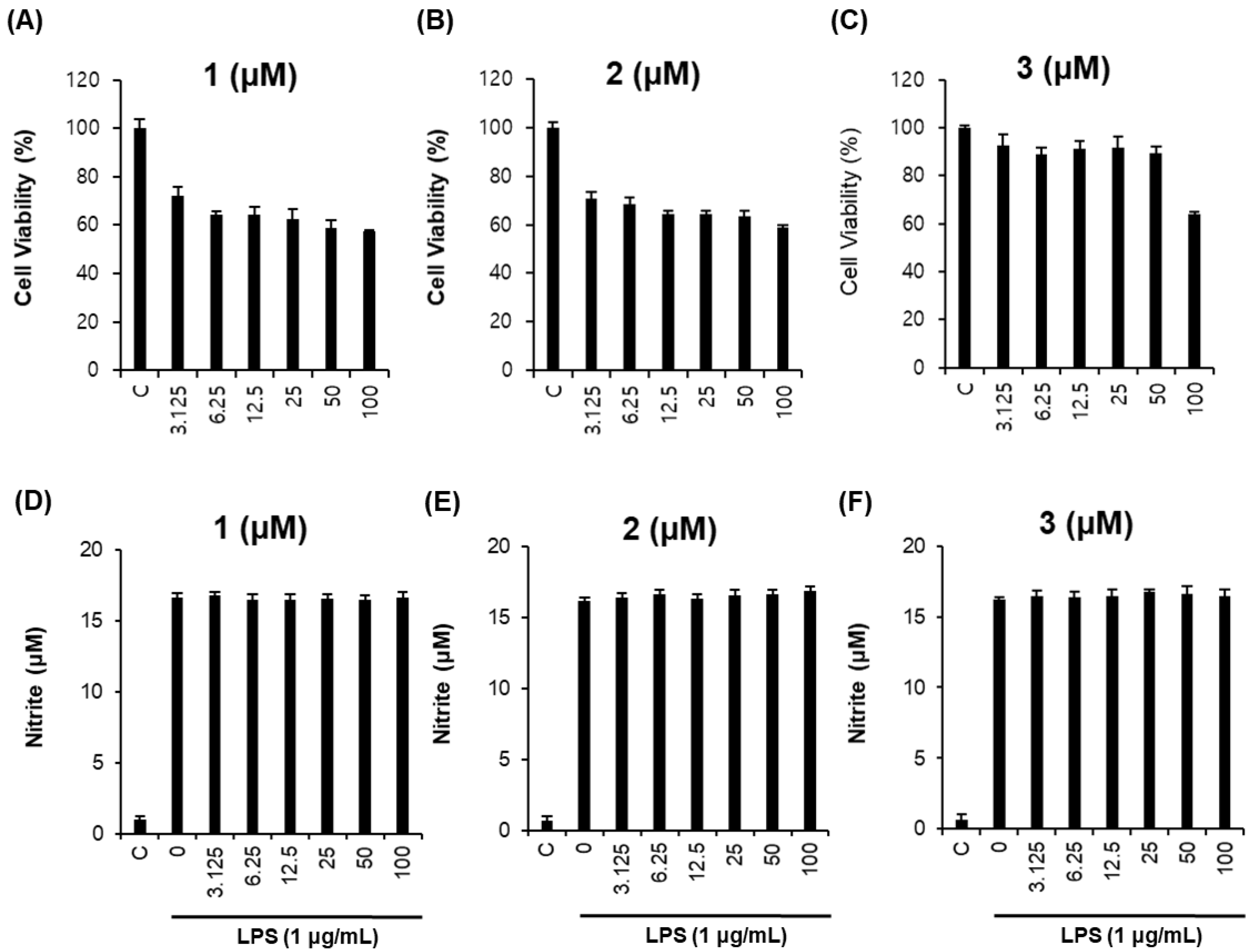
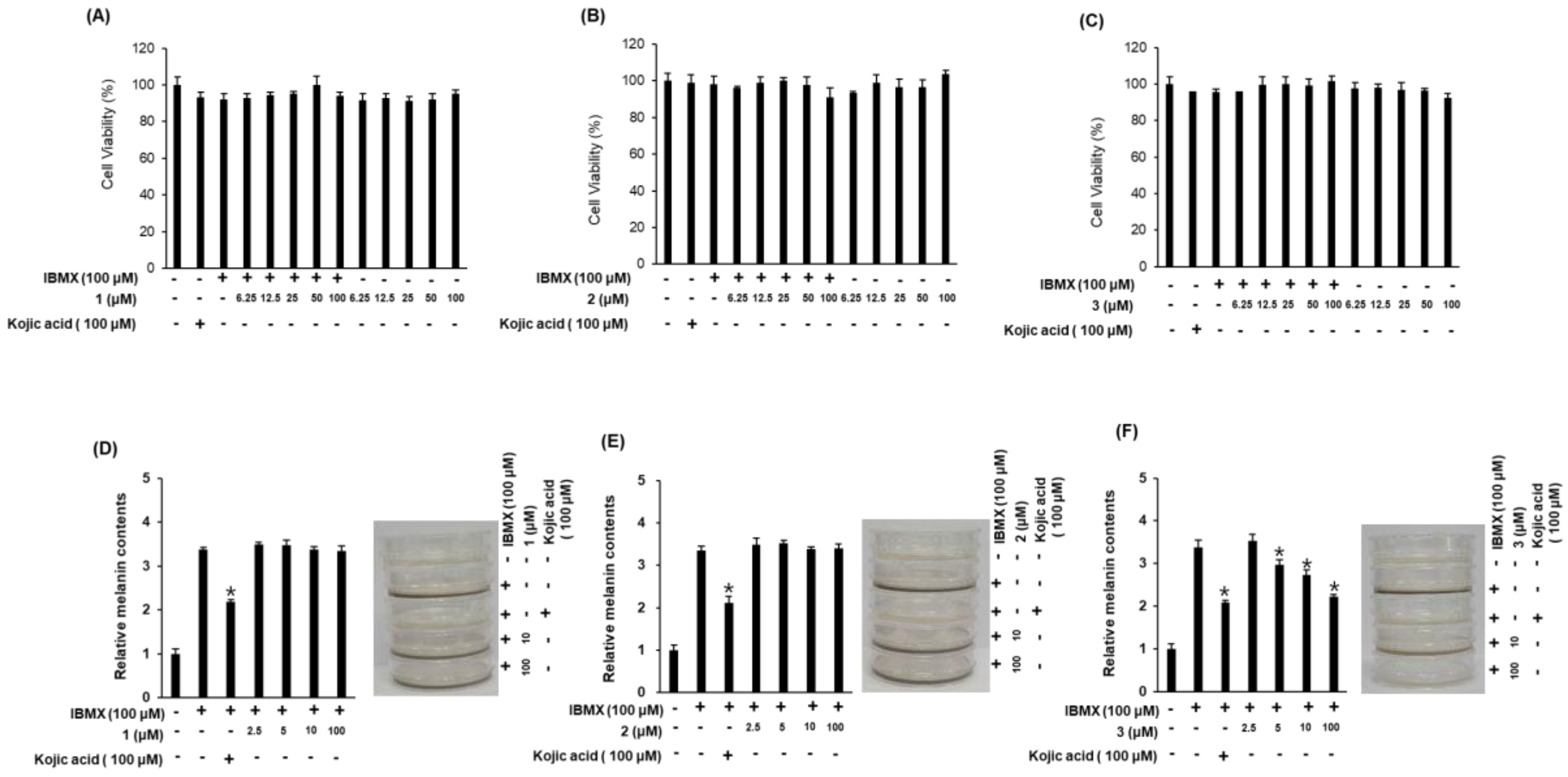
2.3. Biological Activities of the Compounds 1–3
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Microbial Material
3.3. DNA Extraction and Polymerase Chain Reaction Amplification
3.4. Sequencing and Species Identification
3.5. Extraction and Isolation
3.5.1. Natalenamide A (1)
3.5.2. Natalenamide B (2)
3.5.3. Natalenamide C (3)
3.6. Acid Hydrolysis of Compounds 1–3
3.7. Determination of the Absolute Configuration of Amino Acids in 1–3
3.8. Cytotoxic Assays Using Cancer Cells
3.9. Anti-Inflammatory Activity
3.10. Measurement of Melanin Content
3.11. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [PubMed]
- Mishra, B.B.; Tiwari, V.K. Natural products: An evolving role in future drug discovery. Eur. J. Med. Chem. 2011, 46, 4769–4807. [Google Scholar] [CrossRef] [PubMed]
- Beemelmanns, C.; Guo, H.; Rischer, M.; Poulsen, M. Natural products from microbes associated with insects. Beilstein J. Org. Chem. 2016, 12, 314–327. [Google Scholar] [CrossRef] [PubMed]
- Ramadhar, T.R.; Beemelmanns, C.; Currie, C.R.; Clardy, J. Bacterial symbionts in agricultural systems provide a strategic source for antibiotic discovery. J. Antibiot. 2014, 67, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Harvey, A.L.; Edrada-Ebel, R.; Quinn, R.J. The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discov. 2015, 14, 111–129. [Google Scholar] [CrossRef] [PubMed]
