Flubendiamide Enhances Adipogenesis and Inhibits AMPKα in 3T3-L1 Adipocytes
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. 3T3-L1 Culture
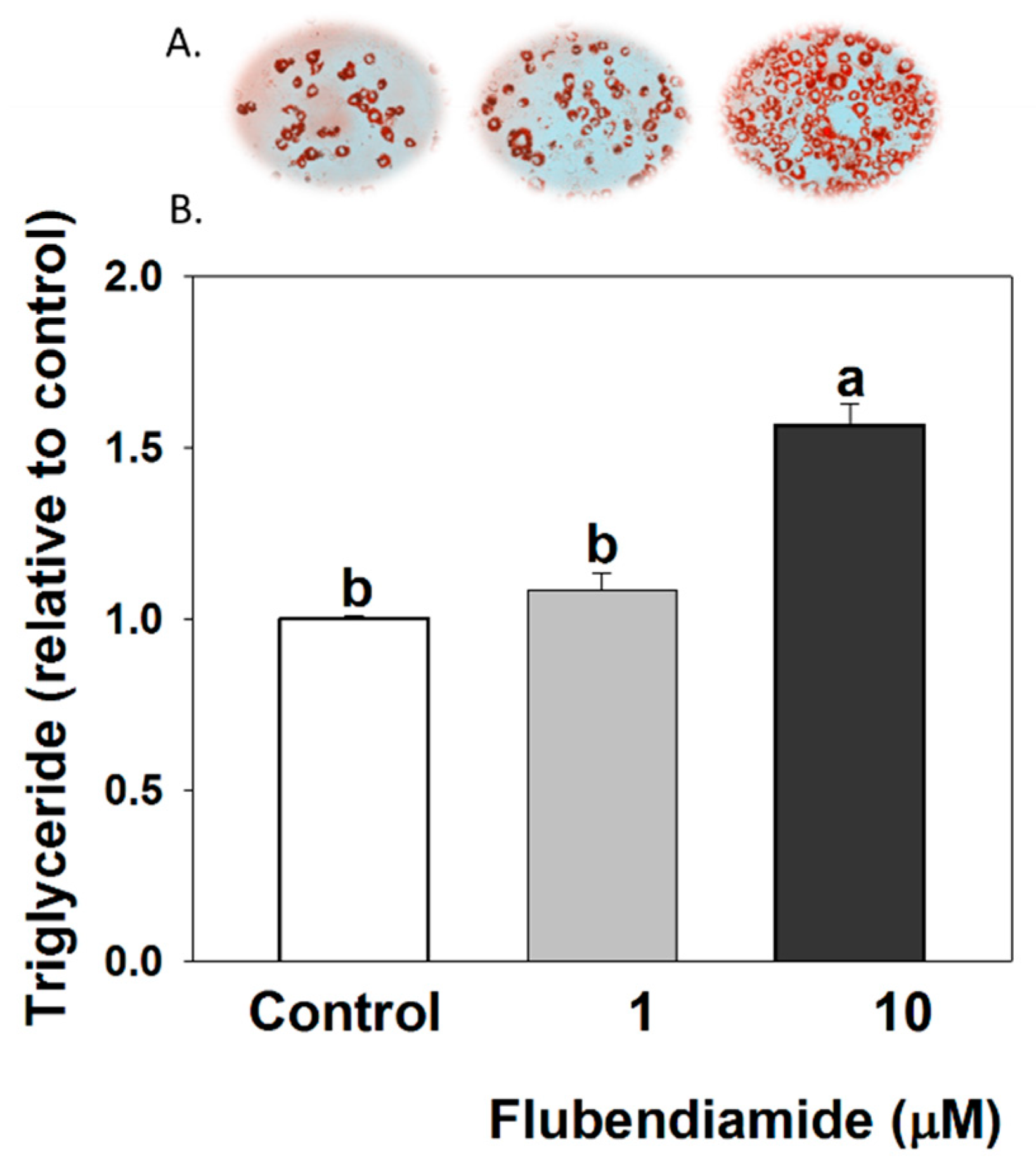
2.3. Oil Red O Staining
2.4. Triglyceride Quantification
2.5. Immunoblotting
2.6. Statistical Analyses
3. Results
3.1. Triglyceride Measurements in 3T3-L1 Adipocytes
3.2. Influence of Flubendiamide on the Protein Expression of Regulators Controlling Adipocyte Differentiation and Lipid Metabolism
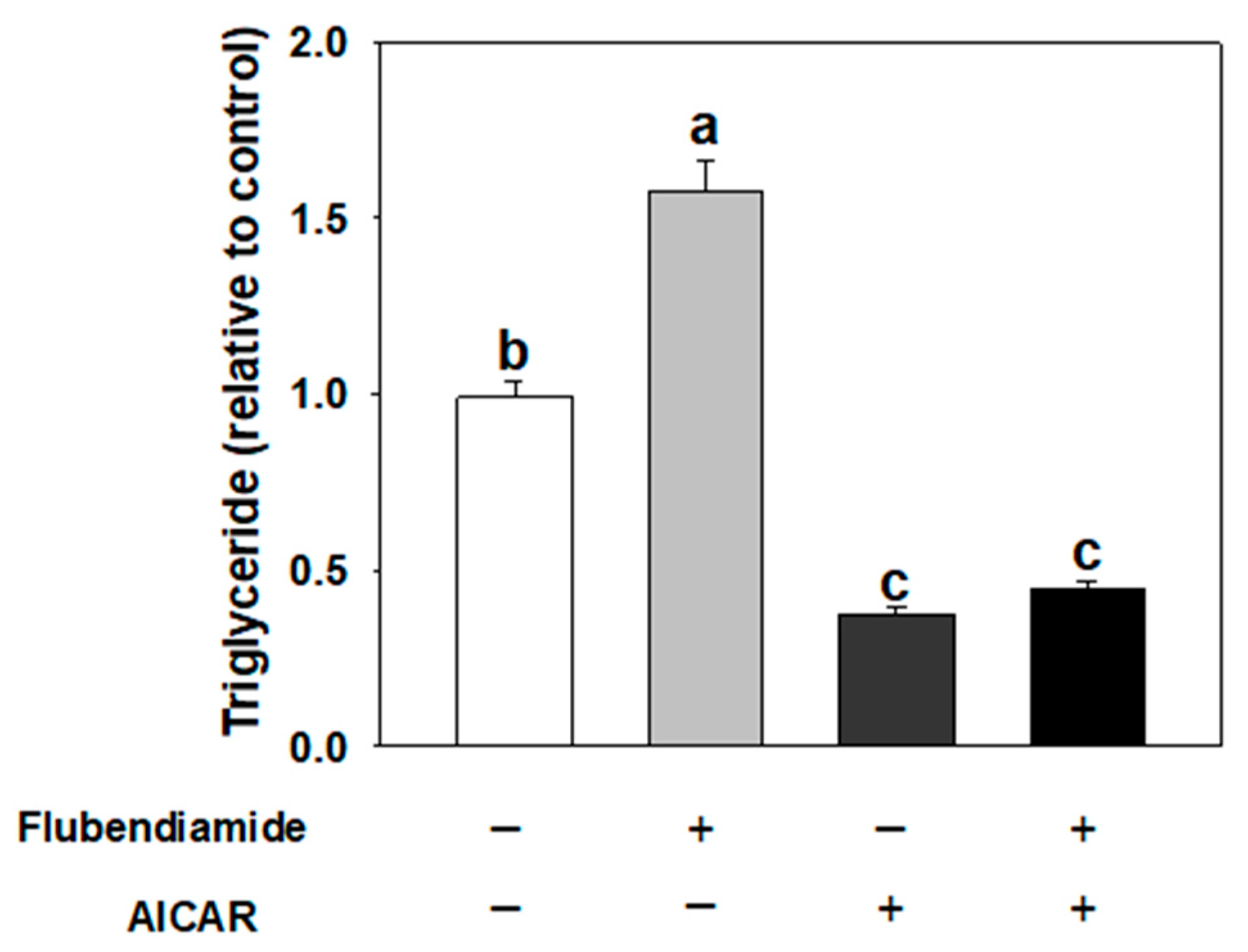
3.3. Effect of AICAR on Adipogenesis Induced by Flubendiamide
3.4. Influence of AICAR on Protein Expression of Regulators Controlling Adipogenesis
3.5. Influence of A769662 on Enhanced Adipogenesis Induced by Flubendiamide
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hotamisligil, G.S. Inflammation and metabolic disorders. Nature 2006, 444, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Mangum, L.H.; Howell, G.E., 3rd; Chambers, J.E. Exposure to p,p’-dde enhances differentiation of 3t3-l1 preadipocytes in a model of sub-optimal differentiation. Toxicol. Lett. 2015, 238, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Longnecker, M.P.; Daniels, J.L. Environmental contaminants as etiologic factors for diabetes. Environ. Health Persp. 2001, 109, 871–876. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.H.; Jacobs, D.R.; Porta, M. Could low-level background exposure to persistent organic pollutants contribute to the social burden of type 2 diabetes? J. Epidemiol. Community Health 2006, 60, 1006–1008. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Roy, S. Monitoring the effects of a lepidopteran insecticide, flubendiamide, on the biology of a non-target dipteran insect, drosophila melanogaster. Environ. Monit. Assess. 2017, 189, 14. [Google Scholar] [CrossRef] [PubMed]
- United States Environmental Protection Agency. Flubendiamide—Notice of Intent to Cancel and Other Supporting Documents. 2016. Available online: https://www.epa.gov/ingredients-used-pesticide-products/flubendiamide-notice-intent-cancel-and-other-supporting (accessed on 11 November 2018).
