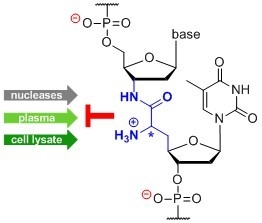
Enhanced Stability of DNA Oligonucleotides with Partially Zwitterionic Backbone Structures in Biological Media †
Abstract
1. Introduction
2. Results
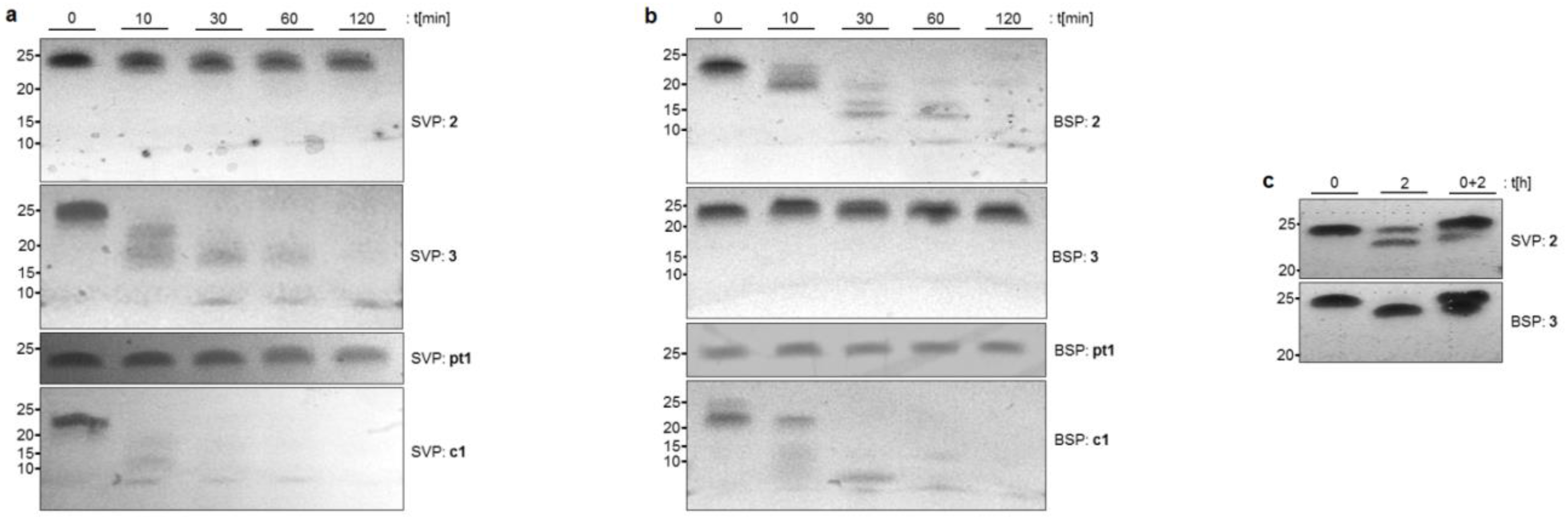
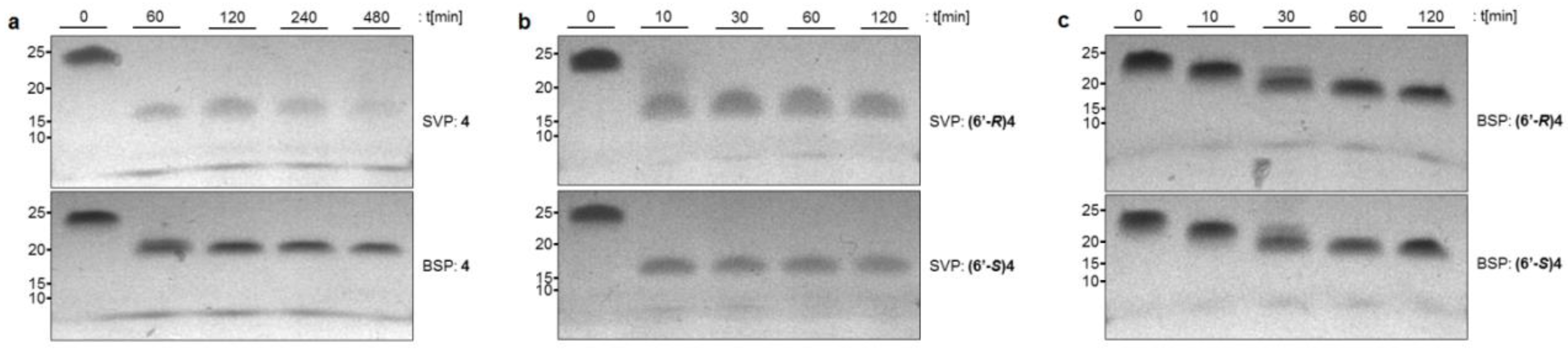
2.1. Stability of NAA-modified Oligonucleotides towards Nuclease-mediated Cleavage
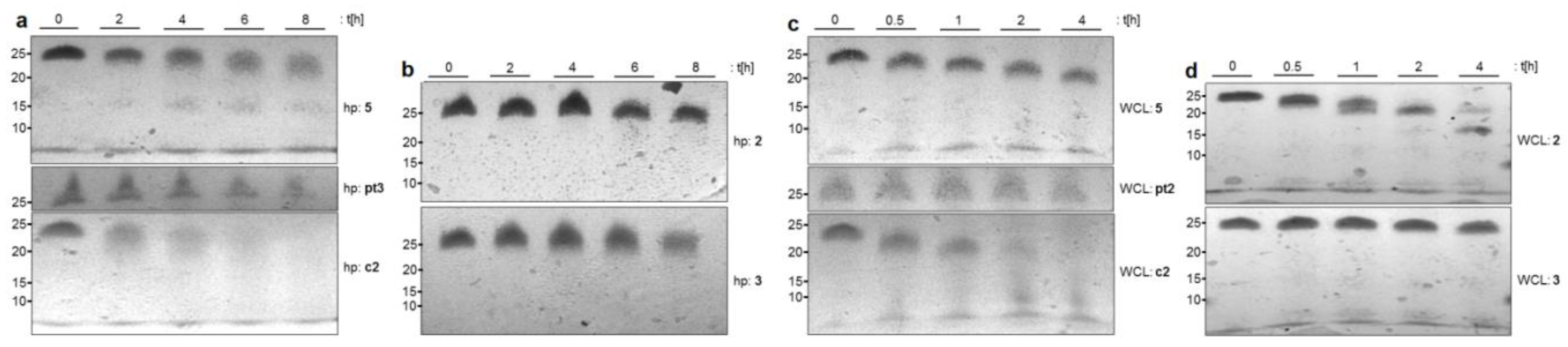
2.2. Stability of NAA-modified Oligonucleotides in Complex Biological Media
3. Discussion
4. Materials and Methods
4.1. Synthesis of NAA-Modified Oligonucleotides
4.2. Stability Assay with 3′-exonuclease
4.3. Stability Assay with 5′-exonuclease
4.4. Stability Assay with Human Plasma
4.5. Stability Assay with Whole Cell Lysate
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sharma, V.K.; Rungta, P.; Prasad, A.K. Nucleic acid therapeutics: basic concepts and recent developments. RSC Adv. 2014, 4, 16618–16631. [Google Scholar] [CrossRef]
- Cobb, A.J.A. Recent highlights in modified oligonucleotide chemistry. Org. Biomol. Chem. 2007, 5, 3260–3275. [Google Scholar] [CrossRef] [PubMed]
- Deleavey, G.F.; Damha, M.J. Designing chemically modified oligonucleotides for targeted gene silencing. Chem. Biol. 2012, 19, 937–954. [Google Scholar] [CrossRef] [PubMed]
- Eckstein, F. Nucleoside phosphorothioates. Ann. Rev. Biochem. 1985, 54, 367–402. [Google Scholar] [CrossRef] [PubMed]
- El-Sagheer, A.H.; Brown, T. Synthesis and Polymerase Chain Reaction Amplification of DNA Strands Containing an Unnatural Triazole Linkage. J. Am. Chem. Soc. 2009, 131, 3958–3964. [Google Scholar] [CrossRef] [PubMed]
- El-Sagheer, A.H.; Sanzone, A.P.; Gao, R.; Tavassoli, A.; Brown, T. Biocompatible artificial DNA linker that is read through by DNA polymerases and is functional in Escherichia coli. Proc. Natl. Acad. Sci. USA 2011, 108, 11338–11343. [Google Scholar] [CrossRef] [PubMed]