- Sattely, E.S.; Fischbachb, M.A.; Walsh, C.T. Total biosynthesis: In vitro reconstitution of polyketide and nonribosomal peptide pathways. Nat. Prod. Rep. 2008, 25, 757–793. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.; Scott, J.J.; Currie, C.R.; Clardy, J. Mycangimycin, a polyene peroxide from a mutualist Streptomyces sp. Org. Lett. 2009, 11, 633–636. [Google Scholar] [CrossRef] [PubMed]
- Sit, C.S.; Ruzzini, A.C.; Van Arnam, E.B.; Ramadhar, T.R.; Currie, C.R.; Clardy, J. Variable genetic architectures produce virtually identical molecules in bacterial symbionts of fungus-growing ants. Proc. Natl. Acad. Sci. USA 2015, 112, 13150–13154. [Google Scholar] [CrossRef] [PubMed]
- Low, K.E.; Ler, S.; Chen, K.J.; Campbell, R.L.; Hickey, J.L.; Tan, J.; Scully, C.C.G.; Davies, R.L.; Yudin, A.K.; Zaretsky, S. Rational design of calpain inhibitors based on calpastatin peptidomimetics. J. Med. Chem. 2016, 59, 5403–5415. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Xu, Z.; Wang, Y.; Hong, K.; Liu, P.; Zhu, W. Cyclic Tripeptides from the halotolerant fungus Aspergillus sclerotiorum PT06-1. J. Nat. Prod. 2010, 73, 1133–1137. [Google Scholar] [CrossRef] [PubMed]