- University of Hertfordshire. Flubendiamide. Available online: https://sitem.herts.ac.uk/aeru/ppdb/en/Reports/1132.htm (accessed on 11 November 2018).
- Arpad, A.; Denis, H. Food safety assessment of pesticide residues. J. Environ. Sci. Heal. 2017, 52, 842. [Google Scholar]
- Park, Y.; Kim, Y.; Kim, J.; Yoon, K.S.; Clark, J.; Lee, J.; Park, Y. Imidacloprid, a neonicotinoid insecticide, potentiates adipogenesis in 3t3-l1 adipocytes. J. Agric. Food Chem. 2013, 61, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Park, Y.; Yoon, K.S.; Clark, J.M.; Park, Y. Permethrin alters adipogenesis in 3t3-l1 adipocytes and causes insulin resistance in c2c12 myotubes. J. Biochem. Mol. Toxicol. 2014, 28, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Sun, Q.; Yue, Y.; Yoon, K.S.; Whang, K.-Y.; Clark, J.M.; Park, Y. 4,4′-dichlorodiphenyltrichloroethane (ddt) and 4,4′-dichlorodiphenyldichloroethylene (dde) promote adipogenesis in 3tl1 adipocyte cell culture. Pestic. Biochem. Phys. 2016, 131, 40–45. [Google Scholar] [CrossRef] [PubMed]
- MacDougald, O.A.; Cornelius, P.; Liu, R.; Lane, M.D. Insulin regulates transcription of the ccaat/enhancer binding protein (c/ebp) alpha, beta, and delta genes in fully-differentiated 3t3-l1 adipocytes. J. Biol. Chem. 1995, 270, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Qi, W.; Yang, J.J.; Yoon, K.S.; Clark, J.M.; Park, Y. Fipronil promotes adipogenesis via ampkalpha-mediated pathway in 3t3-l1 adipocytes. Food Chem. Toxicol. 2016, 92, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Mao, L.; Ji, F.; Wang, S.; Xie, Y.; Fei, H.; Wang, X.D. Activating amp-activated protein kinase by an alpha1 selective activator compound 13 attenuates dexamethasone-induced osteoblast cell death. Biochem. Biophys. Res. Commun. 2016, 471, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Gerlier, D.; Thomasset, N. Use of mtt colorimetric assay to measure cell activation. J. Immunol. Methods 1986, 94, 57–63. [Google Scholar] [CrossRef]
- Liu, Z.; Qiao, Q.; Sun, Y.; Chen, Y.; Ren, B.; Liu, X. Sesamol ameliorates diet-induced obesity in c57bl/6j mice and suppresses adipogenesis in 3t3-l1 cells via regulating mitochondria-lipid metabolism. Mol. Nutr. Food Res. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. Nih image to imagej: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Gammon, S.R.; Knippers, J.D.; Paulsen, S.R.; Rubink, D.S.; Winder, W.W. Phosphorylation-activity relationships of ampk and acetyl-coa carboxylase in muscle. J. Appl. Physiol. 2002, 92, 2475–2482. [Google Scholar] [CrossRef] [PubMed]
- Willows, R.; Sanders, M.J.; Xiao, B.; Patel, B.R.; Martin, S.R.; Read, J.; Wilson, J.R.; Hubbard, J.; Gamblin, S.J.; Carling, D. Phosphorylation of ampk by upstream kinases is required for activity in mammalian cells. Biochem. J. 2017, 474, 3059–3073. [Google Scholar] [CrossRef] [PubMed]
- Stein, S.C.; Woods, A.; Jones, N.A.; Davison, M.D.; Carling, D. The regulation of amp-activated protein kinase by phosphorylation. Biochem. J. 2000, 345 Pt 3, 437–443. [Google Scholar] [CrossRef]
- Merrill, G.F.; Kurth, E.J.; Hardie, D.G.; Winder, W.W. Aica riboside increases amp-activated protein kinase, fatty acid oxidation, and glucose uptake in rat muscle. Am. J. Physiol. 1997, 273, E1107–E1112. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.B.; Zhou, G.; Li, C. Ampk: An emerging drug target for diabetes and the metabolic syndrome. Cell Metab. 2009, 9, 407–416. [Google Scholar] [CrossRef] [PubMed]