- Sanzone, A.P.; El-Sagheer, A.H.; Brown, T.; Tavassoli, A. Assessing the biocompatibility of click-linked DNA in Escherichia coli. Nucleic Acids Res. 2012, 40, 10567–10575. [Google Scholar] [CrossRef] [PubMed]
- Birts, C.N.; Sanzone, A.P.; El-Sagheer, A.H.; Blaydes, J.P.; Brown, T.; Tavassoli, A. Transcription of Click-Linked DNA in Human Cells. Angew. Chem. Int. Ed. 2014, 53, 2362–2365. [Google Scholar] [CrossRef] [PubMed]
- Kukwikila, M.; Gale, N.; El-Sagheer, A.H.; Brown, T.; Tavassoli, A. Assembly of a biocompatible triazole-linked gene by one-pot click-DNA ligation. Nat. Chem. 2017, 9, 1089–1098. [Google Scholar] [CrossRef] [PubMed]
- Lebreton, J.; De Mesmaeker, A.; Waldner, A.; Fritsch, V.; Wolf, R.M.; Freier, S.M. Synthesis of thymidine dimer derivatives containing an amide linkage and their incorporation into oligodeoxyribonucleotides. Tetrahedron Lett. 1993, 34, 6383–6386. [Google Scholar] [CrossRef]
- De Mesmaeker, A.; Waldner, A.; Lebreton, J.; Hoffmann, P.; Fritsch, V.; Wolf, R.M.; Freier, S.M. Amides as Substitute for the Phosphodiester Linkage in Antisense Oligonucleotides. Synlett 1993, 733–736. [Google Scholar] [CrossRef]
- De Mesmaeker, A.; Waldner, A.; Lebreton, J.; Hoffmann, P.; Fritsch, V.; Wolf, R.M.; Freier, S.M. Amides as a New Type of Backbone Modification in Oligonucleotides. Angew. Chem. Int. Ed. 1994, 33, 226–229. [Google Scholar] [CrossRef]
- Selvam, C.; Thomas, S.; Abbott, J.; Kennedy, S.D.; Rozners, E. Amides as Excellent Mimics of Phosphate Linkages in RNA. Angew. Chem. Int. Ed. 2011, 50, 2068–2070. [Google Scholar] [CrossRef]
- Tanui, P.; Kennedy, S.D.; Lunstad, B.D.; Haas, A.; Leake, D.; Rozners, E. Synthesis, biophysical studies and RNA interference activity of RNA having three consecutive amide linkages. Org. Biomol. Chem. 2014, 12, 1207–1210. [Google Scholar] [CrossRef] [PubMed]