- Weerakkody, D.; Moshnikova, A.; El-Sayed, N.S.; Adochite, R.C.; Slaybaugh, G.; Golijanin, J.; Tiwari, R.K.; Andreev, O.A.; Parang, K.; Reshetnyak, Y.K. Novel pH-sensitive cyclic peptides. Sci. Rep. 2016, 6, 31322. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Lin, H.C.; Tang, Y. Biosynthesis of the α-nitro-containing cyclic tripeptide psychrophilin. J. Antibiot. 2016, 69, 571–573. [Google Scholar] [CrossRef] [PubMed]
- Zorzi, A.; Deyle, K.; Heinis, C. Cyclic peptide therapeutics: Past, present and future. Curr. Opin. Chem. Biol. 2017, 38, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Ramadhar, T.R.; Beemelmanns, C.; Cao, S.; Poulsen, M.; Currie, C.R.; Clardy, J. Natalamycin A, an ansamycin from a termite-associated Streptomyces sp. Chem. Sci. 2014, 5, 4333–4338. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.R.; Lee, D.; Benndorf, R.; Jung, W.H.; Beemelmanns, C.; Kang, K.S.; Kim, K.H. Termisoflavones A–C, isoflavonoid glycosides from termite-associated Streptomyces sp. RB1. J. Nat. Prod. 2016, 79, 3072–3078. [Google Scholar] [CrossRef] [PubMed]
- Beemelmanns, C.; Ramadhar, T.R.; Kim, K.H.; Klassen, J.L.; Cao, S.; Wyche, T.P.; Hou, Y.; Poulsen, M.; Bugni, T.S.; Currie, C.R.; et al. Macrotermycins A–D, glycosylated macrolactams from a termite-associated Amycolatopsis sp. M39. Org. Lett. 2017, 19, 1000–1003. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Benndorf, R.; Leichnitz, D.; Klassen, J.L.; Vollmers, J.; Görls, H.; Steinacker, M.; Weigel, C.; Dahse, H.M.; Kaster, A.K.; de Beer, Z.W.; et al. Isolation, biosynthesis and chemical modifications of rubterolones A–F: Rare tropolone alkaloids from Actinomadura sp. 5–2. Chem. Eur. J. 2017, 23, 9338–9345. [Google Scholar] [CrossRef] [PubMed]