- Bijland, S.; Mancini, S.J.; Salt, I.P. Role of amp-activated protein kinase in adipose tissue metabolism and inflammation. Clin. Sci. 2013, 124, 491–507. [Google Scholar] [CrossRef] [PubMed]
- Habinowski, S.A.; Witters, L.A. The effects of aicar on adipocyte differentiation of 3t3-l1 cells. Biochem. Biophys. Res. Commun. 2001, 286, 852–856. [Google Scholar] [CrossRef] [PubMed]
- Daval, M.; Diot-Dupuy, F.; Bazin, R.; Hainault, I.; Viollet, B.; Vaulont, S.; Hajduch, E.; Ferre, P.; Foufelle, F. Anti-lipolytic action of amp-activated protein kinase in rodent adipocytes. J. Biol. Chem. 2005, 280, 25250–25257. [Google Scholar] [CrossRef] [PubMed]
- Ebbinghaus-Kintscher, U.; Luemmen, P.; Lobitz, N.; Schulte, T.; Funke, C.; Fischer, R.; Masaki, T.; Yasokawa, N.; Tohnishi, M. Phthalic acid diamides activate ryanodine-sensitive Ca2+ release channels in insects. Cell Calcium 2006, 39, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Scheffler, T.L.; Gunawan, A.M.; Shi, H.; Zeng, C.; Hannon, K.M.; Grant, A.L.; Gerrard, D.E. Chronic elevated calcium blocks ampk-induced glut-4 expression in skeletal muscle. Am. J. Physiol. Cell Physiol. 2009, 296, C106–C115. [Google Scholar] [CrossRef] [PubMed]
- Gormand, A.; Henriksson, E.; Strom, K.; Jensen, T.E.; Sakamoto, K.; Goransson, O. Regulation of amp-activated protein kinase by lkb1 and camkk in adipocytes. J. Cell Biochem. 2011, 112, 1364–1375. [Google Scholar] [CrossRef] [PubMed]
- United States Environmental Protection Agency. Pesticide Fact Sheet. 2008. Available online: https://www3.epa.gov/pesticides/chem_search/reg_actions/registration/fs_PC-027602_01-Aug-08.pdf (accessed on 11 November 2018).
- Nareshkumar, B.; Akbar, S.M.; Sharma, H.C.; Jayalakshmi, S.K.; Sreeramulu, K. Evaluation of flubendiamide-induced mitochondrial dysfunction and metabolic changes in helicoverpa armigera (hubner). Arch. Insect Biochem. Physiol. 2017, 96, e21401. [Google Scholar] [CrossRef] [PubMed]
- Cui, F.; Chai, T.; Qian, L.; Wang, C. Effects of three diamides (chlorantraniliprole, cyantraniliprole and flubendiamide) on life history, embryonic development and oxidative stress biomarkers of daphnia magna. Chemosphere 2017, 169, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Wolterink, G.; Moretto, A.A. Flubendiamide. Available online: http://docplayer.net/62293224-Flubendiamide-first-draft-prepared-by-g-wolterink-1-and-a-moretto-2.html (accessed on 11 November 2018).
Sample Availability: Samples of the flubendiamide are available from the authors. |





© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Q.; Lin, J.; Peng, Y.; Gao, R.; Peng, Y. Flubendiamide Enhances Adipogenesis and Inhibits AMPKα in 3T3-L1 Adipocytes. Molecules 2018, 23, 2950. https://doi.org/10.3390/molecules23112950
Sun Q, Lin J, Peng Y, Gao R, Peng Y. Flubendiamide Enhances Adipogenesis and Inhibits AMPKα in 3T3-L1 Adipocytes. Molecules. 2018; 23(11):2950. https://doi.org/10.3390/molecules23112950
Chicago/Turabian StyleSun, Quancai, Jie Lin, Yukui Peng, Ruichang Gao, and Ye Peng. 2018. "Flubendiamide Enhances Adipogenesis and Inhibits AMPKα in 3T3-L1 Adipocytes" Molecules 23, no. 11: 2950. https://doi.org/10.3390/molecules23112950
APA StyleSun, Q., Lin, J., Peng, Y., Gao, R., & Peng, Y. (2018). Flubendiamide Enhances Adipogenesis and Inhibits AMPKα in 3T3-L1 Adipocytes. Molecules, 23(11), 2950. https://doi.org/10.3390/molecules23112950