- Mutisya, D.; Selvam, C.; Lunstad, B.D.; Pallan, P.S.; Haas, A.; Leake, D.; Egli, M.; Rozners, E. Amides are excellent mimics of phosphate internucleoside linkages and are well tolerated in short interfering RNAs. Nucleic Acids Res. 2014, 42, 6542–6551. [Google Scholar] [CrossRef] [PubMed]
- Moody, H.M.; van Genderen, M.H.P.; Koole, L.H.; Kocken, H.J.M.; Meijer, E.M.; Buck, H.M. Regiospecific inhibition of DNA duplication by antisense phosphate-methylated oligodeoxynucleotides. Nucleic Acids Res. 1989, 17, 4769–4782. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Benner, S.A. Oligodeoxyribonucleotide Analogues with Bridging Dimethylene Sulfide, Sulfoxide, and Sulfone Groups. Toward a Second-Generation Model of Nucleic Acid Structure. J. Org. Chem. 2002, 67, 3996–4013. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, P.E.; Egholm, M.; Berg, R.H.; Buchardt, O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science 1991, 254, 1497–1500. [Google Scholar] [CrossRef] [PubMed]
- Zhilina, Z.V.; Ziemba, A.J.; Ebbinghaus, S.W. Peptide nucleic acid conjugates: synthesis, properties and applications. Curr. Top. Med. Chem. 2005, 5, 1119–1131. [Google Scholar] [CrossRef] [PubMed]
- Strauss, J.K.; Roberts, C.; Nelson, M.G.; Switzer, C.; Maher, L.J., III. DNA bending by hexamethylene-tethered ammonium ions. Proc. Natl. Acad. Sci. USA 1996, 93, 9515–9520. [Google Scholar] [CrossRef] [PubMed]
- Prakash, T.P.; Kawasaki, A.M.; Lesnik, E.A.; Sioufi, N.; Manoharan, M. Synthesis of 2′-O-[2-[(N,N-dialkylamino)oxy]ethyl]-modified oligonucleotides: hybridization affinity, resistance to nuclease, and protein binding characteristics. Tetrahedron 2003, 59, 7413–7422. [Google Scholar] [CrossRef]
- Prhavc, M.; Prakash, T.P.; Minasov, G.; Cook, P.D.; Egli, M.; Manoharan, M. 2′-O-[2-[2-(N,N-Dimethylamino)ethoxy]ethyl] Modified Oligonucleotides: Symbiosis of Charge Interaction Factors and Stereoelectronic Effects. Org. Lett. 2003, 5, 2017–2020. [Google Scholar] [CrossRef] [PubMed]