- Sano, S.; Ikai, K.; Katayama, K.; Takesako, K.; Nakamura, T.; Obayashi, A.; Ezure, Y.; Enomoto, H. OF4949, new inhibitors of aminopeptidase B. II. Elucidation of Structure. J. Antibiot. 1986, 39, 1685–1696. [Google Scholar] [CrossRef] [PubMed]
- Ciasullo, L.; Casapullo, A.; Cutignano, A.; Bifulco, G.; Debitus, C.; Hooper, J.; Gomez-Paloma, L.; Riccio, R. Renieramide, a cyclic tripeptide from the vanuatu sponge Reniera n. sp. J. Nat. Prod. 2002, 65, 407–410. [Google Scholar] [CrossRef] [PubMed]
- Solano, F.; Briganti, S.; Picardo, M.; Ghanem, G.H. Hypopigmenting agents: An updated review on biological, chemical and clinical aspects. Pigm. Cell Melanoma Res. 2006, 19, 550–571. [Google Scholar] [CrossRef] [PubMed]
- Visser, A.A.; Nobre, T.; Currie, C.R.; Aanen, D.K.; Poulsen, M. Exploring the potential for actinobacteria as defensive symbionts in fungus-growing termites. Microb. Ecol. 2012, 63, 975–985. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequences alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Pruesse, E.; Peplies, J.; Glöckner, F.O. SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 2012, 28, 1823–1829. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |
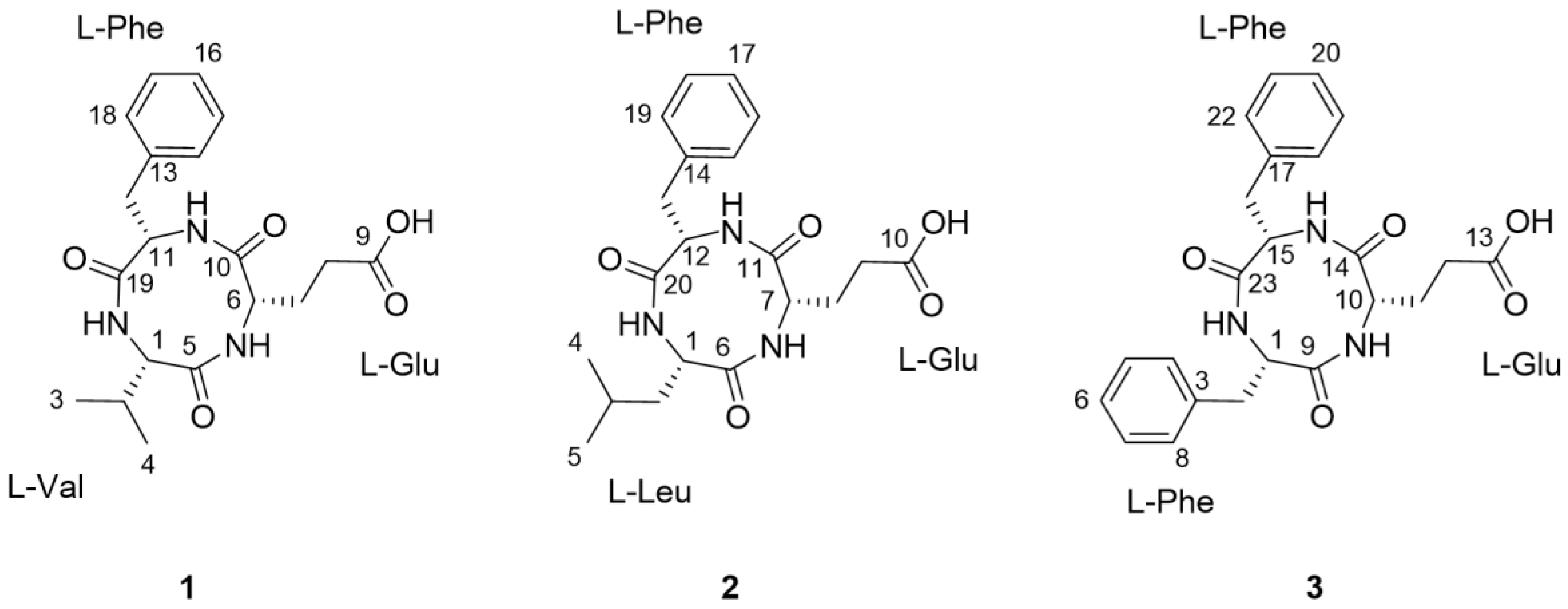
 ) and HMBC (→) correlations for compounds 1–3.
) and HMBC (→) correlations for compounds 1–3.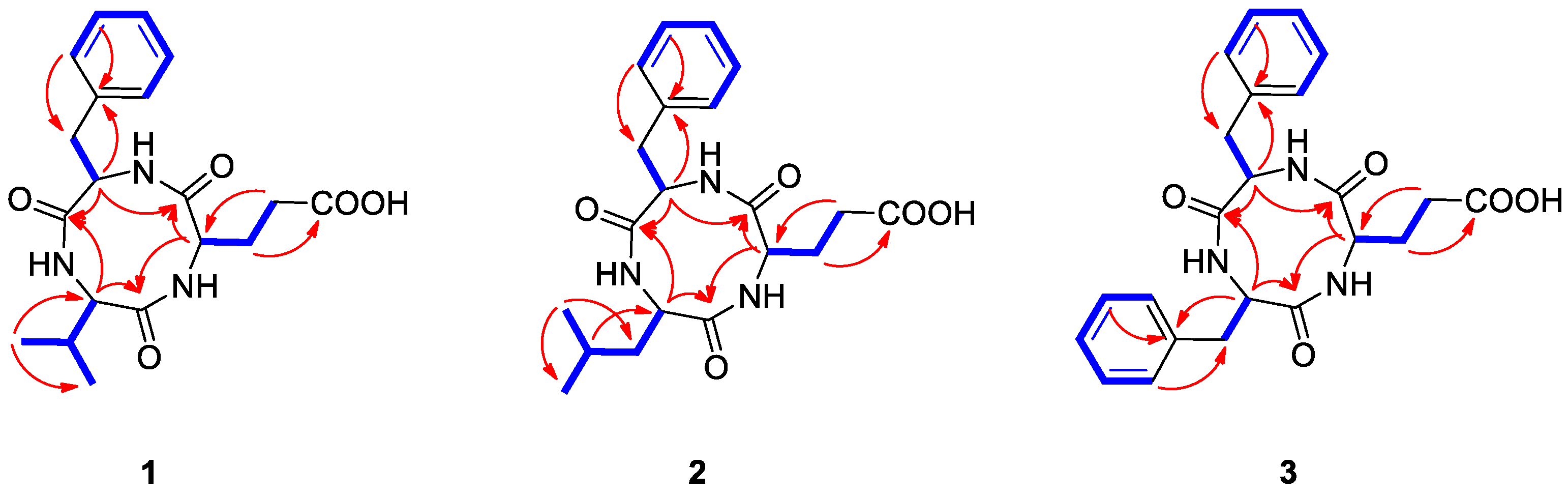
| 1 | 2 | 3 | ||||||
|---|---|---|---|---|---|---|---|---|
| Position | δC | δH (J in Hz) | Position | δC | δH (J in Hz) | Position | δC | δH (J in Hz) |
| 1 | 60.4 d | 4.16 d (7.5) | 1 | 53.1 d | 4.42 m | 1 | 56.3 d | 4.53 dd (7.5, 5.0) |
| 2 | 32.1 d | 2.02 m | 2 | 42.4 t | 1.63 m | 2a | 38.5 t | 3.02 dd (14.0, 7.5) |
| 3 | 18.9 q | 0.91 d (7.0) | 3 | 26.2 d | 1.69 m | 2b | - | 3.24 dd (14.0, 5.0) |
| 4 | 19.9 q | 0.92 d (7.0) | 4 | 22.1 q | 0.92 d (6.5) | 3 | 138.9 s | - |
| 5 | 174.8 s | - | 5 | 23.7 q | 0.95 d (6.5) | 4/8 | 130.7 d | 7.21 m |
| 6 | 58.0 d | 4.20 dd (8.5, 4.5) | 6 | 174.9 s | - | 5/7 | 129.7 d | 7.23 m |
| 7a | 27.0 t | 1.95 m | 7 | 58.1 d | 4.09 dd (9.0, 4.5) | 6 | 127.8 d | 7.17 m |
| 7b | 2.48 m | 8a | 26.9 t | 1.82 m | 9 | 175.0 s | - | |
| 8a | 30.6 t | 2.27 m | 8b | - | 2.34 m | 10 | 57.9 d | 4.04 dd (9.0, 4.5) 8 |
| 8b | 2.36 m | 9a | 30.3 t | 2.16 m | 11a | 26.6 t | 1.74 m | |
| 9 | 181.8 s | - | 9b | - | 2.19 m | 11b | - | 2.29 m |
| 10 | 173.3 s | - | 10 | 181.6 s | - | 12 | 30.0 t | 2.15 m |
| 11 | 55.6 d | 4.63 dd (8.0, 5.0) | 11 | 173.4 s | - | 13 | 181.7 s | - |
| 12a | 38.8 t | 2.97 dd (14.0, 8.0) | 12 | 55.8 d | 4.72 dd (10.0, 5.0) | 14 | 174.0 s | - |
| 12b | - | 3.20 dd (14.0, 5.0) | 13a | 38.9 t | 2.89 dd (14.0, 10.0) | 15 | 55.8 d | 4.63 dd (10.0 4.5) |
| 13 | 138.8 s | - | 13b | - | 3.23 dd (14.0, 5.0) | 16a | 38.4 t | 2.80 dd (14.0, 10.0) |
| 14/18 | 130.6 d | 7.23 m | 14 | 138.6 s | - | 16b | - | 3.23 dd (14.0, 4.5) |