- Milton, S.; Honcharenko, D.; Rocha, C.S.J.; Moreno, P.M.D.; Smith, C.I.E.; Strömberg, R. Nuclease resistant oligonucleotides with cell penetrating properties. Chem. Commun. 2015, 51, 4044–4047. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.L.; Bruice, P.Y.; Szabo, I.E.; Bruice, T.C. Incorporation of Positively Charged Linkages into DNA and RNA Backbones: A Novel Strategy for Antigene and Antisense Agents. Chem. Rev. 2012, 112, 1284–1309. [Google Scholar] [CrossRef] [PubMed]
- Meng, M.; Ducho, C. Oligonucleotide analogues with cationic backbone linkages. Beilstein J. Org. Chem. 2018, 14, 1293–1308. [Google Scholar] [CrossRef] [PubMed]
- Letsinger, R.L.; Singman, C.N.; Histand, G.; Salunkhe, M. Cationic oligonucleotides. J. Am. Chem. Soc. 1988, 110, 4470–4471. [Google Scholar] [CrossRef]
- Blasko, A.; Dempcy, R.O.; Minyat, E.E.; Bruice, T.C. Association of Short-Strand DNA Oligomers with Guanidinium-Linked Nucleosides. A Kinetic and Thermodynamic Study. J. Am. Chem. Soc. 1996, 118, 7892–7899. [Google Scholar] [CrossRef]
- Barawkar, D.A.; Bruice, T.C. Synthesis, biophysical properties, and nuclease resistance properties of mixed backbone oligodeoxynucleotides containing cationic internucleoside guanidinium linkages: deoxynucleic guanidine/DNA chimeras. Proc. Natl. Acad. Sci. USA 1998, 95, 11047–11052. [Google Scholar] [CrossRef] [PubMed]
- Linkletter, B.A.; Szabo, I.E.; Bruice, T.C. Solid-Phase Synthesis of Deoxynucleic Guanidine (DNG) Oligomers and Melting Point and Circular Dichroism Analysis of Binding Fidelity of Octameric Thymidyl Oligomers with DNA Oligomers. J. Am. Chem. Soc. 1999, 121, 3888–3896. [Google Scholar] [CrossRef]
- Arya, D.P.; Bruice, T.C. Replacement of the Negative Phosphodiester Linkages of DNA by Positive S-Methylthiourea Linkers: A Novel Approach to Putative Antisense Agents. J. Am. Chem. Soc. 1998, 120, 6619–6620. [Google Scholar] [CrossRef]