| 15/17 | 129.5 d | 7.24 m | 15/19 | 130.5 d | 7.21 m | 17 | 138.4 s | - |
| 16 | 127.8 d | 7.18 m | 16/18 | 129.4 d | 7.25 m | 18/22 | 130.2 d | 7.21 m |
| 19 | 173.2 s | - | 17 | 127.6 d | 7.19 m | 19/21 | 129.3 d | 7.23 m |
| - | - | - | 20 | 173.3 s | - | 20 | 127.4 d | 7.17 m |
| - | - | - | - | - | - | 23 | 173.8 s | - |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.R.; Lee, D.; Yu, J.S.; Benndorf, R.; Lee, S.; Lee, D.-S.; Huh, J.; De Beer, Z.W.; Kim, Y.H.; Beemelmanns, C.; et al. Natalenamides A–C, Cyclic Tripeptides from the Termite-Associated Actinomadura sp. RB99. Molecules 2018, 23, 3003. https://doi.org/10.3390/molecules23113003
Lee SR, Lee D, Yu JS, Benndorf R, Lee S, Lee D-S, Huh J, De Beer ZW, Kim YH, Beemelmanns C, et al. Natalenamides A–C, Cyclic Tripeptides from the Termite-Associated Actinomadura sp. RB99. Molecules. 2018; 23(11):3003. https://doi.org/10.3390/molecules23113003
Chicago/Turabian StyleLee, Seoung Rak, Dahae Lee, Jae Sik Yu, René Benndorf, Sullim Lee, Dong-Soo Lee, Jungmoo Huh, Z. Wilhelm De Beer, Yong Ho Kim, Christine Beemelmanns, and et al. 2018. "Natalenamides A–C, Cyclic Tripeptides from the Termite-Associated Actinomadura sp. RB99" Molecules 23, no. 11: 3003. https://doi.org/10.3390/molecules23113003
APA StyleLee, S. R., Lee, D., Yu, J. S., Benndorf, R., Lee, S., Lee, D.-S., Huh, J., De Beer, Z. W., Kim, Y. H., Beemelmanns, C., Kang, K. S., & Kim, K. H. (2018). Natalenamides A–C, Cyclic Tripeptides from the Termite-Associated Actinomadura sp. RB99. Molecules, 23(11), 3003. https://doi.org/10.3390/molecules23113003