- Challa, H.; Bruice, T.C. Incorporation of positively charged deoxynucleic S-methylthiourea linkages into oligodeoxyribonucleotides. Bioorg. Med. Chem. Lett. 2001, 11, 2423–2427. [Google Scholar] [CrossRef]
- Schmidtgall, B.; Spork, A.P.; Wachowius, F.; Höbartner, C.; Ducho, C. Synthesis and properties of DNA oligonucleotides with a zwitterionic backbone structure. Chem. Commun. 2014, 50, 13742–13745. [Google Scholar] [CrossRef] [PubMed]
- Schmidtgall, B.; Höbartner, C.; Ducho, C. NAA-modified DNA oligonucleotides with zwitterionic backbones: stereoselective synthesis of A–T phosphoramidite building blocks. Beilstein J. Org. Chem. 2015, 11, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Schmidtgall, B.; Kuepper, A.; Meng, M.; Grossmann, T.N.; Ducho, C. Oligonucleotides with Cationic Backbone and Their Hybridization with DNA: Interplay of Base Pairing and Electrostatic Attraction. Chem. Eur. J. 2018, 24, 1544–1553. [Google Scholar] [CrossRef] [PubMed]
- Spork, A.P.; Ducho, C. Novel 5′-deoxy nucleosyl amino acid scaffolds for the synthesis of muraymycin analogues. Org. Biomol. Chem. 2010, 8, 2323–2326. [Google Scholar] [CrossRef] [PubMed]
- Spork, A.P.; Wiegmann, D.; Granitzka, M.; Stalke, D.; Ducho, C. Stereoselective Synthesis of Uridine-Derived Nucleosyl Amino Acids. J. Org. Chem. 2011, 76, 10083–10098. [Google Scholar] [CrossRef] [PubMed]
- Spork, A.P.; Büschleb, M.; Ries, O.; Wiegmann, D.; Boettcher, S.; Mihalyi, A.; Bugg, T.D.H.; Ducho, C. Lead structures for new antibacterials: stereocontrolled synthesis of a bioactive muraymycin analogue. Chem. Eur. J. 2014, 20, 15292–15297. [Google Scholar] [CrossRef] [PubMed]
- Wiegmann, D.; Koppermann, S.; Wirth, M.; Niro, G.; Leyerer, K.; Ducho, C. Muraymycin nucleoside-peptide antibiotics: uridine-derived natural products as lead structures for the development of novel antibacterial agents. Beilstein J. Org. Chem. 2016, 12, 769–795. [Google Scholar] [CrossRef] [PubMed]
- Williams, E.J.; Sung, S.; Laskowski, M. Action of Venom Phosphodiesterase on Deoxyribonucleic Acid. J. Biol. Chem. 1961, 236, 1130–1134. [Google Scholar] [PubMed]
- Letsinger, R.L.; Bach, S.A.; Eadie, J.S. Effects of pendant groups at phosphorus on binding properties of d-ApA analogues. Nucleic Acids Res. 1986, 14, 3487–3499. [Google Scholar] [CrossRef] [PubMed]
- Saneyoshi, H.; Seio, K.; Sekine, M. A General Method for the Synthesis of 2′-O-Cyanoethylated Oligoribonucleotides Having Promising Hybridization Affinity for DNA and RNA and Enhanced Nuclease Resistance. J. Org. Chem. 2005, 25, 10453–10460. [Google Scholar] [CrossRef] [PubMed]
- Geary, R.S.; Norris, D.; Yu, R.; Bennett, C.F. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv. Drug Deliv. Rev. 2015, 87, 46–51. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |
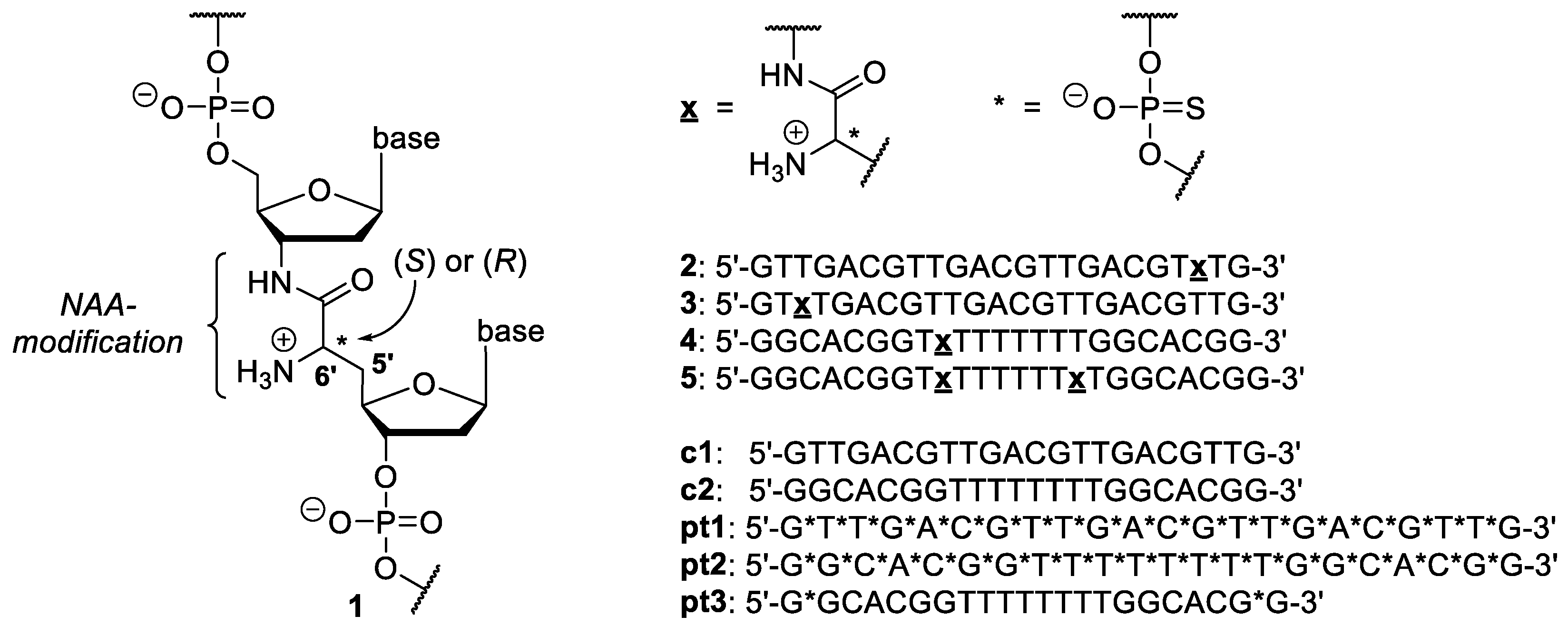
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meng, M.; Schmidtgall, B.; Ducho, C. Enhanced Stability of DNA Oligonucleotides with Partially Zwitterionic Backbone Structures in Biological Media. Molecules 2018, 23, 2941. https://doi.org/10.3390/molecules23112941
Meng M, Schmidtgall B, Ducho C. Enhanced Stability of DNA Oligonucleotides with Partially Zwitterionic Backbone Structures in Biological Media. Molecules. 2018; 23(11):2941. https://doi.org/10.3390/molecules23112941
Chicago/Turabian StyleMeng, Melissa, Boris Schmidtgall, and Christian Ducho. 2018. "Enhanced Stability of DNA Oligonucleotides with Partially Zwitterionic Backbone Structures in Biological Media" Molecules 23, no. 11: 2941. https://doi.org/10.3390/molecules23112941
APA StyleMeng, M., Schmidtgall, B., & Ducho, C. (2018). Enhanced Stability of DNA Oligonucleotides with Partially Zwitterionic Backbone Structures in Biological Media. Molecules, 23(11), 2941. https://doi.org/10.3390/molecules23112941